Open Access Article
Open Access ArticleDifferences between the binding modes of enantiomers S/R-nicotine to acetylcholinesterase
Ji Yangab,
Yongkuan Chen*b,
Zhihua Liub,
Liu Yangb,
Jianguo Tangb,
Mingming Miaob,
Na Gana and
Hui Li *a
*a
aSchool of Chemical Engineering, Sichuan University, Chengdu, Sichuan 610065, China. E-mail: lihuilab@sina.com; Fax: +86 028 85401207; Tel: +86 026 85405220
bR&D Center of China Tobacco Yunnan Industrial Co., Ltd., Kunming, 650231, China. E-mail: cyk1966@163.com
First published on 11th January 2019
Abstract
Nicotine causes neurotoxic effects because it quickly penetrates the blood–brain barrier after entering the human body. Acetylcholinesterase (AChE) is a key enzyme in the central and peripheral nervous system associated with neurotoxicity. In this study, a spectroscopic method and computer simulation were applied to explore the mode of interaction between AChE and enantiomers of nicotine (S/R-nicotine). Fluorescence spectroscopy showed that the quenching mechanism of endogenous fluorescence of AChE by S/R-nicotine was static, as confirmed by the time-resolved steady-state fluorescence. The binding strength of both nicotine to AChE was weak (S-AChE: Ka = 80.06 L mol−1, R-AChE: Ka = 173.75 L mol−1). The main driving forces of S-AChE system interaction process were van der Waals force and hydrogen bonding, whereas that of R-AChE system was electrostatic force. Computer simulations showed that there were other important forces involved. S/R-Nicotine had a major binding site on AChE, and molecular docking showed that they bound mainly to the cavities enclosed by the active sites (ES, PAS, OH, AACS, and AP) in the protein. UV-vis spectroscopy and 3D spectroscopy indicated that nicotine significantly affected the microenvironment of Trp amino acids in AChE. The CD spectra indicated that S-nicotine increased the α-helical structure of AChE, but the overall conformation did not change significantly. By contrast, R-nicotine significantly changed the secondary structure of AChE. 5,5′-Dithiobis-2-nitrobenzoic acid (DTNB) method indicated that S and R nicotine produced different degrees of inhibition on the catalytic activity of AChE. Both experimental methods and computer simulations showed that R-nicotine had a significantly higher effect on AChE than S-nicotine. This research comprehensively and systematically analyzed the mode of interaction between nicotine and AChE for neurotoxicity assessment.
1. Introduction
Nicotine (Fig. 1) is an alkaloid found in Solanaceae species and is an important component in tobacco.1 Nicotine is optically active, with both left-handed and right-handed conformations, namely S/R-nicotine; S-nicotine exists in naturally.2,3 When nicotine enters the body through smoking cigarettes, it quickly passes through the blood–brain barrier into the brain, acting on the nicotine acetylcholine receptor.4,5 Although the neurotoxicity of nicotine is considerable, studies on the interaction between nicotine and proteins are rarely reported. Acetylcholinesterase (AChE) is mainly distributed in the nervous system, particularly the brain, and can rapidly catalyze the hydrolysis of the neurotransmitter acetylcholine, leading to the termination of nerve impulse transfer, thereby ensuring the normal physiological functions of the human body.6,7 AChE is closely related to cell development and maturation, promotes neuronal development and nerve regeneration, and is one of the most important enzymes in the nervous system. Any active substance in the brain that affects the activity of the enzyme may cause neurotoxicity.8–10 Wang et al.11 analyzed the interaction between AChE and enantiomers (BDE-47 and BDE-209) and found that BDE-209 has significant neurotoxic effect to humans. Moreover, BDE-47 is more likely to bind to AChE than BED-209.Common methods used to investigate the binding of small ions or molecules to albumin proteins are fluorescence,12,13 synchronous fluorescence,14 UV-visible,15,16 FTIR17 and circular dichroism (CD)18 spectroscopy, as well as equilibrium dialysis19 and potentiometry.20,21 Among these methods, equilibrium dialysis is used widely; but it requires the analysis of free and total drug concentration and takes a long time. In the potentiometric method, ion selective electrodes are used. Unfortunately, these electrodes are nonselective in case of many ligands, such as e.g. drug molecules.20,21 Fluorescence spectroscopy is very helpful in protein–ligand interactions, because it can provide some useful information about the binding of ligands to protein such as binding mechanism, binding mode, binding constants and number of binding sites.16
Studying the interaction between S/R-nicotine and AChE has important significance for determining the damage mechanism and toxicology of nicotine in humans and also provides experimental and theoretical guidance for the development of new tobacco products. In this study, a variety of spectral methods were used to elucidate the binding mechanism of the AChE-S/R-nicotine systems, including binding strength, number of binding sites, thermodynamic constants, quenching mechanism, and other important parameters. Synchronous fluorescence spectroscopy and three-dimensional fluorescence spectroscopy combined with CD spectroscopy were used to explore the effect of S/R-nicotine on AChE structure. UV-vis spectroscopy was employed to validate the results. Comprehensive molecular docking and molecular dynamics (MD) simulations were applied to study the interaction mode under simulated physiological conditions. The theoretical prediction combined with the experimental results further confirmed the existence of a series of physical–chemical behaviors between the binary systems. The results help us understand the binding pattern between S/R-nicotine and AChE and assess the neurotoxicity of nicotine.
