Open Access Article
Open Access ArticleInactivation of antibiotic-resistant bacteria by chlorine dioxide in soil and shifts in community composition
M. S. Wu *abc and
X. Xud
*abc and
X. Xud
aCollege of Resources and Civil Engineering, Northeastern University, Shenyang 100819, China. E-mail: wumingsong@163.com
bSchool of Resources and Materials, Northeastern University at Qinhuangdao, Qinhuangdao, 066004, China
cQinhuangdao Key Laboratory of Water Conservation and Pollution Control and Ecological Restoration, Qinhuangdao, 066004, China
dTongji Zhejiang College, Jiaxing, 314051, China
First published on 25th February 2019
Abstract
To study the efficacy of chlorine dioxide in the inactivation of antibiotic-resistant bacteria in soil, bacteria resistant to penicillin, amoxicillin or streptomycin were screened out from the soils around a hennery. The effects of dosage, contact time and pH value on the killing rates were investigated by batch experiments. The community composition before and after inactivation was analyzed by high-throughput genetic sequencing. The results showed that antibiotic-resistant bacteria are common and widespread in soil and the most resistant species is Staphylococcus aureus. More than 99% of antibiotic-resistant bacteria could be killed by chlorine dioxide at 5 mg L−1 within 30 min under neutral conditions. The killing log value declined slightly when the pH was changed from 4 to 9. The dominant genus was Sphingomonas, which was sensitive to chlorine dioxide and could be inactivated easily similar to Arthrobacter and Massilia. However, Micromonosporaceae and Thaumarchaeota were more resistant to chlorine dioxide than other species, and their relative abundance increased after disinfection.
Introduction
Antibiotics have an essential role in controlling bacterial diseases in medical treatments and agriculture. They enter water and soil by the spreading of manure1 or by direct excretion from livestock. Antibiotics then enter deeper soil layers by surface runoff, driftage or leaching2 and finally accumulate in plants.3 Many antibiotic-resistant bacteria (ARB) in soil have been found where livestock congregate. Manure application has significantly increased the diversity and abundance of antibiotic resistance genes (ARGs) in soil and also markedly shifted the bacterial composition that was significantly correlated with the ARG profiles.4 Uncontrolled use of antibiotics has led to the enrichment of ARGs in manure5 and affected environments, particularly soils.6 In the U.S., 60% to 80% of all antibiotics were used in animal production, and more than 80% of them were excreted in manure,7 which led to further pollution.ARGs have been widely found in the soils of livestock areas. Zhu5 found 149 kinds of ARGs in the soils around three large farms, which were about 192–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times higher than those found in soils without the use of antibiotics. Although antibiotic resistance may decline after the relaxation of selection pressures, low yet detectable levels of resistance determinants are likely to persist for decades.8 Also, the quantity of ARGs in the soil remains high even when the livestock has been removed from the site for two years.9
000 times higher than those found in soils without the use of antibiotics. Although antibiotic resistance may decline after the relaxation of selection pressures, low yet detectable levels of resistance determinants are likely to persist for decades.8 Also, the quantity of ARGs in the soil remains high even when the livestock has been removed from the site for two years.9
These ARB and ARGs from agricultural settings can be transferred to humans and become a critical health concern.10 Completely antibiotic-resistant tuberculosis cases were reported in Iran in 2009 and India in 2011, independently. They were resistant to all the first- and second-line drugs.11 An analysis of 264 soil isolates obtained from different natural habitats in and around Hyderabad has identified 5 isolates that are resistant to as many as 10 antibiotics.12 Increased consumption of antibiotics may produce not only greater resistance at the individual patient level, but also greater resistance at community, country, and regional levels, which nevertheless can harm individual patients.13
