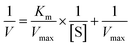Open Access Article
Open Access ArticlePurification and characterization of alkaline phosphatase from lactic acid bacteria
Yu-Hao Chuab,
Xin-Xin Yuab,
Xing Jinab,
Yu-Tang Wangab,
Duo-Jia Zhaoab,
Po Zhangab,
Guang-Mei Sunab and
Ying-Hua Zhang *ab
*ab
aKey Laboratory of Dairy Science, Ministry of Education, Northeast Agricultural University, China. E-mail: yinghuazhang@neau.edu.cn; Fax: +86 451 5519 0340; Tel: +86 451 55190479
bDepartment of Food Science, Northeast Agricultural University, Harbin 150030, P. R. China
First published on 2nd January 2019
Abstract
Alkaline phosphatase (ALP) excreted from lactic acid bacteria (LAB) showed the ability to degrade organophosphorus pesticides. This study reported the first purification and characterization of ALP from LAB. The molecular weight of ALP was estimated to be 43 kDa measured by SDS-PAGE. The activity of purified enzyme was determined with the binding of p-nitrophenyl phosphate as the substrate. The results showed that the optimal temperature for ALP activity was 37 °C, and the optimal pH was 8.5. But ALP was stable at temperatures below 32 °C. The ALP activity remained at 80% when the pH was 8–9.5. The enzyme activity could be activated by Mg2+, Ca2+, and inhibited by Cu2+, Zn2+, and EDTA. The Michaelis–Menten constant was 6.05 mg kg−1 with dimethoate as the substrate according to the Lineweaver–Burk plots. These results highlight an important potential use of ALP from LAB for the cleanup of pesticide pollution in raw materials for the food industry.
1 Introduction
Organophosphorus compounds are widely used as pesticides in agriculture. Organophosphorus pesticides (OPPs) are essential to modern agriculture, and extensively used to control pests and weeds in order to improve the agricultural harvest. Their persistent consumption worldwide has led to the accumulation of these compounds in water and fertile land. This exposure presents a major hazard to flora, aquatic life, agricultural products, and eventually human beings.1,2 Over the past decade, numerous ways have been developed to degrade OPPs, including physical degradation, chemical oxidation and biological methods. Recent interest has focused on the use of microbial organisms and enzymes as an efficient method. In this scenario, OPPs-degrading enzymes have attracted more attention as promising candidates due to their high efficiency compared to other methods.3,4 The identification of microbial enzymes that have the ability to degrade OPPs has resulted in encouraging biotechnology platforms with noteworthy applications.Enzyme based pesticide degradation is an innovative method for removal of pesticide from polluted environments and raw material. There are classes of microbial enzymes that have the ability to degrade harmful OPPs. The most studied and potential OPPs-degrading enzymes are organophosphorus hydrolase and organophosphorus acid anhydrolase, which have both been characterized from a number of organisms, such as fungi, bacteria, actinomycetes and viruses.5 An enzyme capable of degrading OPPs was produced by Flavobacterium sp. and identified as Sphingobium fuliginis.6 Spirulina platensis was capable of excreting alkaline phosphatase showed the ability to degrade chlorpyrifos.7 With recent advances in molecular biology, microbial systems and enzymes can be characterized and utilized more efficiently. Organophosphate hydrolase encoding gene has been isolated from different species, and has been sequenced, cloned in different organisms. Engineered microorganisms have been tested for their ability to degrade different specific pesticide. But challenge is that they cannot introduce in the field and industry because they might cause some other environmental problems. Limitations of using microbial enzyme for OPPs degradation in applications consisted of the high cost of preparing pure enzyme due to low yields and also poor enzyme stability.
Lactic acid bacteria (LAB), as a functional class of microaerophilic Gram-positive bacteria, ferment lactose to primarily produce lactic acid. LAB has been of major concern to microbiologists since human beings began eating fermented foods via the traditional biotechnology.8,9 LAB plays crucial roles in manufacturing of fermented milk products, vegetables, meat and wine.