2. Reagents and methods
2.1 Reagents
AChE was purchased from Dalian Meilun Biotechnology Co., Ltd. (article number: MB0301); S-nicotine and R-nicotine were purchased from Mansite Biotech Co., Ltd. and Canadian TRC Chemical Co., Ltd.; 5,5′-disulfide (2-nitrophenylmethyl ester) (DTNB) and thioacetylcholine iodide (ATCh) were purchased from Tianjin Xiens Biochemical Technology Co., Ltd. The AChE mother liquor was dissolved in fresh Tris–HCl (pH 7.4) to a concentration of 0.5 mg L−1, the nicotine mother liquor and the initial concentration at the time of purchase. All liquors were stored in a refrigerator at 277 K in the dark and diluted to desired concentration with Tris–HCl. All reagents were of analytical grade and triple distilled water was used throughout the experiment.2.2 Methods
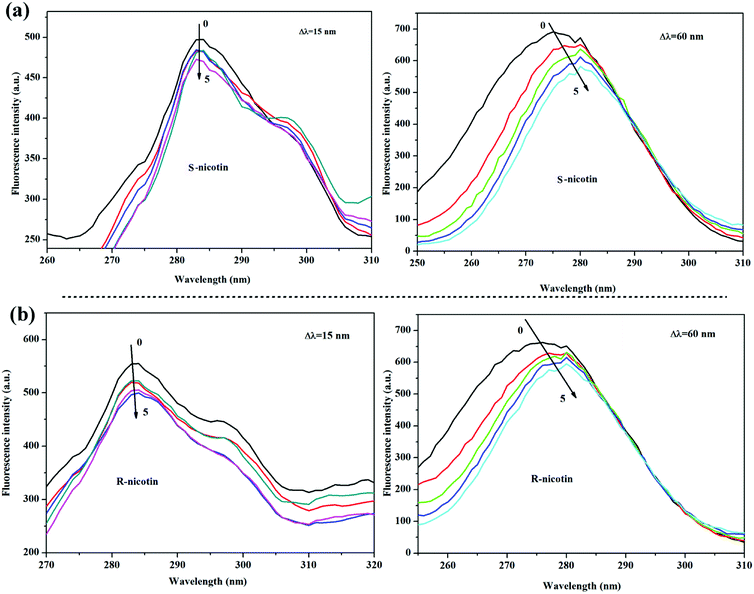
In the synchronous fluorescence spectroscopy, the excitation wavelength was 280 nm, the excitation and the emission slits were set to 20/10 nm, respectively, and the fluorescence emission spectra were scanned from 290 nm to 500 nm. The concentration of AChE was fixed at 0.075 mg L−1. The concentrations of S/R-nicotine were 0.124, 0.249, 0.373, 0.498, 0.622, and 0.746 mM.
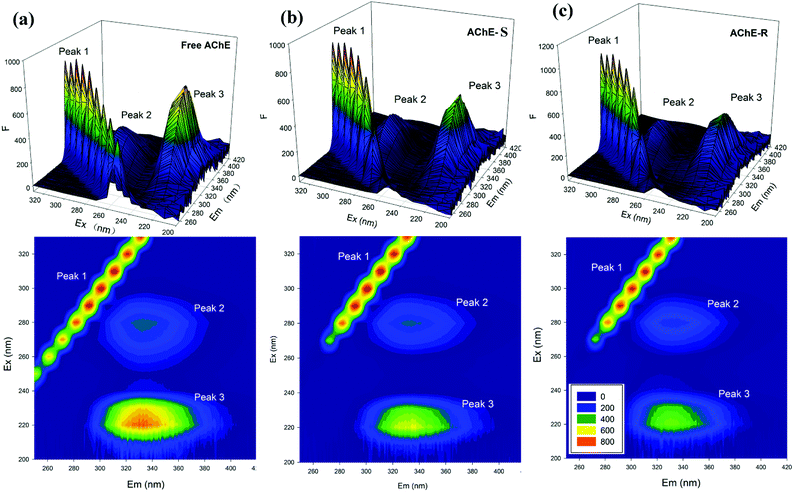
In the three-dimensional fluorescence spectroscopy, the photomultiplier tube voltage was 700 V, and the excitation and emission slits were both 10 nm. Scanning was performed at excitation wavelength range of 200–330 nm and emission wavelength range of 250–470 nm. The concentration of AChE was fixed at 0.075 mg L−1 and the concentration of S/R-nicotine was 0.746 mM.