It has been a hot topic to find out methods to slow or restrain spreading of drug resistance.14 Most studies have reported the effect of various disinfection methods on ARB inactivation in water, and these methods include the use of ozone,15 chlorine,16 and UV. However, UV and ozone disinfections result in apoptosis, and the bacterial DNA is released into the environment; then, ARGs are mostly found as free DNA in the treated wastewater.17 Very few studies are available about the effects of the disinfection process on the inactivation of ARB and ARGs in soils. Drug resistance to traditional disinfectants used in farms, such as phenol, formaldehyde,18 sodium hypochlorite, calcium hypochlorite,19 and quaternary ammonium compounds,20 has been common due to the extensive application for many years. As the native soil bacterial species play a role in inhibiting the survival of ARB or dissemination of ARGs,21 the restoration of the microbial ecosystem after disinfection is also very important not only for soil function but also for reducing antibiotic resistance. Calcined eggshell amendment mitigated mixed pollutant accumulation in bell pepper significantly and enhanced the dissipation of soil tetracycline, sulfadiazine, roxithromycin, and chloramphenicol; it also decreased the water-soluble fractions of antibiotics and the diversity of ARB/ARGs inside the vegetables and contributed to the significant restoration of microbial biodiversity and stability.22
Chlorine dioxide (ClO2) has been widely used for the inactivation of microorganisms and the removal of organic compounds as a substitute of chlorine for henhouse and cowshed disinfection and for other purposes in animal husbandry.23 After soaking in ClO2, the hatchabilities of eggs and duck eggs increase by 2% and 4%, respectively, compared to that observed for fumigation with potassium permanganate and formaldehyde,24 and the incidence of zoonosis decreases.25 The Ct 99 values of ClO2 and chlorine are similar for ARB, and the effects of ClO2 disinfection are not affected by ammonia nitrogen.14 Chlorine dioxide also has an excellent effect on the inactivation of intracellular ARGs,26 which indicates that fewer ARGs would be released into the environment. Truchado27 found that the use of low residual ClO2 concentrations (approx. 0.25 mg L−1) to treat irrigation water decreases the relative abundance of Pseudomonadaceae (2.28-fold) and Enterobacteriaceae (2.5-fold) when comparing treated versus untreated baby spinach samples. Members of these two bacterial families are responsible for food spoilage and foodborne illnesses.28 There is still no report on ClO2 killing antibiotic-resistant bacteria directly in soil.
In this paper, the effect of ClO2 on the inactivation of ARB in soil near a henhouse was investigated, and the differences in the bacteria community before and after treatment were compared to provide a reference for the application of ClO2 to reduce ARB and ARGs in the soil.
Materials and methods
Soil samples
Soil samples were collected at 4 places in the gardens and vegetable fields that were 2 km away from a hennery located at 119.241672E and 39.871822N in Qinhuangdao City in China. All the soil samples were collected 10–15 cm underground and then screened through a 35 mesh sieve (with a particle size of 425 μm) after drying.In order to investigate the antibiotic resistance under severe contamination pressure, the natural soil obtained above was mixed equally and then contaminated by antibiotics in the laboratory. Then, an antibiotic solution (penicillin, amoxicillin or streptomycin) was added into a 50 mL centrifuge tube (filled with 25 mL sterile water and 1 g mixed soil sample) at the concentration of 1, 2.5, 5.0, 10, or 50 mg L−1. The control group was prepared by the same procedure using sterilized water instead of antibiotics.
Chemicals
Chlorine dioxide was prepared by the reaction between H2SO4 and NaClO2 (eqn (1)) and absorbed by pure water.| 5NaClO2 + 2H2SO4 = 4ClO2 + 2Na2SO4 + 11NaCl + 2H2O | (1) |
Then, it was diluted and calibrated before use by sequential iodometry.29 Sodium thiosulfate solution (0.05 mol L−1), which was used to neutralize ClO2 at the end of the treatment process, was prepared by dissolving 7.9 g sodium thiosulfate in 1 L pH 7 buffer solution. All the pH buffer solutions (pH 4–9) were prepared by using KH2PO4 and Na2HPO4. All the solutions were sterilized by autoclaving and stored at 4 °C before use.
Screening of antibiotic-resistant bacteria
Antibiotic-resistant bacteria were screened from natural soil and contaminated soil samples by using the plate streaking method. At first, 1 g of dried soil was soaked by 25 mL of sterile water. Then, it was centrifuged at 4000 rpm for 5 min. The bacterial supernatant prepared from different soils was then diluted 100-fold and spread onto 3 plates filled with nutrient agar medium. Three parallel plates were prepared for each sample. All the plates were incubated for 24 h at 37 °C in an HPS-400 biochemical incubator (Guowang Instruments Co., Ltd., Changzhou, China).Antibiotic susceptibility was tested by the CLSI M100-S26 method using a drug resistance paper disc30 (Hangzhou Microbial Agent Ltd., product no. S1001 for penicillin, no. S1079 for amoxicillin and no. S1031 for streptomycin). The results were reported as sensitive (S), intermediate (I) and resistant (R). All the resistant strains were identified by culture and biochemical tests31 and the most resistant ARBs were further identified by 16SrDNA sequences.
Disinfection
The effects of chlorine dioxide (ClO2) on ARB inactivation were investigated by the suspension quantitative germicidal test. An ARB suspension was quantified by spectrophotometry at a wavelength of 530 nm. Then, it was diluted to an absorbance of 0.350 before disinfection to keep the cell concentration at about 1 × 108 CFU mL−1. The calibrated ClO2 solution was added by 1.0 mL pipette to a 10 mL centrifuge tube filled with 4.0 mL of different diluted ARB suspensions. Sterile water was used as a positive control. The tubes were put into a shaking table at a constant temperature. After a certain time, 1 mL of 0.05 mol L−1 buffered sodium thiosulfate was added to neutralise the remaining ClO2. Then, the plate counting method was used to count the residual bacterial after ten-fold serial dilution. Three parallel samples were made for each antibiotic-resistant strain, and the thalli concentration was reported (N). The killing log value (KL) was calculated by log(N0/Nx). The killing rate (KR) was calculated as KR = (1 − 10KL) × 100%.Sequencing of 16S rRNA gene and bioinformatics analysis
Disinfection was performed by adding 25 mL 15 mg L−1 ClO2 to 50 g of the mixed soil sample collected in the wild. Then, it was centrifuged for 10 min at 4000 rpm. Total genomic DNA from the disinfected and raw soil samples was extracted using the CTAB/SDS method. DNA concentration and purity were monitored on 1% agarose gels. The 16S rRNA genes of distinct regions (16SV4) were amplified using a specific primer (515F-806R) with the barcode.All PCR reactions were carried out with Phusion® High-Fidelity PCR Master Mix (New England Biolabs). We mixed the same volume of 1× loading buffer (containing SYB green) with PCR products and operated electrophoresis on 2% agarose gel for detection. Samples with a bright main strip between 400 and 450 bp were chosen for further experiments. PCR products were mixed in equidensity ratios. Then, the mixture of PCR products was purified with a Qiagen Gel Extraction Kit (Qiagen, Germany). Sequencing libraries were generated using a TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA) following the manufacturer's recommendations and index codes were added. The library quality was assessed on a Qubit@2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. At last, the library was sequenced on an Illumina HiSeq2500 platform and 250 bp paired-end reads were generated. Paired-end reads were merged using V1.2.7 FLASH.32 Quality filtering on the raw tags was performed under specific filtering conditions to obtain the high-quality clean tags33 according to QIIME.34
Sequence analysis was performed by using Uparse software v7.0.1001.35 Sequences with ≥97% similarity were assigned to the same Operational Taxonomic Units (OTUs). For each representative sequence, the Green Gene Database36 was used based on an RDP classifier (version 2.2)37 algorithm to annotate the taxonomic information. The OTU abundance information was normalized using a standard of sequence number corresponding to the sample with the least sequences.
Results
Antibiotic susceptibility of ARB
ARB are found in every sample taken in different places irrespective of whether it is a natural or contaminated soil sample, and there is no significant difference. Bacteria that showed resistance to 3 kinds of antibiotics were found in all the samples, which indicated that resistance can be widely transferred. Moreover, 24 ARB strains were isolated from the soils (Table 1) altogether. Cross drug resistance was ubiquitous. All the penicillin-resistant strains were resistant to amoxicillin as amoxicillin is a kind of semi-synthetic penicillin.| Sensitive | Intermediate | Resistant | ||||
|---|---|---|---|---|---|---|
| Natural soil | Contaminated soil | Natural soil | Contaminated soil | Natural soil | Contaminated soil | |
| Penicillin | 0 | 4 | 0 | 2 | 6 | 4 |
| Amoxicillin | 0 | 0 | 2 | 1 | 6 | 1 |
| Streptomycin | 2 | 5 | 3 | 2 | 3 | 2 |
The 7 strongest antibiotic resistant isolates were chosen for the disinfection experiment. P3, P4, and N6 isolates showed the strongest resistance to penicillin; A2 and N3 showed the strongest resistance to amoxicillin and S8 and N4 showed the strongest resistance to streptomycin. Their cross drug resistance is shown in Table 2. The results of the bacterial morphological examination indicated that all the colonies were round, smooth, non-transparent, faint yellow, neatly edged and wet. P3 and P4 were about 4–5 mm in diameter, and the others were about 3 mm in diameter. Based on 16S rDNA sequencing, all of the 7 isolates were found to be Staphylococcus aureus (SA).
| Strains | P3 | P4 | N6 | A2 | N3 | S8 | N4 | |
|---|---|---|---|---|---|---|---|---|
| Antibiotic susceptibility | Penicillin | R | R | R | R | R | R | R |
| Amoxicillin | R | I | R | R | R | S | I | |
| Streptomycin | I | R | R | I | I | R | R | |
Disinfection efficacy of chlorine dioxide on ARB
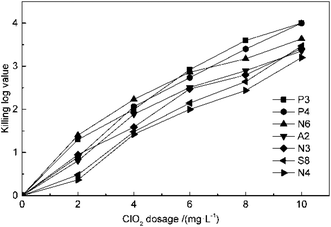
The effect of ClO2 dosage on disinfection was investigated at concentrations of 2, 4, 6, 8 and 10 mg L−1 at 25 °C and pH 7.2 for 30 min. From the result in Fig. 1, it can be seen that the killing log values for all the antibiotic-resistant bacteria increase along with ClO2 dosages. Streptomycin-resistant bacteria were more resistant to ClO2 than penicillin- or amoxicillin-resistant bacteria. At 4 mg L−1 dosage of ClO2, KL values were 2.6, 2.7, 2.8 and 2.5 for P3, P4, N6, and A2, while they were only 2.2, 2.1 and 2.0 for N3, S8, and N4, respectively. The KL values increased to 2.0 and 2.1 for streptomycin-resistant bacteria (S8, N4) at a dosage of 6 mg L−1.The kinetics of ClO2 inactivation of antibiotic-resistant bacteria was investigated by fitting the killing number using the Chick–Watson law (eqn (2)). The Ct values and k values at 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log killing rate are listed in Table 3. The Ct values were in the range of 23–28, which indicated that all the antibiotic-resistant bacteria could be easily killed by ClO2.
log killing rate are listed in Table 3. The Ct values were in the range of 23–28, which indicated that all the antibiotic-resistant bacteria could be easily killed by ClO2.
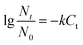 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KL values
KL values
| P3 | P4 | N6 | A2 | N3 | S8 | N4 | |
|---|---|---|---|---|---|---|---|
Ct (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log) log) |
23.65 | 25.83 | 23.33 | 27.06 | 26.05 | 24.36 | 23.71 |
| k | 0.127 | 0.116 | 0.129 | 0.111 | 0.115 | 0.123 | 0.127 |
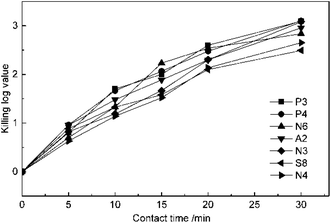
The experiments of the effect of contact time on disinfection were conducted at 5 mg L−1 ClO2 dosage and pH of 7.2. The suspension quantitative germicidal test was terminated by sodium thiosulfate after contacting for 5, 10, 15, 20 and 30 min. From the results in Fig. 2, it can be seen that the KL values of all the antibiotic-resistant bacteria increase for a contact time of 30 min. P3, P4, and A2 were killed by ClO2 quickly. The KL values of P4 and A2 almost reached 1.0 after contacting for 5 min, while those of N3 and N4 were just beyond 1.0 after 10 min. The slopes of the KL value and contact time were all slightly down after 20 min, which may be because ClO2 decreased with time.
The reactivation of ARB after disinfection was investigated at room temperature and under light conditions. The plates with 1 × 108 CFU mL−1 ARBs were treated by 5 mg L−1 ClO2 for 30–50 min. The KL values of the 3 kinds of ARBs were all about 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log. Then, they were calculated every 12 hours; the results are listed in Table 4. Twenty-four hours after ClO2 disinfection, all the KL values of ARBs were maintained at 3
log. Then, they were calculated every 12 hours; the results are listed in Table 4. Twenty-four hours after ClO2 disinfection, all the KL values of ARBs were maintained at 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log. Isolate A2 showed reactivation and regrowth at 36 h, and its KL value decreased to 2.7. Then, P3 and P4 started reactivation and regrowth at 48 h; their KL values were also 2.7 and that of A2 continued to decrease to 2.5 at the same time.
log. Isolate A2 showed reactivation and regrowth at 36 h, and its KL value decreased to 2.7. Then, P3 and P4 started reactivation and regrowth at 48 h; their KL values were also 2.7 and that of A2 continued to decrease to 2.5 at the same time.
| Time/h | Strains | ||||||
|---|---|---|---|---|---|---|---|
| P3 | P4 | N6 | A2 | N3 | S8 | N4 | |
| 12 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| 24 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| 36 | 3 | 3 | 3 | 2.7 | 3 | 3 | 3 |
| 48 | 2.7 | 2.7 | 3 | 2.5 | 3 | 3 | 3 |
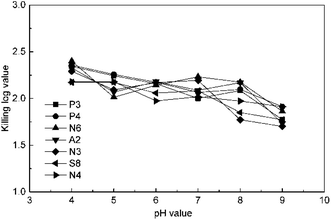
The effect of pH value was studied at 4 mg L−1 ClO2 and 15 min by changing the pH value to 4, 5, 6, 7, 8, and 9 using a phosphate buffer solution. The results in Fig. 3 indicate that the KL values slightly decrease in general in the pH range of 4–9. This may be because chlorite, a common by-product in ClO2 disinfection, is a kind of oxoacid group and has a higher redox potential under low pH conditions. The KL values for P3, P4, and N6 were higher than the others at pH 4; then, those of P3, P4, and N3 decreased along with the increase in pH value, whereas the value for N6 first increased and then decreased. The KL values of S8 and N4 were not significantly affected until the pH value was raised to 9, which is due to the disproportionation occurring for ClO2 beyond pH 9. However, the effect of pH on ClO2 killing antibiotic-resistant bacteria was not significant in soil because the pH of common soils is between 4 and 9.
Analysis of bacterial community shift
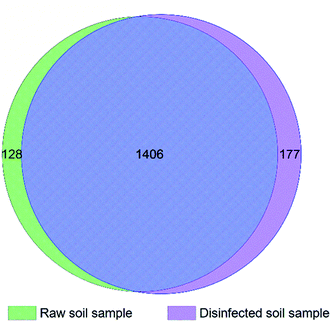
Total 45754 and 66360 effective tags were collected in the extracts of disinfected and raw soil samples, respectively, from which 1608 and 1654 OTUs were clustered and annotated. From the relationship of OTUs between the two samples as the Venn graph demonstrates in Fig. 4 and the relative abundance of the species described in Fig. 5, it can be seen that there is no great difference between the species in the soil before and after ClO2 disinfection. Proteobacteria dominated in both samples, followed by Actinobacteria. The relative abundances of Proteobacteria, Acidobacteria, Bacteroidetes, and Thaumarchaeota significantly decreased after disinfection, while those of Actinobacteria and Firmicutes increased.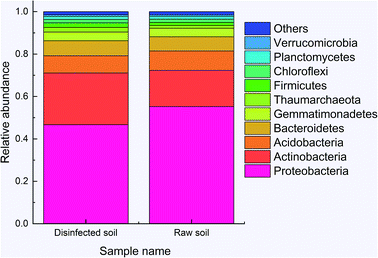 | ||
| Fig. 5 The relative abundance of the species between disinfected and raw samples at the kingdom level. | ||
The 50 most abundant OTUs in the heat map of Fig. 6 illustrate highly similar profiles of OTU frequencies for both samples, in which S1 represents a disinfected soil sample and S2 represents a raw sample. The dominant species in both samples were Arthrobacter oxydans and Sphingomonas, whose relative abundances decreased by 48% and 55%, respectively, after ClO2 disinfection (from 1035 sequences to 636 and 346 + 990 + 461 + 320 to 188 + 502 + 285 + 185, respectively). The amounts of Paucimonas, Massilia timonae, and other species belonging to Sphingomonadales and Piscinibacter were also reduced. Furthermore, the relative abundance of Micromonosporaceae increased mostly (from 98 to 494) and that of Blastococcus also increased significantly (from 330 to 547). Other major species that increased after disinfection were Steroidobacter (from 225 to 396), Microvirga (from 241 to 390), and Thaumarchaeota (from 181 to 321).
Discussion
From the results mentioned above, we inferred that all the strains found in soil and having the strongest resistance were Staphylococcus aureus and not Escherichia coli, as reported before.38–40 Staphylococcus aureus is a major pathogenic microorganism responsible for a series of infections. In particular, methicillin-resistant Staphylococcus aureus (MRSA) exists all around the world and has become a major pathogenic microorganism in hospitals and community.41 Penicillin, amoxicillin, and streptomycin are 3 kinds of antibiotics that are commonly used in animal husbandry in China. Therefore, this proves that antibiotics are no longer as effective as before, and their long-term use has led to serious drug resistance.42Chlorine dioxide can inactivate ARBs effectively and is an effective agent for soil disinfection. Although there have been many reports about ClO2 inactivation of ARB and ARGs in water and wastewater in recent years, there is no such study about soil disinfection and no mention of multi-drug resistant MRSA. Judging by the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ct values, ClO2 is a highly efficient disinfectant not only for water but also for soil. Chlorine dioxide is more suitable for soil disinfection than NaClO.43 It evaporates rapidly from an aqueous solution as a gas and then permeates the soil to contact and kill bacteria. However, HOCl and other liquid disinfectants have less permeability in the soil.44 As the pH range of common soil is about 5–8, the effect of pH on the disinfection is negligible.
Ct values, ClO2 is a highly efficient disinfectant not only for water but also for soil. Chlorine dioxide is more suitable for soil disinfection than NaClO.43 It evaporates rapidly from an aqueous solution as a gas and then permeates the soil to contact and kill bacteria. However, HOCl and other liquid disinfectants have less permeability in the soil.44 As the pH range of common soil is about 5–8, the effect of pH on the disinfection is negligible.
There is no great difference between the species in the soil before and after ClO2 disinfection; this is consistent with the results of Truchado's study,44 in which the abundance of Proteobacteria (the major genus), Pseudomonadaceae and Enterobacteriaceae decreased when comparing samples that were treated and untreated with ClO2. It has also been reported that Proteobacteria, Firmicutes, and Planctomycetes are tolerant of chlorine in the secondary effluent of wastewater treatment plants.45 Also, the relative abundances of Pseudomonas and Sphingomonas increase in the drinking water disinfection process after chlorination.46 This is similar to the results observed for the use of ClO2 in this study as phylum Planctomycetes is a kind of aquatic bacterium. Sphingomonas is sensitive to ClO2 but not to chlorine.47 This may be due to the good penetrating ability of chlorine dioxide, which can penetrate the fatty acid structure of the cell membrane of Sphingomonas.48 Thus, the result suggests that ClO2 is a better disinfectant for soil disinfection treatments than chlorine.
It should be noted that in this study, we only investigated the characteristics and changes in ARB in the soil during ClO2 disinfection. The ability of ClO2 to destroy ARGs still needs to be studied further.
Conclusion
In this study, we identified bacterial species that are resistant to penicillin, amoxicillin, and streptomycin in the soil around a hennery and the disinfection efficiency of ClO2 on them. Bacteria resistant to penicillin, amoxicillin and streptomycin are common in natural soils. Staphylococcus aureus exhibited the strongest resistance. All the bacteria resistant to penicillin were also resistant to amoxicillin. ClO2 could inactivate ARB in soils effectively, and the effect of pH value was not significant. Micromonosporaceae and the identified species in Thaumarchaeota were more resistant to ClO2 than Sphingomonas, Arthrobacter and Massilia. At the phylum and class levels, no significant differences in the bacterial communities were observed between the untreated and ClO2-treated soil samples. Sphingomonas was the dominant genus in the soil before and after treatment by ClO2. Based on the results obtained, ClO2 could be considered as a suitable disinfectant for ARB in soil.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge funding support by the Key Research and Development Plan of Jiangxi of China (20171BBG70035) and the Chinese Universities Scientific Fund (N172304046).Notes and references
- S. Peng, Y. Wang, B. Zhou and X. Lin, Sci. Total Environ., 2015, 506–507, 279–286 CrossRef CAS PubMed.
- N. Kemper, Ecol. Indic., 2008, 8, 1–13 CrossRef CAS.
- X. Hu, Q. Zhou and Y. Luo, Environ. Pollut., 2010, 158, 2992–2998 CrossRef CAS PubMed.
- Q. L. Chen, X. L. An, H. Li, Y. G. Zhu, J. Q. Su and L. Cui, Soil Biol. Biochem., 2017, 114, 229–237 CrossRef CAS.
- Y. G. Zhu, T. A. Johnson, J. Q. Su, M. Qiao, G.-X. Guo, R. D. Stedtfeld, S. A. Hashsham and J. M. Tiedje, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 3435–3440 CrossRef CAS PubMed.
- N. Wu, M. Qiao, B. Zhang, W. D. Cheng and Y. G. Zhu, Environ. Sci. Technol., 2010, 44, 6933–6939 CrossRef CAS PubMed.
- E. Cytryn, Soil Biol. Biochem., 2013, 63, 18–23 CrossRef CAS.
- P. Amy, D. G. Larsson Joakim, A. Alejandro, C. Peter, K. Brandt Kristian, D. W. Graham, M. Lazorchak James, S. Suzuki, S. Peter, R. Snape Jason, E. Topp, T. Zhang and Y.-G. Zhu, Environ. Health Perspect., 2013, 121, 878–885 CrossRef PubMed.
- L. M. Durso and K. L. Cook, Curr. Opin. Microbiol., 2014, 19, 37–44 CrossRef PubMed.
- V. Economou and P. Gousia, Infect. Drug Resist., 2015, 8, 49 CrossRef CAS PubMed.
- Z. F. Udwadia, R. A. Amale, K. K. Ajbani and C. Rodrigues, Clin. Infect. Dis., 2012, 54, 579–581 CrossRef PubMed.
- S. Sengupta and M. K. Chattopadhyay, Resonance, 2012, 17, 177–191 CrossRef CAS.
- B. G. Bell, F. Schellevis, E. Stobberingh, H. Goossens and M. Pringle, BMC Infect. Dis., 2014, 14, 13 CrossRef PubMed.
- Y. Zhang, Y. Zhuang, J. Geng, H. Ren, Y. Zhang, L. Ding and K. Xu, Sci. Total Environ., 2015, 512–513, 125–132 CrossRef CAS PubMed.
- N. Czekalski, S. Imminger, E. Salhi, M. Veljkovic, K. Kleffel, D. Drissner, F. Hammes, H. Bürgmann and U. von Gunten, Environ. Sci. Technol., 2016, 50, 11862–11871 CrossRef CAS PubMed.
- J. J. Huang, H. Y. Hu, F. Tang, Y. Li, S. Q. Lu and Y. Lu, Water Res., 2011, 45, 2775–2781 CrossRef CAS PubMed.
- J. Zheng, C. Su, J. Zhou, L. Xu, Y. Qian and H. Chen, Chem. Eng. J., 2017, 317, 309–316 CrossRef CAS.
- P. Gilbert and A. J. McBain, Clin. Microbiol. Rev., 2003, 16, 189–208 CrossRef CAS PubMed.
- H. F. Ridgway and B. H. Olson, Appl. Environ. Microbiol., 1982, 44, 972–987 CAS.
- S. Buffet-Bataillon, P. Tattevin, M. Bonnaure-Mallet and A. Jolivet-Gougeon, Int. J. Antimicrob. Agents, 2012, 39, 381–389 CrossRef CAS PubMed.
- S. Peng, B. Zhou, Y. Wang, X. Lin, H. Wang and C. Qiu, Biol. Fertil. Soils, 2016, 52, 655–663 CrossRef CAS.
- M. Ye, M. Sun, Y. Feng, X. Li, A. P. Schwab, J. Wan, M. Liu, D. Tian, K. Liu, J. Wu and X. Jiang, J. Agric. Food Chem., 2016, 64, 5446–5453 CrossRef CAS PubMed.
- C. N. Cutter and W. J. Dorsa, J. Food Prot., 1995, 58, 1294–1296 CrossRef.
- P. H. Patterson, S. C. Ricke, M. L. Sunde and D. M. Schaefer, Avian Dis., 1990, 34, 1–6 CrossRef CAS PubMed.
- M. E. Berrang, N. A. Cox, J. F. Frank, R. J. Burh and J. S. Bailey, J. Appl. Poult. Res., 2000, 9, 279–284 CrossRef.
- P. Zhou, S. Li and M. C. Dodd, Proceedings of the Water Environment Federation, 2013, 2013, 131–135 CrossRef.
- P. Truchado, M. I. Gil, T. Suslow and A. Allende, PLoS One, 2018, 13, e0199291 CrossRef PubMed.
- J. Mansfield, S. Genin, S. Magori, V. Citovsky, M. Sriariyanum, P. Ronald, M. Dow, V. Verdier, S. V. Beer, M. A. Machado, I. Toth, G. Salmond and G. D. Foster, Mol. Plant Pathol., 2012, 13, 614–629 CrossRef PubMed.
- M. Wu, J. Liu, S. You, L. Wang, J. Huang and Y. Tian, Environ. Eng. Sci., 2012, 29, 133–138 CrossRef CAS.
- M. J. Ferraro, Performance standards for antimicrobial susceptibility testing, Clinical and Laboratory Standards Institute, Wayne, PA, 26th edn, 2016 Search PubMed.
- G. Garrity, Bergey's Manual of Systematic Bacteriology Volume 2: The Proteobacteria, Springer US, New York, 2005, vol. 2 Search PubMed.
- T. Magoč and S. L. Salzberg, Bioinformatics, 2011, 27, 2957–2963 CrossRef PubMed.
- N. A. Bokulich, S. Subramanian, J. J. Faith, D. Gevers, J. I. Gordon, R. Knight, D. A. Mills and J. G. Caporaso, Nat. Methods, 2013, 10, 57–59 CrossRef CAS PubMed.
- J. G. Caporaso, J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, N. Fierer, A. G. Peña, J. K. Goodrich, J. I. Gordon, G. A. Huttley, S. T. Kelley, D. Knights, J. E. Koenig, R. E. Ley, C. A. Lozupone, D. McDonald, B. D. Muegge, M. Pirrung, J. Reeder, J. R. Sevinsky, P. J. Turnbaugh, W. A. Walters, J. Widmann, T. Yatsunenko, J. Zaneveld and R. Knight, Nat. Methods, 2010, 7, 335–336 CrossRef CAS PubMed.
- R. C. Edgar, Nat. Methods, 2013, 10, 996–998 CrossRef CAS PubMed.
- T. Z. DeSantis, P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu and G. L. Andersen, Appl. Environ. Microbiol., 2006, 72, 5069–5072 CrossRef CAS PubMed.
- Q. Wang, G. M. Garrity, J. M. Tiedje and J. R. Cole, Appl. Environ. Microbiol., 2007, 73, 5261–5267 CrossRef CAS PubMed.
- C. S. Riesenfeld, R. M. Goodman and J. Handelsman, Environ. Microbiol., 2004, 6, 981–989 CrossRef CAS PubMed.
- S. Demanèche, H. Sanguin, J. Poté, E. Navarro, D. Bernillon, P. Mavingui, W. Wildi, T. M. Vogel and P. Simonet, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 3957–3962 CrossRef PubMed.
- R. Marti, A. Scott, Y.-C. Tien, R. Murray, L. Sabourin, Y. Zhang and E. Topp, Appl. Environ. Microbiol., 2013, 79, 5701–5709 CrossRef CAS PubMed.
- S. R. Harris, E. J. Feil, M. T. G. Holden, M. A. Quail, E. K. Nickerson, N. Chantratita, S. Gardete, A. Tavares, N. Day, J. A. Lindsay, J. D. Edgeworth, H. de Lencastre, J. Parkhill, S. J. Peacock and S. D. Bentley, Science, 2010, 327, 469–474 CrossRef CAS PubMed.
- A.-P. Magiorakos, A. Srinivasan, R. B. Carey, Y. Carmeli, M. E. Falagas, C. G. Giske, S. Harbarth, J. F. Hindler, G. Kahlmeter, B. Olsson-Liljequist, D. L. Paterson, L. B. Rice, J. Stelling, M. J. Struelens, A. Vatopoulos, J. T. Weber and D. L. Monnet, Clin. Microbiol. Infect., 2012, 18, 268–281 CrossRef CAS PubMed.
- A. Hinenoya, S. P. Awasthi, N. Yasuda, A. Shima, H. Morino, T. Koizumi, T. Fukuda, T. Miura, T. Shibata and S. Yamasaki, Jpn. J. Infect. Dis., 2015, 68, 276–279 CrossRef CAS PubMed.
- Z. Noszticzius, M. Wittmann, K. Kály-Kullai, Z. Beregvári, I. Kiss, L. Rosivall and J. Szegedi, PLoS One, 2013, 8, e79157 CrossRef CAS PubMed.
- Y. C. Pang, J. Y. Xi, Y. Xu, Z. Y. Huo and H. Y. Hu, Appl. Microbiol. Biotechnol., 2016, 100, 6435–6446 CrossRef CAS PubMed.
- S. Jia, P. Shi, Q. Hu, B. Li, T. Zhang and X. X. Zhang, Environ. Sci. Technol., 2015, 49, 12271–12279 CrossRef CAS PubMed.
- W. Sun, W. Liu, L. Cui, M. Zhang and B. Wang, Sci. Total Environ., 2013, 458–460, 169–175 CrossRef CAS PubMed.
- Y. Q. Chen, X. D. Duan, P. P. Lu, Q. Wang, X. J. Zhang and C. Chen, Huanjing Kexue, 2012, 33, 104–109 Search PubMed.
| This journal is © The Royal Society of Chemistry 2019 |