Nowadays, LAB was investigated for the effects on OPPs degradation. For example, Lactobacillus brevis WCP902 and Lactobacillus plantarum were studied for their abilities to degrade OPPs in kimchi and wheat dough.10,11 Researchers measured the degradation kinetics of seven OPPs in yoghurt fermentation and in skimmed milk cultured with Lactobacillus sp.12,13 In addition, researchers began to pay more attention to phosphatase extracted from bacteria to degrade pesticides.14–16 Spirulina platensis capable of excreting alkaline phosphatase showed ability to degrade chlorpyrifos.17 The phosphatase production of the inoculated LAB might be one of the key factors responsible for the accelerated OPPs degradation.18 However, the purification of alkaline phosphatase from LAB has not been reported.
Alkaline phosphatase (EC 3.1.3.1, ALP) is hydrolase that has been implemented to remove phosphate groups from many types of molecules, such as nucleotides, proteins, and alkaloids. The process of removing the phosphate group is dephosphorylation. Many bacteria produce ALP, such as Escherichia coli, Bacillus species, Mycobacterium smegmatis, Thermotogamaritime, Haloarcula marismortui, especially Gram-negative bacteria starved of inorganic phosphate.19 LAB is permitted for processing in food raw materials.
In order to investigate the fundamental basis of OPPs biodegradation by the phosphatase from LAB, the purification of ALP from LAB was developed in this study. More knowledge about the physicochemical and kinetic characteristics of the enzyme is necessary before it can be employed as OPPs biodegradation tool. The purity of the products was analyzed by electrophoresis, and some characteristics of the purified enzyme were determined. The research targeted critical issues of industrial applications including enzymatic catalysis, thermo-stability, the ability to function at high temperature and extreme pH. The aim of the research provides more cost-effective and efficient processes, and ultimately allows for successful implementation of OPPs degradation systems under a variety of field conditions. Characterizing the enzyme involved in the application represents new areas for food industry.
2 Experimental
2.1 Materials
The lyophilized Lactobacillus casei. 355 was obtained from the Centre of Key Laboratory of Dairy Science (Harbin, China). p-Nitrophenyl phosphate was from Sigma Chemical Co. (Saint Louis, MO, USA). DEAE Sepharose Fast Flow and Superdex 75 were purchased from GE Healthcare Bio-Sciences (Connecticut, Fairfield, USA). Dimethoate standard was purchased from Sigma Chemical Co. (Saint Louis, MO, USA). All chemicals used were of analytical agents.2.2 Determination of ALP activity
The activity of ALP was assayed using a colourless substrate p-nitrophenyl phosphate (p-NPP) as per the method, which yielded yellow p-nitrophenol (p-NP).17 The reaction mixture consisted of 1.0 mL 5 mmol L−1 p-NPP (prepared in the carbonate buffer containing 1 mmol L−1 MgCl2), 1.0 mL carbonate buffer (10 mmol L−1, pH 10.5), 1.0 mL crude enzyme solution and distilled water, with a fixed final volume of 5.0 mL. After a reaction time of 60 min at 37 °C, the reaction was stopped by adding 3.0 mL termination buffer (0.1 mol L−1 NaOH and 5 mmol L−1 EDTA). The generated p-NP was measured by UV-2401 PC spectrophotometer (Shimadzu Corporation, Kyoto, Japan) at 405 nm.One unit (U) of phosphatase activity is defined as the amount of enzyme liberating 1 μmol of p-NP per minute at 37 °C.
2.3 Preparation of crude enzyme
The medium was inoculated with Lactobacillus casei. 355 at 37 °C for 24 h in Erlenmeyer flasks, and harvested by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min at 4 °C. The cell pellet was washed three times with carbonate–bicarbonate buffer (10 mmol L−1, pH 10.5), resuspended in carbonate–bicarbonate buffer and agitated for 1 min. The mixture was sonicated 10 times (10 s at 15 s interval), as described in a previous study, and centrifuged at 10
000 × g for 10 min at 4 °C. The cell pellet was washed three times with carbonate–bicarbonate buffer (10 mmol L−1, pH 10.5), resuspended in carbonate–bicarbonate buffer and agitated for 1 min. The mixture was sonicated 10 times (10 s at 15 s interval), as described in a previous study, and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g and 4 °C for 15 min.20 The supernatant was collected for subsequent enzyme purification.