For AChE in the presence of S/R-nicotine: 500 μL of 500 U L−1 AChE and 50 μL of 31 mM nicotine were added in 2.95 mL of Tris–HCl buffer solution (50 mM, pH = 7.4). The procedure for determination of free AChE was the same as described above.
3. Results and discussion
3.1 Quenching mechanism research
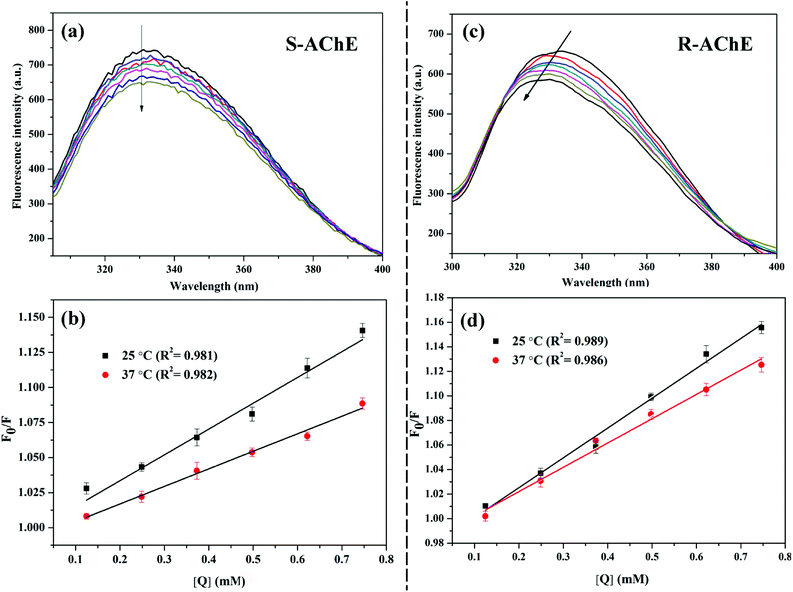
AChE is a macromolecule protein with endogenous fluorescence. The fluorescence intensity of AChE decreases upon interaction with a small molecule; this phenomenon is called fluorescence quenching.26 The quenching mechanism is divided into static and dynamic quenching, or a combination of two quenching mechanisms.27 The most common way to determine the mechanism of quenching is by examining the dependence of the quenching constant on temperature and the change in the fluorescence lifetime of the protein in the presence of small molecules. As shown in Fig. 2(a and c), as the concentration of S/R-nicotine gradually increases, the fluorescence intensity of AChE gradually decreases. The quenching constant Ksv was calculated by using the Stern–Volmer equation:28| F0/F = 1 + Ksv [Q] | (1) |
To verify this result, we examined the fluorescence lifetime of AChE with different S/R-nicotine concentrations. The average life 〈τ〉 was calculated by the following formula:29
| 〈τ〉 = α1τ1 + α2τ2 + α3τ3 | (2) |
The results are summarized in Table 1. The fitting results were evaluated with χ2, and after three fittings, χ2 ≈ 1, indicating that the fitting results were credible. The fluorescence lifetime map did not change significantly after adding different concentrations of S/R-nicotine (Fig. 3). The fluorescence lifetime of free AChE was approximately 3.659 ns. The addition of S-nicotine and R-nicotine caused the fluorescence lifetime of the protein to fluctuate at around 3.659 ns. However, the magnitude of the float was small, which may be caused by instrumental errors and differences in sample preparation. The results of fluorescence lifetime mapping were consistent with those obtained by the Stern–Volmer equation. Thus, the quenching mechanism of nicotine on the endogenous fluorescence of AChE was static quenching, and a ground-state complex is formed between the two molecules.30
| System | CT6 (mM) | τ1 (ns) | τ2 (ns) | τ3 (ns) | α1 | α2 | α3 | 〈τ〉 (ns) | χ2 |
|---|---|---|---|---|---|---|---|---|---|
| AChE | 0 | 2.295 | 0.532 | 5.669 | 0.428 | 0.110 | 0.462 | 3.659 | 1.088 |
| AChE-S | 0.4 | 2.423 | 0.539 | 5.811 | 0.452 | 0.121 | 0.427 | 3.641 | 1.119 |
| 0.8 | 2.317 | 0.456 | 5.797 | 0.457 | 0.113 | 0.429 | 3.597 | 1.130 | |
| AChE-R | 0.4 | 2.861 | 0.510 | 5.721 | 0.460 | 0.107 | 0.433 | 3.747 | 1.205 |
| 0.8 | 2.209 | 0.471 | 5.598 | 0.472 | 0.112 | 0.416 | 3.524 | 1.179 |
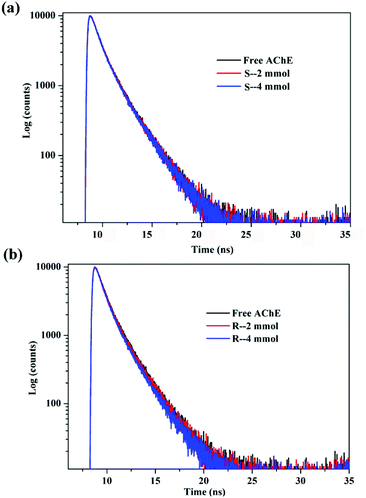 | ||
| Fig. 3 Time-resolved fluorescence spectra of the system in a 298 K environment (a) AChE-S-nicotine (b) AChE-R-nicotine. | ||
3.2 Analysis of binding stoichiometry, strength, and stability
The binding strength of ligand to protein can be judged by the binding constant between two molecules. Given that AChE and nicotine undergo static quenching, the modified Stern–Volmer was used to calculate the binding stoichiometry (n) and binding constants (Ka):31
log (F0 − F)/F = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka + n Ka + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[Q] log[Q]
| (3) |
| System | T/K | Ksv (L mol−1) | Ka (L mol−1) | n | ΔG (kJ mol−1) | ΔH (kJ mol−1) | ΔS (J mol−1K−1) |
|---|---|---|---|---|---|---|---|
| S-AChE | 298 | 183.96 ± 5.21 | 80.06 ± 5.00 | 0.90 ± 0.11 | −15.18 | −82.62 | −232.80 |
| 310 | 107.65 ± 1.22 | 22.63 ± 4.35 | 0.74 ± 0.04 | −15.45 | |||
| R-AChE | 298 | 242.11 ± 10.10 | 173.75 ± 2.44 | 0.99 ± 0.04 | −15.76 | −12.19 | 11.17 |
| 310 | 170.76 ± 5.34 | 144.65 ± 1.89 | 0.90 ± 0.07 | −16.02 |
Fig. 4(a and b) show the 100 conformations of the molecular docking results, which indicated the formation of clusters mainly at the same sites on the protein. Cluster analysis of all conformations yielded four major conformations for both S- and R-nicotine, and the first ones were combined in an active pocket surrounded by specific active sites as described above. Small molecules bind to the active site attachments and may interact with the amino acids that make up the active site, which in turn affects the related functional activities of proteins [Fig. 4(c and d)].
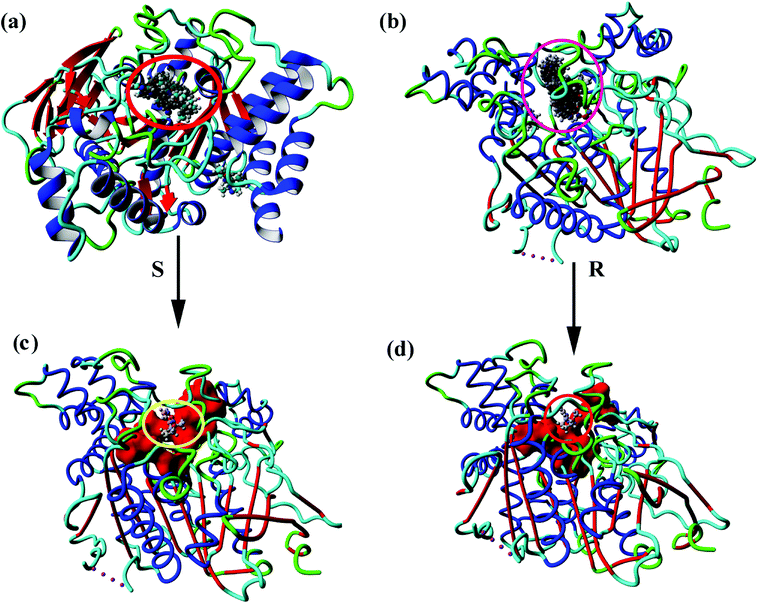 | ||
| Fig. 4 Molecular docking of all 100 conformational maps for (a) AChE-S-nicotine and (b) AChE-R-nicotine. Optimal conformation of (c) AChE-S-nicotine and (d) AChE-R-nicotine. | ||
Fig. 5(a and b) showed that as the temperature increased, the binding constant between the two configurations of nicotine and protein decreased slightly (Table 2). The binding strength of R-AChE system was about an order of magnitude stronger than that of the S-AChE system, indicating that R-nicotine bound to AChE more strongly, but both systems were weakly bound.1
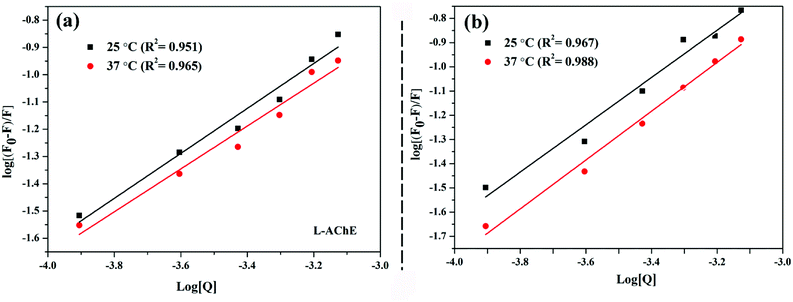 | ||
| Fig. 5 Double logarithmic plot of fluorescence quenching at different temperatures: (a) AChE-S-nicotine system and (b) AChE-R-nicotine system. | ||
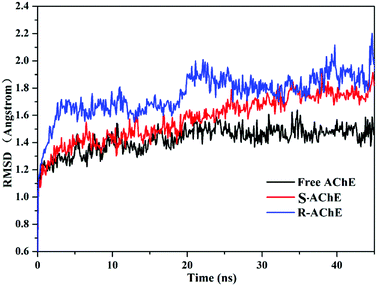
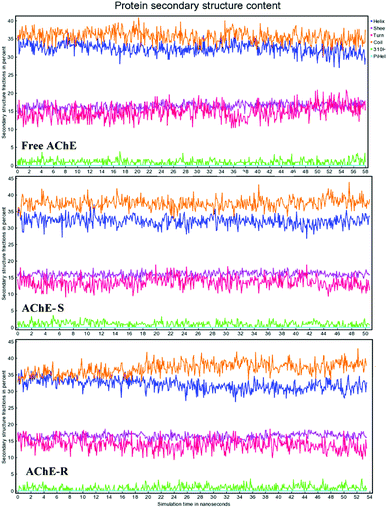
To investigate the binding stability, we selected the conformation scored first in the molecular docking to perform MD analysis. Fig. 6 shows the time-dependent root mean square deviation (RMSD)36 of the three systems. The average order of the RMSD values of the three systems was R-AChE > S-AChE > free AChE, indicating that nicotine had a negative effect on the stability of the protein backbone. In addition, in the R-AChE complex system, the numerical fluctuation of RMSD was very obviously, this proved that R-nicotine induced the deviation of the protein skeleton of AChE seriously from the original position. Compared with R-nicotine, although S-nicotine increased the RMSD value of AChE, it generally showed a stable state, and the system tends to balance at around 35 ns.
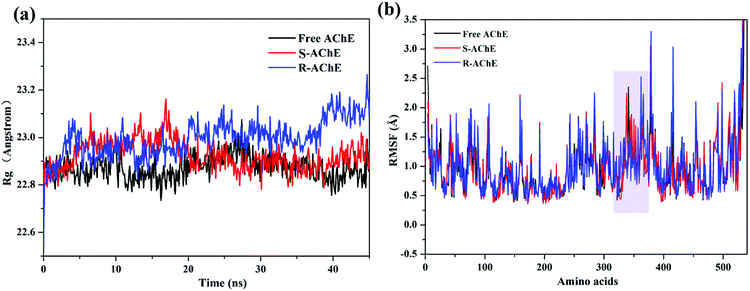
The radius of gyration value (Rg)37 of the three systems is shown in Fig. 7(a). The values of free AChE and S-AChE differed greatly at the initial phase, but they remained stable and started to overlap together at approximately 20 ns. Thus, S-nicotine did not affect the looseness of the AChE protein structure. The Rg value of R-AChE system deviated from the initial by about 0.4 angstrom, and the fluctuations were significant in the whole process, which suggested that R-nicotine made the structure of AChE looser and more unstable.
Comparison of the root mean square fluctuations (RMSF)27 of the three systems [Fig. 7(b)] indicated that the coincidence degree between the free AChE and the S-AChE system was higher than that of AChE and R-AChE. In the latter system, the RMSF value of the residues with amino acid numbers 310–330 were significantly lower than that of free protein, which may be due to the hydrogen bonding (will be discussed later) between R-nicotine and protein during the simulation to make the conformation more mechanically stable.
3.3 Interaction force analysis
The following two formulas were used to obtain the thermodynamic constants and estimate the main interaction forces in the system:38
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka = −ΔH/RT + ΔS/R Ka = −ΔH/RT + ΔS/R
| (4) |
| ΔG = ΔH − TΔS | (5) |
The interaction forces were further analyzed using the simulation trajectory of different stages in the MD simulation process. As shown in the upper panel of Fig. 8, in the S-AChE system, S-nicotine has hydrophobic interaction with Trp84, Tyr121, Phe331, Phe330, Tyr334, and Glu199 amino acids near the binding site at 0 ns. Over time, the interaction between S-nicotine and surrounding amino acids changed, but Glu199, Phe330, and Trp84 interacted with S-nicotine throughout the process. The amino acid residues that participated in the second half of the simulation remained essentially unchanged. Thus, S-nicotine maintained a constant interaction with surrounding amino acid residues within the binding site, thereby maintaining the conformation in a relatively stable state.
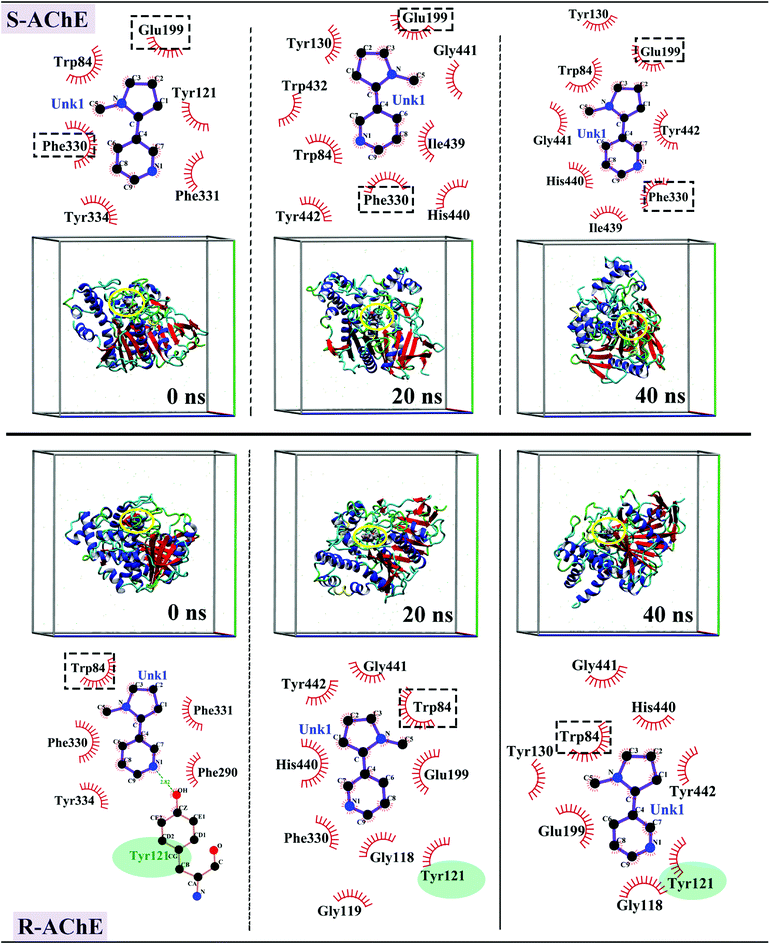 | ||
| Fig. 8 Interaction force map and three-dimensional conformation of S-AChE complex (top) and R-AChE complex (bottom) of optimal conformation in MD simulation process. | ||
The force interactions in R-AChE system presented a different situation. As shown in the lower panel of Fig. 8, at 0 ns, R-nicotine formed a hydrophobic interaction with Trp84, Phe331, PhE290, Tyr334, and Phe330, and there was a hydrogen bond of 2.82 Å between the hydrogen atom in the hydroxyl group of Tyr121 and the N atom in the nicotine molecule. Tyr121 and Trp84 interacted with R-nicotine in the entire simulation process. Tyr121 amino acid is an important component of the active site of the PAS and has a negative charge.
This observation was consistent with the conclusion that the R-AChE binding process was dominated by electrostatic interaction in the experiment. Moreover, Glu199 and His440 interacted with AChE in the latter half of the simulation, and these two amino acids are important constituent amino acids of OH and ES. Therefore, the binding of R-nicotine at this site may have a certain effect on the enzymatic hydrolysis function of AChE, which agreed with the experimental results of the enzyme activity test. Moreover, the simulated three-dimensional map showed that S/R-nicotine reversed the spatial conformation of AChE and changed the secondary structure.
The number of hydrogen bonds between small molecules and proteins is shown in Fig. 9(a). There was almost no hydrogen bond involved in the S-AChE system. However, according to the fluorescence experiment, we cannot rule out the existence of hydrogen bonding in the system. The reason for this discrepancy may be due to the limitations of computer algorithms, and it is impossible to simulate accurately the actual interactions. The experiment and computer simulation complement each other and can more fully analyze the forces that exist in the system. Hydrogen bond plays an important role in the R-AChE system. The above results showed that the interaction between small molecules and proteins is very complicated, and multiple binding forces may exist between the molecules owing to the huge molecular network of proteins.
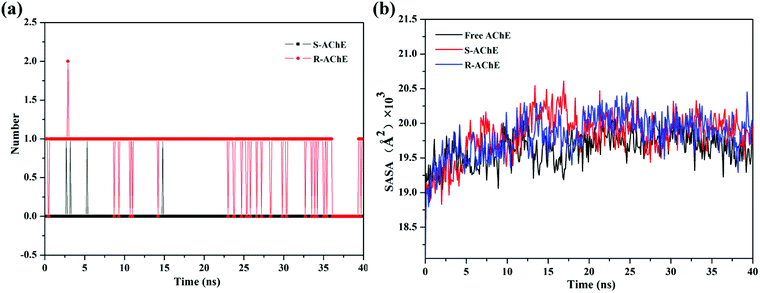 | ||
| Fig. 9 (a) Number of hydrogen bonds in the simulation process and (b) SASA values in the simulation process. | ||
The variation in solvent accessible surface area (SASA)37 of the system was also analyzed, as shown in Fig. 9(b). The SASA values of both complex systems were increased slightly, indicating that AChE will be more exposed to the solvent than the free one, thus prompting the quencher (S/R-nicotine) to better contact the AChE and then quench the protein. The results further demonstrated that the S/R-nicotine interacted with AChE and affected the spatial conformation of amino acids.
3.4 Effect of S/R-nicotine on AChE enzyme activity
5,5′-Dithiobis-2-nitrobenzoic acid (DTNB) spectrophotometry is a sensitive method for detecting AChE activity.42–45 Ultraviolet spectroscopy was used to detect the rate of catalytic production, and the effects of two configurations of nicotine on the enzymatic activity of AChE were qualitatively investigated. Both S- and R-nicotine reduced the catalytic activity of AChE (Fig. 10), but the latter's effect intensity was significantly greater than the former. According to the results of MD simulation, this phenomenon may be closely related to the Trp121 amino acid near the R-nicotine binding site, based on the hydrogen bond and hydrophobic interaction between the two molecules. Combined with the results of UV-vis spectroscopy and CD spectroscopy, R-nicotine had a greater effect on the structure and amino acid microenvironment of AChE than S-nicotine, and it had a greater degree of inhibition on the catalytic function of AChE.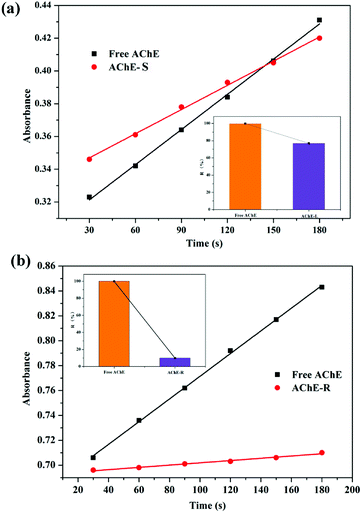 | ||
| Fig. 10 Determination of enzyme catalytic activity of the (a) AChE-S-nicotine and (b) AChE-R-nicotine systems at 298 K. | ||
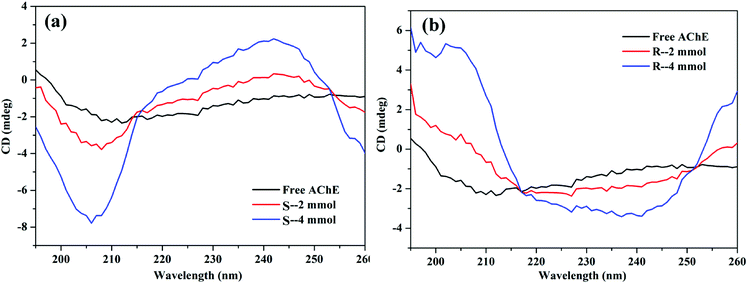
3.5 Effect of nicotine on AChE conformation
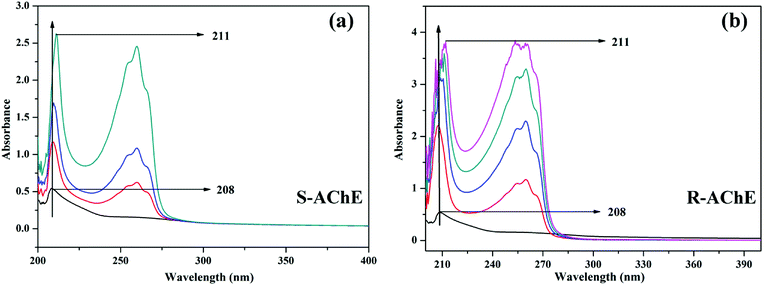 | ||
| Fig. 11 UV-vis spectra of the (a) AChE-S-nicotine and (b) AChE-R-nicotine systems at 298 K. The concentration of the T6 in the direction of the arrow gradually increases. | ||
When S- or R-nicotine combines with AChE, the absorption spectrum of AChE changed; not only its maximum absorption intensity was significantly enhanced, but also the peak position of its maximum absorption wavelength was redshifted. This phenomenon indicated that S/R-nicotine changed the microenvironment of AChE amino acid residues, causing a change in AChE conformation.
4. Conclusion
In this study, the interaction mode between S/R-nicotine and AChE was explored by experiments and computer simulation. Spectroscopic analysis showed that S/R-nicotine and AChE formed a ground-state complex with very weak binding strength with different driving forces, and there was only one major binding site. R-nicotine had a much greater effect on the conformation of AChE than S-nicotine, which may be due to the difference in the activity of S/R-nicotine. This finding was also verified by studying the effect of S/R-nicotine on enzyme activity of AChE. The results of computer simulation were consistent with the experimental results. MD simulation further confirmed that Tyr121 amino acid played an important role in the interaction, and the fluctuation of secondary structure content in the simulation process was also more obvious for R-nicotine than S-nicotine. In general, R-nicotine may have a more adverse effect on AChE than S-nicotine. The results provided reference for the commercialization and industrialization of nicotine products, and also presented a basis for analyzing the toxicological characteristics and biochemical behavior of nicotine.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the tobacco corporation Key technology research on the quality stability of colloidal smokeless tobacco products from major science and technology special projects of China [110201601005(2016xx-05)], and the study on key technologies of tobacco in mouth based on comfort and oral health from Yunnan China tobacco industry company technology development program project (2018CP07).References
- J. N. Langley and W. L. Dickinson, J. Physiol., 1890, 11, 265–306 CrossRef CAS PubMed.
- J. D. Pilcher and T. Sollmann, J. Pharmacol. Exp. Ther., 1915, 6, 369–372 Search PubMed.
- E. Asmus and D. Papenfuss, Fresenius' Z. Anal. Chem., 1962, 185, 201–211 CrossRef CAS.
- M. A. Abbassy, M. E. Eldefrawi and A. T. Eldefrawi, Pestic. Biochem. Physiol., 1983, 19, 299–308 CrossRef CAS.
- B. Schafer, Chem. Unserer Zeit, 2008, 42, 408–424 CrossRef.
- A. Das, M. Dikshit and C. Nath, Pharmacol., Biochem. Behav., 2005, 81, 89–99 CrossRef CAS PubMed.
- J. Massoulie, L. Pezzementi, S. Bon, E. Krejci and F. M. Vallette, Prog. Neurobiol., 1993, 41, 31–91 CrossRef CAS PubMed.
- C. Scheffel, H. Thiermann and F. Worek, Toxicol. Lett., 2015, 232, 557–565 CrossRef CAS PubMed.
- H. Gnahn, F. Hefti, R. Heumann, M. E. Schwab and H. Thoenen, Dev. Brain Res., 1983, 9, 45–52 CrossRef CAS.
- M. Wang, G. Zhang, D. Zhang, D. Zhu and B. Z. Tang, J. Mater. Chem., 2010, 20, 1858–1867 RSC.
- S. Wang, C. Wu, Z. Liu and H. You, Toxicol. Lett., 2018, 287, 42–48 CrossRef CAS PubMed.
- E. Ayranci and O. Duman, Protein Pept. Lett., 2004, 11, 331–337 CrossRef CAS PubMed.
- E. Ayranci and O. Duman, Food Chem., 2004, 84, 539–543 CrossRef CAS.
- M. Tanaka, Y. Asahi and S. Masuda, J. Macromol. Sci., Part A: Pure Appl.Chem., 1995, 32, 339–347 CrossRef.
- B. K. Bozoglan, S. Tunc and O. Duman, J. Lumin., 2014, 155, 198–204 CrossRef CAS.
- J. H. Tang, F. Luan and X. G. Chen, Bioorg. Med. Chem., 2006, 14, 3210–3217 CrossRef CAS PubMed.
- S. Tunc, A. Cetinkaya and O. Duman, J. Photochem. Photobiol., B, 2013, 120, 59–65 CrossRef CAS PubMed.
- S. Tunc, O. Duman and B. K. Bozoglan, J. Lumin., 2013, 140, 87–94 CrossRef CAS.
- S. Tunc, O. Duman, I. Soylu and B. K. Bozoglan, J. Lumin., 2014, 151, 22–28 CrossRef CAS.
- S. Tunc, O. Duman, I. Soylu and B. K. Bozoglan, J. Hazard. Mater., 2014, 273, 36–43 CrossRef CAS PubMed.
- O. Duman, S. Tunc and B. K. Bozoglan, J. Fluoresc., 2013, 23, 659–669 CrossRef CAS PubMed.
- M. Rarey, B. Kramer, T. Lengauer and G. Klebe, J. Mol. Biol., 1996, 261, 470–489 CrossRef CAS PubMed.
- E. Krieger and G. Vriend, Bioinformatics, 2014, 30, 2981–2982 CrossRef CAS PubMed.
- J. Park, J. J. McDonald, R. C. Petter and K. N. Houk, J. Chem. Theory Comput., 2016, 12, 2066–2078 CrossRef CAS PubMed.
- A. Jakalian, D. B. Jack and C. I. Bayly, J. Comput. Chem., 2002, 23, 1623–1641 CrossRef CAS PubMed.
- P. L. T, M. Mondal, K. Ramadas and S. Natarajan, Spectrochim. Acta, Part A, 2017, 183, 90–102 CrossRef PubMed.
- N. Gan, Q. Sun, P. Tang, D. Wu, T. Xie, Y. Zhang and H. Li, Spectrochim. Acta, Part A, 2018, 206, 126–134 CrossRef.
- M. Pathak, D. Sharma, N. Sharma and M. Sharma, J. Mol. Struct., 2018, 1166, 183–189 CrossRef CAS.
- G. G. Ariga, P. N. Naik, S. A. Chimatadar and S. T. Nandibewoor, J. Mol. Struct., 2017, 1137, 485–494 CrossRef CAS.
- D. Wu, D. Liu, Y. Zhang, Z. Zhang and H. Li, Eur. J. Med. Chem., 2018, 146, 245–250 CrossRef CAS PubMed.
- E. T.-L. L. Karami and A. A. Saboury, Phys. Chem. