Structural complementarity from DNA for directing two-dimensional polydopamine nanomaterials with biomedical applications†
Tao
Ding
,
Yuxin
Xing
,
Zhenqiang
Wang
,
Haidi
Guan
,
Liucan
Wang
,
Jixi
Zhang
 * and
Kaiyong
Cai
* and
Kaiyong
Cai

Key Laboratory of Biorheological Science and Technology, Ministry of Education, College of Bioengineering, Chongqing University, No. 174 Shazheng Road, Chongqing 400044, China. E-mail: jixizhang@cqu.edu.cn
First published on 7th December 2018
Abstract
Mussel-inspired polydopamine (PDA) has great potential in constructing 2D nanomaterials under mild synthesis conditions (weakly alkaline pH and room temperature), due to the unique-layered structure during its self-polymerization. However, the manipulation of PDA oligomer's aggregation remains a great challenge. Motivated by the similarity in the inter-plane spacings (∼3.4–3.6 Å) of the subunits (base pairs for DNA and oligomers for PDA), we first demonstrate the synthesis of free-standing PDA nanoplatelets/sheets through solution-based supramolecular interactions guided by the structural complementarity of the subunit arrangements. The vertical interdigitation and lateral packing of PDA's primary planar structures were involved in the formation process directed by DNA. This system provided us a new platform for understanding the self-assembly interactions between DNA and PDA and the construction of sophisticated 2D nanomaterials with excellent photothermal conversion properties for cancer inhibition applications.
Conceptual insightsPolydopamine (PDA) possesses a great potential in constructing advanced 2D nanomaterials because of the layered structures of its subunits, i.e. rigid disc-like aggregates consisting of four to five rigid planar sheets stacking with a graphite-like stacking spacing (∼3.4–3.7 Å). Motivated by the structural complementarity in the inter-plane distance of PDA oligomers and adjacent nucleotide pairs along the duplex axis of DNA (i.e. the axial rise, 3.4 Å for B-type helices), we demonstrated for the first time that free-standing PDA nanoplatelets/nanosheets can be synthesized through the solution-based supramolecular self-assembly of PDA oligomers directed by DNA. During the formation of nanoplatelets/nanosheets, PDA oligomers were packed through the vertical interdigitation and lateral π–π stacking. The successful synthesis not only solves the challenge in organizing the orientations of the hierarchical structures of PDA via directing agents DNA, but also provides us with a great opportunity to explore the nanoplatelets as potential photothermal conversion agents for combined cancer therapy. |
Two-dimensional (2D) nanomaterials possess enhanced physicochemical attributes owing to their high surface-to-volume ratios and high degree of anisotropy. These features make them suitable for a wide range of biomedical applications, including drug/gene delivery, bioimaging, tissue engineering, and photothermal therapy,1 among others. Increasing efforts have been made to exploit new free-standing 2D nanomaterials with layered structures under mild synthesis conditions (weakly alkaline pH and room temperature).2 Among them, mussel-inspired polydopamine (PDA) has great potential, due to its great biocompatibility, versatile functionalization ability, and unique adhesive properties for building multifunctional nanocarriers.3–6 As seen from the structural elucidation of polydopamine, dopamine oligomers (mainly tetramers) self-organize to form 1–2 nm sized rigid disc-like aggregates consisting of four to five rigid planar sheets stacking along the z direction with a graphite-like stacking spacing of ∼3.4–3.7 Å.7,8 These primary nanoplates π-stack to second and third level aggregates that are tens to hundreds of nanometers in diameter. The manipulation of supramolecular nanostructures of PDA by organizing the orientation of the hierarchical structures via small molecules with aromatic similarity has been illustrated in 1D nanofiber systems with properties that outperform those of their bulk counterparts.9,10 Up until now, however, there has been no report on the regulated arrangement of PDA subunits to anisotropic 2D nanostructures, where guided growth with additional structural complementarity must be satisfied.
Because of their biocompatibility, inherent spatial properties in guiding/organizing the orientation and periodicity of hierarchical structures, biomolecular building blocks are widely used for building state-of-the-art nanomaterials in aqueous-solution-based syntheses.11,12 DNA offers a unique opportunity for addressing the necessary features toward this goal. Despite the elegancy of DNA origami as templating agents for polydopamine nanostructures13 and 2D nanomaterials,14,15 the complicated scaffold design, the extraordinary synthetic precision, and the time-consuming annealing protocols represent limitations in generality. As a cost-effective and more realistic template, common DNA usually possesses a right-handed double-helical B-type conformation with a cross-sectional diameter of ∼2.0 nm, which has been beneficial for the synthesis of different nanomaterials.16–19 Heretofore, the construction of 2D nanostructures, driven by the interactions and guided by the spatial complementarity20 between DNA and the material microstructures, however, has rarely been reported.
It is noteworthy that the distance between adjacent nucleotide pairs along the duplex axis of DNA (i.e. the axial rise, 3.4 Å for B-type helices) is comparable to the inter-plane spacing of PDA. We demonstrate in this work the synthesis of free-standing PDA nanoplatelets and nanosheets with lateral sizes up to the micrometer range through the solution-based supramolecular self-assembly of PDA and DNA via π–π stacking interactions under the regulation of spatial/geometrical matching. Herein, the packing behavior of DNA and PDA substructures was investigated by varying both the DNA concentration and the strand length under ambient conditions of weakly alkaline pH and room temperature. The successful synthesis not only greatly advances our ability to control the assembly of sophisticated and hierarchical PDA materials, but also provides us with a great opportunity to explore their potential application as photothermal conversion agents for combined cancer therapy.
In a typical synthesis, herring sperm DNA (hsDNA, ∼50 bp) and dopamine reacted in an aqueous solution of tris(tris (hydroxymethyl)aminomethane) at a hsDNA/dopamine weight ratio of 0.025![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
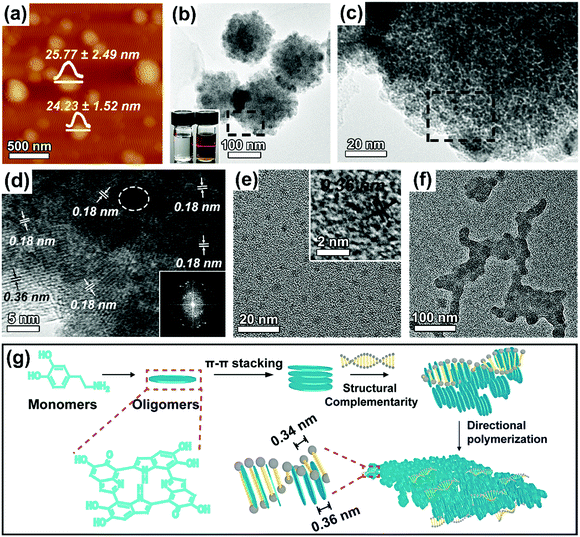
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The structure and morphology of the purified 2D DNA–PDA nanomaterials were then characterized using atomic force microscopy (AFM, Fig. 1a) and transmission electron microscopy (TEM, Fig. 1b), respectively. Both characterization methods indicated that DNA–PDA nanoplatelets with lateral sizes around 162 ± 17 nm were obtained. The thickness of the nanoplatelets was measured to be ∼25 nm. A typical Tyndall light-scattering effect was observed by irradiating the aqueous suspension of the materials with a laser beam, confirming the colloidal sizes (inset of Fig. 1b). The corresponding close-up TEM image showed that the nanoplatelets were assemblies of dark domains with sizes around 4.55 ± 1.15 nm (Fig. 1c). In the high resolution TEM (HRTEM) image (Fig. 1d), ordered domains consisting of several tens (up to ∼25) of stacking layers, were observed, together with randomly distributed amorphous regions (highlighted by white circles). Interestingly, the domain orientation was random in spite of the consistent in-plane orientation (with the normal direction perpendicular to that of the platelet plane, i.e., edge-on orientation). In addition, HRTEM of the flat-lying nanoplatelets (Fig. 1d and Fig. S1, ESI†) revealed two sets of interplanar distances (representing a two-fold relationship), i.e. 0.18 nm throughout the platelets and 0.36 nm on the periphery, which were further confirmed by FFT studies. The intrinsic layer distance of 0.36 nm from the π–π stacking spacing of PDA's primary structure of planar tetramers was widely observed in previous reports.9,21 By using the powder X-ray diffraction (XRD) pattern (Fig. S2a, ESI,† with a broad peak at a 2θ value of 24.6°), this distance was also confirmed in our PDA nanoparticles synthesized without DNA (Fig. S3, ESI†). However, the diffraction peak disappeared in the DNA–PDA nanoplatelets' pattern. We thus proposed that the unique half interplanar distance may result from a vertically interdigitated stacking22 of neighboring nanodiscs from PDA's primary structure. In the Raman spectrum (inset of Fig. S2b, ESI†), the nanoplatelets exhibited two dominating peaks (1596 cm−1, 1350 cm−1), the former of which was remarkably blue/red-shifted from that of PDA nanoparticles (1578 cm−1)/DNA (1601 cm−1). As the peaks should be assigned to the stretching and deformation of aromatic π-planes,23 this result revealed the π–π stacking interactions between the aromatic bases of DNA and the tetramer planes of PDA, as well as the interlocking inside PDA.
1. The structure and morphology of the purified 2D DNA–PDA nanomaterials were then characterized using atomic force microscopy (AFM, Fig. 1a) and transmission electron microscopy (TEM, Fig. 1b), respectively. Both characterization methods indicated that DNA–PDA nanoplatelets with lateral sizes around 162 ± 17 nm were obtained. The thickness of the nanoplatelets was measured to be ∼25 nm. A typical Tyndall light-scattering effect was observed by irradiating the aqueous suspension of the materials with a laser beam, confirming the colloidal sizes (inset of Fig. 1b). The corresponding close-up TEM image showed that the nanoplatelets were assemblies of dark domains with sizes around 4.55 ± 1.15 nm (Fig. 1c). In the high resolution TEM (HRTEM) image (Fig. 1d), ordered domains consisting of several tens (up to ∼25) of stacking layers, were observed, together with randomly distributed amorphous regions (highlighted by white circles). Interestingly, the domain orientation was random in spite of the consistent in-plane orientation (with the normal direction perpendicular to that of the platelet plane, i.e., edge-on orientation). In addition, HRTEM of the flat-lying nanoplatelets (Fig. 1d and Fig. S1, ESI†) revealed two sets of interplanar distances (representing a two-fold relationship), i.e. 0.18 nm throughout the platelets and 0.36 nm on the periphery, which were further confirmed by FFT studies. The intrinsic layer distance of 0.36 nm from the π–π stacking spacing of PDA's primary structure of planar tetramers was widely observed in previous reports.9,21 By using the powder X-ray diffraction (XRD) pattern (Fig. S2a, ESI,† with a broad peak at a 2θ value of 24.6°), this distance was also confirmed in our PDA nanoparticles synthesized without DNA (Fig. S3, ESI†). However, the diffraction peak disappeared in the DNA–PDA nanoplatelets' pattern. We thus proposed that the unique half interplanar distance may result from a vertically interdigitated stacking22 of neighboring nanodiscs from PDA's primary structure. In the Raman spectrum (inset of Fig. S2b, ESI†), the nanoplatelets exhibited two dominating peaks (1596 cm−1, 1350 cm−1), the former of which was remarkably blue/red-shifted from that of PDA nanoparticles (1578 cm−1)/DNA (1601 cm−1). As the peaks should be assigned to the stretching and deformation of aromatic π-planes,23 this result revealed the π–π stacking interactions between the aromatic bases of DNA and the tetramer planes of PDA, as well as the interlocking inside PDA.
The ability of DNA at this small amount to manipulate the assembly structure and the formation of 2D morphology was never found in previous reports on DNA templated nanomaterials. Instead, a precursor nucleation on the DNA backbone, followed by reconfiguration and elongation along the template axis (1D) or lateral packing with interaxial separations (2D), were generally observed.16,18 To elucidate the formation mechanism of the nanoplatelets, aliquots of the reaction solution were taken out at different reaction durations in the early stage. The product collected at 0.5 h was dominantly composed of tiny nanoparticles with sizes of ∼3.85 ± 0.50 nm and a layer distance of 0.36 nm inside (Fig. 1e). No DNA contour-following structures11 were observed, possibly because the rigid primary structures could not encompass the DNA backbone. At 1 h, these small nanoparticles started to grow into planar aggregates (Fig. 1f), wherein the primary particles acted as building blocks. The evolution clearly indicates that self-assembly occurs during the growth of the nanoplatelets.24,25 The aggregates evolved into well-defined nanoplatelets after 2 h (Fig. S4, ESI†) because of the lateral extension26,27 by continuous attachment of PDA, where the rearrangement of the orientation of the oligomer planes may occur. The absorbance increase at 400 nm with time (Fig. S5, ESI†) indicated a slower polymerization rate of dopamine with the presence of DNA. In addition, the mass spectra indicated that the cyclic tetramer was the important structural motif in the DNA–PDA nanoplatelets/sheets (Fig. S6, ESI†). Thus, the formation mechanism of the DNA–PDA nanoplatelets was proposed, as outlined in Fig. 1g. The self-nucleated primary PDA nanoparticles, consisting of π–π stacked oligomer discs, interfacially interacted with DNA through directional component intercalation via the guidance of the spatial complementarity, which thereby led to the lateral aggregation from two directions and the subsequent extension for the nanoplatelets.
The regulation of the directional interaction is of great importance for dictating the structures produced. The axial rise of DNA is 0.2 Å smaller than the interplane distance of the PDA nanodiscs, implying that mismatching may happen after a certain distance along the axial direction of hsDNA (∼17 nm in length). Higher hsDNA/dopamine weight ratios from 0.125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 2
1 to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
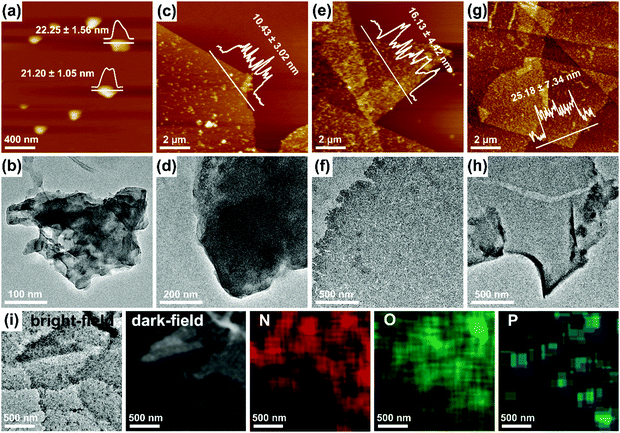
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were therefore employed to compensate for the effect of mismatching on the lateral packing. AFM, as well as TEM, images of the DNA–PDA nanoplatelets/sheets are shown in Fig. 2a–h. The sample at an hsDNA/dopamine weight ratio of 0.125
1 were therefore employed to compensate for the effect of mismatching on the lateral packing. AFM, as well as TEM, images of the DNA–PDA nanoplatelets/sheets are shown in Fig. 2a–h. The sample at an hsDNA/dopamine weight ratio of 0.125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 had an increased size (∼300 nm) and a slightly smaller thickness (∼21 nm, Fig. 2a). Surprisingly, higher concentrations of DNA (1–2
1 had an increased size (∼300 nm) and a slightly smaller thickness (∼21 nm, Fig. 2a). Surprisingly, higher concentrations of DNA (1–2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Fig. 2c–h) led to the generation of nanosheets with a micro-scale lateral size (up to ∼10 μm), indicating a stronger lateral assembly directed by DNA. Furthermore, the sheet thickness significantly declined to ∼10 nm at a DNA/dopamine weight ratio of 1
1, Fig. 2c–h) led to the generation of nanosheets with a micro-scale lateral size (up to ∼10 μm), indicating a stronger lateral assembly directed by DNA. Furthermore, the sheet thickness significantly declined to ∼10 nm at a DNA/dopamine weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, then gradually increased to ∼16 nm (1.5
1, then gradually increased to ∼16 nm (1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and ∼25 nm (2
1) and ∼25 nm (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). A peak (at a 2θ value of 0.8°) in the small-angle XRD pattern (Fig. S7, ESI†), which is related to the thickness of the lamellar nanosheets,28 can even be observed for the DNA–PDA nanosheets at the weight ratio of 1.5
1). A peak (at a 2θ value of 0.8°) in the small-angle XRD pattern (Fig. S7, ESI†), which is related to the thickness of the lamellar nanosheets,28 can even be observed for the DNA–PDA nanosheets at the weight ratio of 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Moreover, the Ra roughness from AFM grew from 3.02 nm to 11.88 nm (Fig. S8, ESI†), while the surface of the 2D platelets/sheets became rough and loose. Some pores or even cracks could be observed at high hsDNA/dopamine weight ratios (Fig. 2e–h), which may result from the electrostatic repulsion between DNA molecules residing on the surface of the nanosheets at a high density. The scanning TEM (STEM) images, the corresponding element mapping (Fig. 2i) and the XPS spectrum (Fig. S9, ESI†) of the nanosheets (at the hsDNA/dopamine weight ratio of 1.5
1. Moreover, the Ra roughness from AFM grew from 3.02 nm to 11.88 nm (Fig. S8, ESI†), while the surface of the 2D platelets/sheets became rough and loose. Some pores or even cracks could be observed at high hsDNA/dopamine weight ratios (Fig. 2e–h), which may result from the electrostatic repulsion between DNA molecules residing on the surface of the nanosheets at a high density. The scanning TEM (STEM) images, the corresponding element mapping (Fig. 2i) and the XPS spectrum (Fig. S9, ESI†) of the nanosheets (at the hsDNA/dopamine weight ratio of 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) proved the presence of DNA in the DNA–PDA nanosheets. The scattered P signals on the nanosheets verified that the DNA molecules with surface-normal orientations and spacing distances can efficiently direct the formation of nanosheets.
1) proved the presence of DNA in the DNA–PDA nanosheets. The scattered P signals on the nanosheets verified that the DNA molecules with surface-normal orientations and spacing distances can efficiently direct the formation of nanosheets.
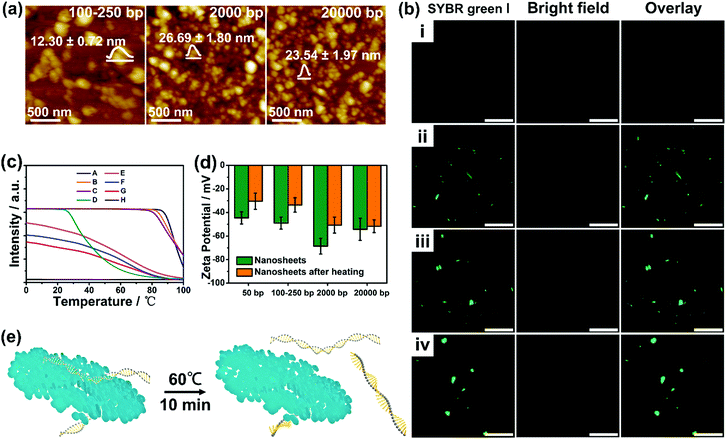
The results above indicated that the directional arrangement of the PDA discs possesses an effective length along the DNA backbone. To gain further insight into the role of the effective length in directing the supramolecular interaction, DNA with different lengths (sonicated salmon sperm DNA: ∼100–250 bp (Fig. S10, ESI†), salmon sperm DNA: ∼2000 bp, calf thymus DNA: ∼20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 bp) were further employed for the synthesis (at a DNA/dopamine weight ratio of 1.5
000 bp) were further employed for the synthesis (at a DNA/dopamine weight ratio of 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Surprisingly, nanosheets with large lateral sizes could not be obtained (Fig. 3a and Fig. S11, ESI†), on account of the possible disruption of lateral interactions during formation. It is hard to distinguish the existence of nanoplatelets in the case of the calf thymus DNA (ctDNA) owing to the further reduction in the particle size. Nanoplatelets with diameters of 100–500 nm (Fig. S12, ESI†) can only be obtained at a low ctDNA/dopamine weight ratio of 0.025
1). Surprisingly, nanosheets with large lateral sizes could not be obtained (Fig. 3a and Fig. S11, ESI†), on account of the possible disruption of lateral interactions during formation. It is hard to distinguish the existence of nanoplatelets in the case of the calf thymus DNA (ctDNA) owing to the further reduction in the particle size. Nanoplatelets with diameters of 100–500 nm (Fig. S12, ESI†) can only be obtained at a low ctDNA/dopamine weight ratio of 0.025![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. In addition, the CLSM (confocal laser scanning microscope) images, shown in Fig. 3b, revealed a gradual increase of the green fluorescence emitted from the SYBR green I (a minor groove binder) stained DNA duplex in the nanostructures. These results suggest that an appropriate DNA length (∼50 bp) is the prerequisite for the formation of well-defined nanoplatelets/sheets.
1. In addition, the CLSM (confocal laser scanning microscope) images, shown in Fig. 3b, revealed a gradual increase of the green fluorescence emitted from the SYBR green I (a minor groove binder) stained DNA duplex in the nanostructures. These results suggest that an appropriate DNA length (∼50 bp) is the prerequisite for the formation of well-defined nanoplatelets/sheets.
Considering the mutual interactions between DNA and PDA, the melting temperatures (Tm, the temperature at which half of the DNA strands are in the random coil or single-stranded state) of DNA were then determined by running a heating program via quantitative fluorescence PCR (qf-PCR). As shown in Fig. 3c and Fig. S13 (ESI†), clear shifts of Tm from 90–95° to ∼60° and wider ranges of melting were observed after integrating longer DNA into the nanomaterials. In contrast, the melting curve of the DNA–PDA nanoplatelets templated by hsDNA showed negligible fluorescence, which may be attributed to the difficulty in staining the surface-bound DNA by SYBR green I resulting from the strongest lateral interactions between DNA and PDA. Furthermore, DNA–PDA nanosheets exhibited a significant increase in the zeta potential (−43 mV to −30 mV) after heating, implying an easier detachment of short-length DNA from the surface (Fig. 3d and e). The fluorescence signals in the supernatant (by staining with SYBR Green I or SYBR Green II) of the DNA–PDA nano-plates/sheets were detected and analyzed (Fig. S14, ESI†). All the solutions emitted strong fluorescence, which demonstrated the presence of both double- and single-stranded DNA in the supernatant after the heat treatment.
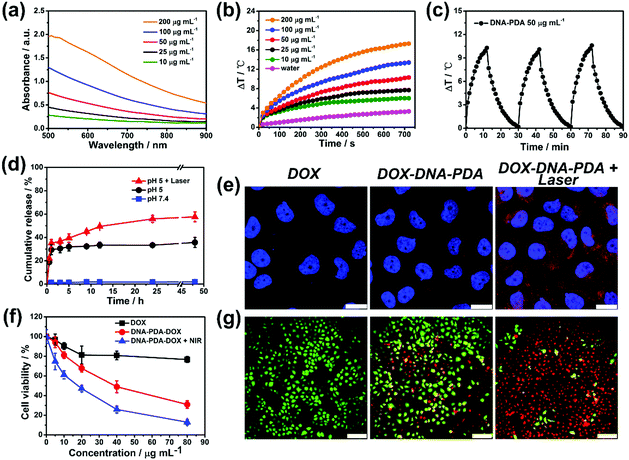
PDA-based nanomaterials are considered efficient photothermal conversion agents and drug delivery carriers.29,30 Motivated by the intriguing 2D morphology and the interdigitated nanostructures of the synthesized PDA materials, we therefore investigated the application properties of the DNA–PDA nanoplatelets/sheets (hsDNA/dopamine weight ratio of 0.025![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The Vis-NIR absorption spectra of DNA–PDA nanoplatelets aqueous solutions at various concentrations indicated that their absorbance at 808 nm linearly correlated with their concentrations (Fig. 4a). The aqueous suspensions of the DNA–PDA nanoplatelets presented a significant concentration-dependent temperature increase (6 °C–17 °C, Fig. 4b) after NIR irradiation (808 nm, 0.5 W cm−2). The photothermal conversion efficiency (η, Fig. S15, ESI†) of the DNA–PDA nanoplatelets was calculated to be 51%, which is remarkedly larger than spherical polydopamine nanoparticles (36%) and other previously reported 2D nanomaterials.31,32 No obvious deterioration was observed after three cycles of temperature evolution by laser irradiation (Fig. 4c), highlighting a good reusability and stability of the nanoplatelets as a durable photothermal agent. The DNA–PDA nanosheets (DNA/dopamine weight ratio of 1.5
1). The Vis-NIR absorption spectra of DNA–PDA nanoplatelets aqueous solutions at various concentrations indicated that their absorbance at 808 nm linearly correlated with their concentrations (Fig. 4a). The aqueous suspensions of the DNA–PDA nanoplatelets presented a significant concentration-dependent temperature increase (6 °C–17 °C, Fig. 4b) after NIR irradiation (808 nm, 0.5 W cm−2). The photothermal conversion efficiency (η, Fig. S15, ESI†) of the DNA–PDA nanoplatelets was calculated to be 51%, which is remarkedly larger than spherical polydopamine nanoparticles (36%) and other previously reported 2D nanomaterials.31,32 No obvious deterioration was observed after three cycles of temperature evolution by laser irradiation (Fig. 4c), highlighting a good reusability and stability of the nanoplatelets as a durable photothermal agent. The DNA–PDA nanosheets (DNA/dopamine weight ratio of 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) also showed attractive photothermal properties (Fig. S16, ESI†). What's more, the photothermal conversion efficiency reached 86% (Fig. S17 and S18, ESI†), because of the large lateral sizes and efficient in-sheet packing of PDA. Benefiting from the large surface area-to-volume ratios of the PDA nanoplatelets, a typical adsorption isotherm of the 2D nanomaterials and an extremely high DOX loading capacity (557 μg mg−1) were obtained (Fig. S19 and S20, ESI†). The premature DOX release can be efficiently prohibited at a neutral condition, while an acidic pH (5.0) and NIR irradiation (Fig. 4d) can serve as triggers to stimulate continuous drug release, in agreement with previous reports on PDA nanocarriers.33,34
1) also showed attractive photothermal properties (Fig. S16, ESI†). What's more, the photothermal conversion efficiency reached 86% (Fig. S17 and S18, ESI†), because of the large lateral sizes and efficient in-sheet packing of PDA. Benefiting from the large surface area-to-volume ratios of the PDA nanoplatelets, a typical adsorption isotherm of the 2D nanomaterials and an extremely high DOX loading capacity (557 μg mg−1) were obtained (Fig. S19 and S20, ESI†). The premature DOX release can be efficiently prohibited at a neutral condition, while an acidic pH (5.0) and NIR irradiation (Fig. 4d) can serve as triggers to stimulate continuous drug release, in agreement with previous reports on PDA nanocarriers.33,34
The in vitro toxicities of DNA–PDA nanoplatelets to a model drug-resistant cell line (MCF-7/ADR cells) were evaluated by a standard CCK-8 assay which indicated a good biocompatibility (Fig. S21, ESI†). The nanoplatelets could be efficiently uptaken by cells and release DOX with the assistance of NIR laser treatment (Fig. 4e). Compared with free drug treatment, the platelets induced drug delivery significantly enhanced the cytotoxicity which can be further improved by applying PTT. Calcein-AM/PI double staining also intuitively evidenced that the combination of photothermal/chemo therapy had a high efficiency in destroying cancer cells (Fig. 4f, g and Fig. S22, ESI†). Furthermore, as expected, the 808 nm NIR at this low power density was biocompatible (Fig. S23, ESI†). These results clearly demonstrate the great potential of the platelets in combined therapy.
Conclusions
In summary, we herein reported the first synthesis of 2D polydopamine nanostructures. It was illustrated that a specific structural complementarity between the base pairs of DNA and the oligomers of dopamine and the supramolecular assembly contribute to the formation of the nanoplatelets/sheets by vertical interdigitation and lateral packing. The morphology and size could be controlled by regulating the weight ratio of DNA/dopamine and the length of the DNA templates. This system features excellent photothermal performance (an extremely high photothermal conversion efficiency up to 51%) and a drug delivery property for cancer cell ablation. Our findings offer improved control of DNA as building blocks for the assembly of sophisticated 2D nanomaterials and provide a framework to understand complementarity and directional interactions in DNA-mediated PDA polymerization.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (NSFC, Grant No. 51773022, 51502027, 21734002), 100 Talents Program of Chongqing University (J. Z.), Project Supported by Graduate Research and Innovation Foundation of Chongqing, China (Grant No. CYB18028) and Innovation Team in University of Chongqing Municipal Government (CXTDX201601002).Notes and references
- Y. Chen, L. Wang and J. Shi, Nano Today, 2016, 11, 292–308 CrossRef CAS.
- X. Zhang, H. Cheng and H. Zhang, Adv. Mater., 2017, 29, 1701704 CrossRef PubMed.
- Y. Xing, J. Zhang, F. Chen, J. Liu and K. Cai, Nanoscale, 2017, 9, 8781–8790 RSC.
- B. K. Ahn, J. Am. Chem. Soc., 2017, 139, 10166–10171 CrossRef CAS.
- W. Cheng, C. Liang, L. Xu, G. Liu, N. Gao, W. Tao, L. Luo, Y. Zuo, X. Wang, X. Zhang, X. Zeng and L. Mei, Small, 2017, 13, 1700623 CrossRef.
- H. Lee, S. M. Dellatore, W. M. Miller and P. B. Messersmith, Science, 2007, 318, 426–430 CrossRef CAS.
- X. Yu, H. Fan, Y. Liu, Z. Shi and Z. Jin, Langmuir, 2014, 30, 5497–5505 CrossRef CAS.
- M. d'Ischia, A. Napolitano, A. Pezzella, P. Meredith and T. Sarna, Angew. Chem., Int. Ed., 2009, 48, 3914–3921 CrossRef PubMed.
- X. Yu, H. Fan, L. Wang and Z. Jin, Angew. Chem., Int. Ed., 2014, 53, 12600–12604 CAS.
- V. Ball, Front. Bioeng. Biotechnol., 2018, 6, 109 CrossRef PubMed.
- X. Ma, J. Huh, W. Park, L. P. Lee, Y. J. Kwon and S. J. Sim, Nat. Commun., 2016, 7, 12873 CrossRef CAS.
- J. Wu, L. H. Tan, K. Hwang, H. Xing, P. Wu, W. Li and Y. Lu, J. Am. Chem. Soc., 2014, 136, 15195–15202 CrossRef CAS.
- Y. Tokura, S. Harvey, C. Chen, Y. Wu, D. Y. W. Ng and T. Weil, Angew. Chem., Int. Ed., 2018, 57, 1587–1591 CrossRef CAS.
- Y. Tian, T. Wang, W. Liu, H. L. Xin, H. Li, Y. Ke, W. M. Shih and O. Gang, Nat. Nanotechnol., 2015, 10, 637 CrossRef CAS PubMed.
- C. Zhou, Y. Zhang, Y. Dong, F. Wu, D. Wang, L. Xin and D. Liu, Adv. Mater., 2016, 28, 9819–9823 CrossRef CAS PubMed.
- S. M. D. Watson, M. A. Galindo, B. R. Horrocks and A. Houlton, J. Am. Chem. Soc., 2014, 136, 6649–6655 CrossRef CAS PubMed.
- X. Ma, W. Hu, C. Guo, L. Yu, L. Gao, J. Xie and C. Li, Adv. Funct. Mater., 2014, 24, 5897–5903 CrossRef CAS.
- B. Liu, Y. Yao and S. Che, Angew. Chem., Int. Ed., 2013, 52, 14186–14190 CrossRef CAS.
- A. Stern, G. Eidelshtein, R. Zhuravel, G. I. Livshits, D. Rotem, A. Kotlyar and D. Porath, Adv. Mater., 2018, 30, 1800433 CrossRef.
- M. N. O’Brien, M. R. Jones, B. Lee and C. A. Mirkin, Nat. Mater., 2015, 14, 833–839 CrossRef PubMed.
- H. Fan, X. Yu, Y. Liu, Z. Shi, H. Liu, Z. Nie, D. Wu and Z. Jin, Soft Matter, 2015, 11, 4621–4629 RSC.
- J. A. Lim, F. Liu, S. Ferdous, M. Muthukumar and A. L. Briseno, Mater. Today, 2010, 13, 14–24 CrossRef CAS.
- X. Zheng, J. Zhang, J. Wang, X. Qi, J. M. Rosenholm and K. Cai, J. Phys. Chem. C, 2015, 119, 24512–24521 CrossRef CAS.
- Z. Liu, H. Pan, G. Zhu, Y. Li, J. Tao, B. Jin and R. Tang, Angew. Chem., Int. Ed., 2016, 55, 12836–12840 CrossRef CAS PubMed.
- W. J. E. M. Habraken, J. Tao, L. J. Brylka, H. Friedrich, L. Bertinetti, A. S. Schenk, A. Verch, V. Dmitrovic, P. H. H. Bomans, P. M. Frederik, J. Laven, P. van der Schoot, B. Aichmayer, G. de With, J. J. DeYoreo and N. A. J. M. Sommerdijk, Nat. Commun., 2013, 4, 1507 CrossRef PubMed.
- Y. Liu, Z. Wu and H. Zhang, Adv. Colloid Interface Sci., 2014, 207, 347–360 CrossRef CAS PubMed.
- Y. Min, G. Park, B. Kim, A. Giri, J. Zeng, J. W. Roh, S. I. Kim, K. H. Lee and U. Jeong, ACS Nano, 2015, 9, 6843–6853 CrossRef CAS PubMed.
- X. Chen, Y. Zhou, Q. Liu, Z. Li, J. Liu and Z. Zou, ACS Appl. Mater. Interfaces, 2012, 4, 3372–3377 CrossRef CAS PubMed.
- R. Lv, P. Yang, B. Hu, J. Xu, W. Shang and J. Tian, ACS Nano, 2017, 11, 1064–1072 CrossRef CAS PubMed.
- Y. Liu, K. Ai and L. Lu, Chem. Rev., 2014, 114, 5057–5115 CrossRef CAS PubMed.
- L. Zong, H. Wu, H. Lin and Y. Chen, Nano Res., 2018, 11, 4149–4168 CrossRef CAS.
- H. Lin, Y. Wang, S. Gao, Y. Chen and J. Shi, Adv. Mater., 2018, 30, 1703284 CrossRef.
- X. Wang, J. Zhang, Y. Wang, C. Wang, J. Xiao, Q. Zhang and Y. Cheng, Biomaterials, 2016, 81, 114–124 CrossRef CAS PubMed.
- Z. Li, Y. Hu, K. A. Howard, T. Jiang, X. Fan, Z. Miao, Y. Sun, F. Besenbacher and M. Yu, ACS Nano, 2016, 10, 984–997 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: The synthesis and characterization methods for the materials, determination of photothermal conversion efficiency and drug loading capacity, as well as supplementary figures. See DOI: 10.1039/c8nh00351c |
| This journal is © The Royal Society of Chemistry 2019 |