000 × g and 4 °C for 15 min.20 The supernatant was collected for subsequent enzyme purification.
2.4 Ammonium sulfate precipitation
The crude enzyme solution was added ammonium sulfate to the degree of 80% saturation, and was kept at 4 °C overnight. Subsequently, the precipitate was then collected by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min at 4 °C. The protein was dissolved in minimal amount of Tris–HCl buffer (50 mmol L−1, pH 7.4), and dialyzed against the same buffer for 24 h at 4 °C.
000 × g for 10 min at 4 °C. The protein was dissolved in minimal amount of Tris–HCl buffer (50 mmol L−1, pH 7.4), and dialyzed against the same buffer for 24 h at 4 °C.
2.5 ALP purification
The concentrated crude enzyme was loaded onto DEAE Sepharose column (16 × 200 mm) pre-equilibrated with 50 mmol L−1 Tris–HCl buffer (pH 7.4). The column was washed with buffer until no protein was detected in the elutriate. Then it was eluted using a gradient of 0–1 mol L−1 NaCl in the same buffer at flow rate as 2 mL min−1. Fractions of 5 mL were collected and analyzed for protein content and enzyme activity. Fractions containing ALP were pooled, dialyzed overnight, and lyophilized.The lyophilized enzyme was dissolved in Tris–HCl buffer (50 mmol L−1, pH 7.4). The enzyme solution from the previous step was processed on a column with Superdex 75 (10 × 300 mm) equilibrated previously with Tris–HCl buffer. Elution was performed with the same buffer at a flow rate 0.8 mL min−1. Fractions containing ALP were pooled, dialyzed against Tris–HCl buffer and lyophilized.
The protein concentration was measured following the method of Bradford, by using bovine serum albumin as standard.21
2.6 Sodium dedecylsulfate polyacrylamide gel electrophoresis
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis was performed as per the method.22 Sample solutions of 10 μL were loaded into each well. Protein markers with molecular weights of 14.4–97.4 kDa were used in analysis.2.7 Enzyme characteristics
where [S] is the concentration of substrate, V and Vmax represented the initial and maximum reactions rate, respectively. Km is the Michaelis–Menten constant, which equals to the substrate concentration when the rate is half of Vmax.
2.8 Degradation of dimethoate by ALP
The degradation of dimethoate was performed in buffer solution. A volume of 50 mL buffer was spiked with dimethoate of different concentrate and ALP. The reaction mixture was stirred at 37 °C for 1, 2, 3, 4 h, respectively. Pesticides were extracted by a liquid extraction method. The conditions and procedures were the same as those previously reported.23 Finally, 1 mL of the extract (7.5 U mg−1) was transferred into sample bottle for GC analysis by Gas Chromatography (Agilent 7890, Agilent Technologies, Inc, Santa Clara, USA) with 30 m × 0.250 mm × 0.25 μm capillary column DB-1701.2.9 Statistical analyses
All the experiments were performed with at least three times. All the results were expressed as means of all trials. Standard deviations were determined and reported.3 Results and discussion
3.1 Purification of ALP from lactic acid bacteria
The supernatant obtained from cell free extract was concentrated after precipitation by ammonium sulfate. The protein solution was then loaded into ion exchange chromatography columns. A peak of the active ALP fraction was collected with NaCl solution as eluate (Fig. 1A). The active ALP fraction was further purified by gel filtration chromatography Superdex 75 (Fig. 1B). The results of the purification were summarized in Table 1. The enzyme was purified 36.07-fold to a specific activity of 123.71 U mg−1 protein with a yield of 23.74%. The relatively low yield is attributed to the fact that only fractions from the centers of chromatographic peaks and active bands were collected to ensure the purification.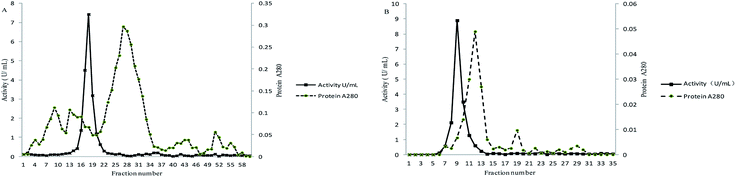 | ||
| Fig. 1 Purification of the ALP from lactic acid bacteria (A) anion exchange chromatography (DEAE-FF); (B) gel filtration molecular sieve Superdex 75. | ||
| Purification step | Total activity (units) | Total protein (mg) | Specific activity (U mg−1) | Purification fold | Recovery (%) |
|---|---|---|---|---|---|
| Crude extract | 1876.00 | 547.55 | 3.43 | 1.00 | 100.00 |
| (NH4)2SO4 | 1432.80 | 239.00 | 5.99 | 1.74 | 76.00 |
| DEAE-Sephadex | 1064.52 | 23.06 | 39.17 | 11.42 | 56.74 |
| Superdex 75 | 445.36 | 3.60 | 123.71 | 36.07 | 23.74 |
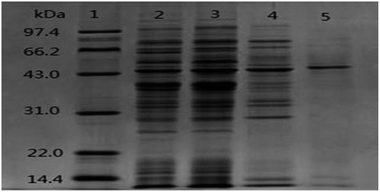
The SDS-PAGE pattern of samples at different purification steps was showed in Fig. 2. After the chromatography process, ALP displayed a single protein band on the electrophoresis gel with a molecular weight of about 43 kDa. The active ALP from LAB appeared on SDS-PAGE gels as a monomer with molecular weight of 43 kDa. The novel alkaline phosphatase, Pi-alkaline phosphatase showed as homodimer with molecular weight of about 54 kDa.24 ALP from B. subtilis is a dimer made up of two subunits with identical molecular weight between 35 to 70 kDa.25 Although most ALP from bacterial and vertebrate are homodimers, monomeric forms may also exist. A monomeric ALP from Thermotoga neapolitana with a molecular mass of 45 kDa has been reported.26 B. cereus produced extracellular and membrane-bound ALP, which molecular weight was estimated to be 44 kDa.27 In the present study, the single predominant band corresponding to active ALP was detected on SDS-PAGE, suggesting that the molecular mass of purified alkaline phosphatase is 43 kDa. The enzyme is comparable with that of microbial enzymes, lower than most mammalian ALP with the molecular weight in the range of 130 to 170 kDa.
3.2 Optimal pH and temperature of ALP from lactic acid bacteria
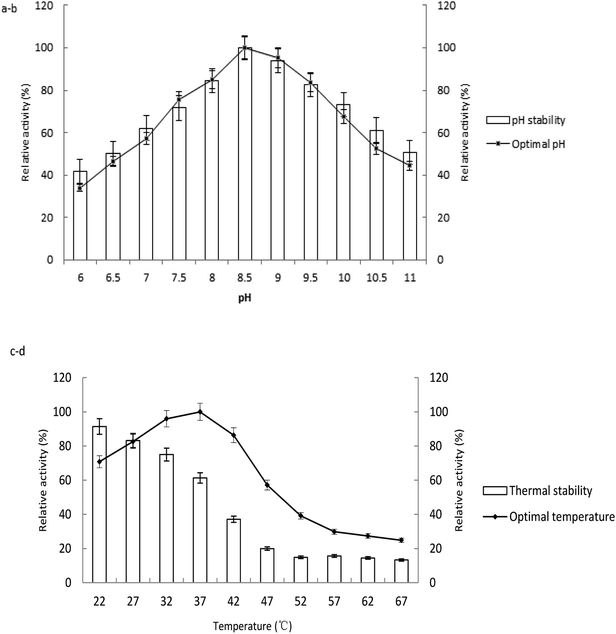
The enzyme activities measured at various temperatures and pH were showed in Fig. 3. The optimum pH and temperature were determined by the samples with the highest enzyme activity. The enzyme activities of the untreated samples were 100% at the pH stability and temperature tolerance tests. The enzyme had optimal activity at pH 8.5. The activity decreased above or below pH 8.5, which confirmed that the enzyme was alkaline. The activity maintained stable at the range of pH 8.5 to 10 after 1 h of incubation in the buffer (Fig. 3a and b).The optimal environmental temperature for application of the enzyme was confirmed as 37 °C (Fig. 3c). The enzyme is sensitive to heating after incubation at various temperatures for 3 h. The enzyme activity was stable below 27 °C, with relative 80% activity compared to the sample at −20 °C. The activity decreased rapidly when the temperature was above 37 °C. 39.0, 80.1 and 84.4% loss in enzymatic activity was observed upon incubation at 37, 47, 57 °C, respectively (Fig. 3d).
The purified ALP from the LAB displayed maximum activity at pH 8.5 and 37 °C with p-NPP as substrate. The optimal pH of ALP from Lactobacillus is similar with those of enzymes from other microbe species such as Rhizopus microsporus,28 Halobacterium halobium,29 filamentous fungus Aspergillus caespitosus.30 The optimal temperature for ALP from LAB corresponded to the growth temperature of the bacterium. The results were similar to those reported by others, such as cyanobacterium Spirulina platensis as 40 °C,17 clam Scrobicularia plana as 30–40 °C,31 Stichopus japonicus as 40 °C,32 but lower than that of the enzyme from Neurospora crassa as 65 °C,33 Thermotoga neapolitana as 85 °C.26 The activity of the purified ALP has shown to be stable during storage, and capable of keeping activity at low temperature and over a wide range of pH. These properties would make it ideal for the enzyme to be immobilized on a matrix. The low temperature stability also indicates that the biomass should be cultivated in low temperature fermentation.
3.3 Effects of EDTA and metal ions on ALP activity
It has certain significance to explore the characteristics of ALP and to investigate the role of ions on the molecular structure and catalytic activity of the enzyme. The effects of metal ions at different concentrations on ALP activity were shown (Table 2). The ALP activity was significantly enhanced by Ca2+ and Mg2+. This enzyme was strongly inhibited by Cu2+, Zn2+ and EDTA. It is clear that ALP from LAB is a metalloenzyme according to these results. Experimental data shows that Ca2+ and Mg2+ has clear activation effect to ALP, which may be a metal ion and enzyme active center union to promote the combination between the joint of the enzyme and substrate, so as to accelerate the enzyme catalytic reaction. Its activity was reduced nearly to zero in the presence of EDTA, which chelate the ions to destroy the active site of ALP resulting in the loss of enzyme activity. The highly sensitive and selective effects of Zn2+, Cu2+, Mg2+, and Ca2+ on the enzyme are based on their inhibiting, activating or reactivating effects on the catalytic activity of ALP. The essential role of Ca2+ and Mg2+ in catalyzing and stabilizing the structure of this enzyme was recognized. As a prosthetic group of ALP, Ca2+ is necessary both for the preservation of the enzyme structure and for the catalytic activity of ALP. Mg2+ controls the occupancy of the catalytic and structural binding sites, and modulates the enzymatic activity.| Metal ions and EDTA | Relative activity (%) | ||||
|---|---|---|---|---|---|
| 1 mmol L−1 | 2 mmol L−1 | 3 mmol L−1 | 4 mmol L−1 | 5 mmol L−1 | |
| a Values are given as the means ± SD. Different letters (A–E, a–e) in the same column and rank indicate significant differences (P < 0.05). | |||||
| Ca2+ | 91.57 ± 0.001Ae | 96.88 ± 0.011Bd | 108.43 ± 0.008Bc | 119.75 ± 0.011Bb | 136.14 ± 0.019Ba |
| Mg2+ | 80.14 ± 0.013Be | 113.74 ± 0.009Ad | 143.76 ± 0.028Ac | 170.21 ± 0.055Ab | 188.00 ± 0.002Aa |
| Zn2+ | 47.23 ± 0.007Ca | 43.76 ± 0.004Cb | 38.11 ± 0.009Cc | 29.91 ± 0.002Cd | 25.29 ± 0.004Ce |
| Cu2+ | 26.10 ± 0.005Ea | 9.00 ± 0.004Db | 0Dc | 0Dc | 0Dc |
| EDTA | 43.88 ± 0.006Da | 2.08 ± 0.002Eb | 0Dc | 0Dc | 0Dc |
It is generally accepted that ions are essential not only for the catalytic activity of ALP, but also for the stability of the enzyme. The similar results were also in agreement with previous studies. The ALPs from caespitosus and S. thermophilum are also activated by Mg2+.34,35 The stimulation by Mg2+ is non-selective, and Mn2+ or Ca2+ can replace Mg2+ to stimulate the enzyme.36 The possible reason of Zn2+ inhibition of the activity of ALP could be attributed to the displacement of Mg2+.37 The key amino acids that facilitate the formation of hydrogen bonds between pesticides and the ALP were as Gln375, Asp55 or Thr413.38 Binding of the Zn2+ and Cu2+ in the active site might impede the hydrogen bonds.
3.4 Kinetic properties of ALP
The method of initial rates was used in determining the kinetic parameters. The Lineweaver–Burk plot for the ALP with p-NPP as substrate was shown in Fig. 4. Km of the enzyme employing p-NPP as substrate was 2.14 mmol L−1, and the Vmax of the enzyme was 0.19 μmol L−1 min−1.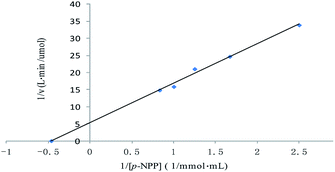 | ||
| Fig. 4 Lineweaver–Burk plot for the Michaelis–Menten constant (Km) and the maximum velocity (Vmax) for the ALP with p-NPP as substrate. | ||
3.5 Accelerated degradation of dimethoate by ALP
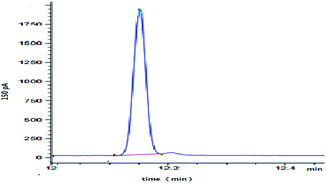
Dimethoate was treated through liquid–liquid extraction. Typical GC profiles for the present study are shown in Fig. 5. The detection of the dimethoate was in the range of 0.005–0.010 mg kg−1, which ensured a precise measurement of the dimethoate in buffer. The samples with different dimethoate concentrations cultured with ALP at 37 °C for 4 h, and the dimethoate concentration in the samples had decreasing trend along with the reaction time (Table 3). Fig. 6 showed the Lineweaver–Burk plot for the ALP with dimethoate as substrate. The Km and the Vmax of the ALP for dimethoate were 6.05 mg kg−1 and 0.74 mg kg−1 h−1, respectively.| Dimethoate samples | The concentration (mg kg−1) at different times (h) | ||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |
| a Values are given as the means ± SD. Different letters (a–e) in the same rank indicate significant differences (P < 0.05). | |||||
| 1 | 1.089 ± 0.032a | 0.992 ± 0.022a | 0.877 ± 0.049b | 0.786 ± 0.038b | 0.662 ± 0.042c |
| 2 | 2.001 ± 0.031a | 1.944 ± 0.016a | 1.713 ± 0.009b | 1.554 ± 0.035c | 1.268 ± 0.023d |
| 3 | 3.127 ± 0.028a | 2.973 ± 0.037a | 2.663 ± 0.021b | 2.446 ± 0.014c | 2.219 ± 0.011d |
| 4 | 4.012 ± 0.017a | 3.838 ± 0.046b | 3.456 ± 0.018c | 3.147 ± 0.019d | 2.974 ± 0.014e |
| 5 | 5.137 ± 0.006a | 4.873 ± 0.036b | 4.464 ± 0.017c | 4.039 ± 0.042d | 3.621 ± 0.034e |
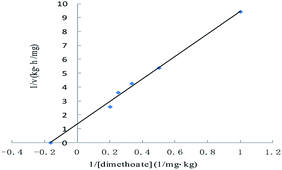 | ||
| Fig. 6 Lineweaver–Burk plot for the Michaelis–Menten constant (Km) and the maximum velocity (Vmax) for the ALP with dimethoate as substrate. | ||
The evidence of OPPs biodegradation by the purified enzyme from an edible microbial organism was provided in this study. Since LAB can be used in the food processing for a variety of raw materials, using LAB for OPPs biodegradation appears to be an inexpensive and ideal option for feed and fodder to degrade OPPs. The results shown in this study provided unequivocal evidences for the involvement of an ALP that can accelerate degradation of OPPs. These findings make LAB as a tool for bioremediation by either employing the whole cells in effluent treatment food or by immobilizing the purified enzyme on a strong matrix to decontaminate polluted raw materials in food industry.
4 Conclusions
In this study we reported the purification of ALP from LAB, and determined some of their molecular and enzymatic properties. Because of the preservation of activity at the relatively broad range of temperature and pH, the isolated ALP from LAB could be of significant biotechnological potential. The results of this paper were to characterize the ALP and to analyze the effect of metals ions on enzyme activity in vitro to estimate its potential use as OPPs cleaner for food process. The present study will help in finding an exact correlation between the pesticide degradation and the ALP in the LAB fermentation.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This work was funded by the National Natural Science Foundation of China (Grant No. 31571930).References
- P. Berny, J. Vet. Pharmacol. Ther., 2007, 30, 93–100 CrossRef CAS PubMed.
- G. M. Calvert, J. Karnik, L. Mehler, J. Beckman, B. Morrissey, J. Sievert, R. Barrett, M. Lackovic, L. Mabee, A. Schwartz, Y. Mitchell and S. Moraga-McHaley, Am. J. Ind. Med., 2008, 51, 883–898 CrossRef PubMed.
- G. Gotthard, J. Hiblot, D. Gonzalez, M. Elias and E. Chabriere, PLoS One, 2013, 8, e7799511 CrossRef PubMed.
- R. Iyer, B. Iken and A. Damania, Environ. Microbiol. Rep., 2013, 5, 787–798 CrossRef CAS PubMed.
- J. P. Verma, D. K. Jaiswal and R. Sagar, Rev. Environ. Sci. Bio/Technol., 2014, 13, 429–466 CrossRef.
- K. Kawahara, A. Tanaka, J. Yoon and A. Yokota, J. Gen. Appl. Microbiol., 2010, 56(3), 249–255 CrossRef CAS PubMed.
- R. R. M. Thengodkar and S. Sivakami, Biodegradation, 2010, 21(4), 637–644 CrossRef CAS PubMed.
- G. Giraffa, FEMS Microbiol. Rev., 2004, 28, 251–260 CrossRef CAS PubMed.
- G. W. Tannock, Appl. Environ. Microbiol., 2004, 70, 3189–3194 CrossRef CAS PubMed.
- T. M. Dordevic, S. S. Siler-Marinkovic, R. D. Durovic-Pejcev, S. I. Dimitrijevic-Brankovic and J. S. G. Umiljendic, Lett. Appl. Microbiol., 2013, 57, 412–419 CrossRef CAS PubMed.
- S. M. A. Islam, R. K. Math, K. M. Cho, W. J. Lim, S. Y. Hong, J. M. Kim, M. G. Yun, J. J. Cho and H. D. Yun, J. Agric. Food Chem., 2010, 58, 5380–5386 CrossRef CAS PubMed.
- L. Bo, Y. Zhang and X. Zhao, J. Serb. Chem. Soc., 2011, 76, 353–362 CrossRef CAS.
- X. Zhao and J. Wang, Food Chem., 2012, 131, 300–304 CrossRef CAS.
- E. S. Gilbert, A. W. Walker and J. D. Keasling, Appl. Microbiol. Biotechnol., 2003, 61, 77–81 CrossRef CAS PubMed.
- Y. Kim, J. Ahn, S. Moon and J. Lee, Chemosphere, 2005, 60, 1349–1355 CrossRef CAS PubMed.
- R. D. Richins, I. Kaneva, A. Mulchandani and W. Chen, Nat. Biotechnol., 1997, 15, 984–987 CrossRef CAS PubMed.
- R. R. M. Thengodkar and S. Sivakami, Biodegradation, 2010, 21, 637–644 CrossRef CAS PubMed.
- Y. Zhang, D. Xu, J. Liu and X. Zhao, Food Chem., 2014, 164, 173–178 CrossRef CAS PubMed.
- B. L. Wanner, J. Cell. Biochem., 1993, 51, 47–54 CrossRef CAS PubMed.
- C. Sasikala, S. Jiwal, P. Rout and M. Ramya, World J. Microbiol. Biotechnol., 2012, 28, 1301–1308 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CrossRef CAS PubMed.
- L. Bo and X. Zhao, Afr. J. Microbiol. Res., 2010, 4, 1171–1179 CAS.
- T. Ansai, S. Awano, X. Chen, T. Fuchi, T. Arimoto, S. Akifusa and T. Takehara, FEBS Lett., 1998, 428, 157–160 CrossRef CAS PubMed.
- F. M. Hulett, E. E. Kim, C. Bookstein, N. V. Kapp, C. W. Edwards and H. W. Wyckoff, J. Biol. Chem., 1991, 266, 1077–1184 CAS.
- G. Dong and J. G. Zeikus, Enzyme Microb. Technol., 1997, 21, 335–340 CrossRef CAS PubMed.
- S. Kostadinova and M. Marhova, Biotechnol. Biotechnol. Equip., 2010, 24, 602–606 CrossRef.
- A. Barbosa Junior, L. H. S. Guimaraes, H. F. Terenzi, J. A. Jorge, F. A. Leone and M. L. T. M. Polizeli, Folia Microbiol., 2008, 53, 509–516 CrossRef PubMed.
- M. L. Bonet, F. I. Llorca and E. Cadenas, Int. J. Biochem., 1992, 24, 839–845 CrossRef CAS PubMed.
- L. Guimaraes, H. F. Terenzi, J. A. Jorge, F. A. Leone and M. Polizeli, Folia Microbiol., 2003, 48, 627–632 CrossRef CAS.
- M. T. Mazorra, J. A. Rubio and J. Blasco, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 2002, 131, 241–249 CrossRef CAS.
- H. Wu, D. Li, B. Zhu, J. Cheng, J. Sun, F. Wang, Y. Yang, Y. Song and C. Yu, Fish. Sci., 2013, 79, 477–485 CrossRef CAS.
- K. R. Bogo, D. C. Masui, F. A. Leone, J. A. Jorge and R. P. M. Furriel, Folia Microbiol., 2006, 51, 431–437 CrossRef CAS.
- L. H. S. Guimaraes, H. F. Terenzi and J. A. Jorge, Folia Microbiol., 2003, 48(5), 627–632 CrossRef CAS.
- L. H. S. Guimaraes, H. F. Terenzi, J. A. Jorge and M. L. T. M. Polizeli, J. Ind. Microbiol. Biotechnol., 2001, 27(4), 265–270 CrossRef CAS PubMed.
- M. Politino, J. Brown and J. J. Usher, Prep. Biochem. Biotechnol., 1996, 26(3–4), 171–181 CrossRef CAS PubMed.
- A. Barbosa, L. H. S. Guimaraes, H. F. Terenzi, J. A. Jorge, F. A. Leone and M. L. T. M. Polizeli, Folia Microbiol., 2008, 53(6), 509–516 CrossRef CAS PubMed.
- Y. H. Chu, Y. Li, Y. T. Wang, B. Li and Y. H. Zhang, Food Chem., 2018, 254, 80–86 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |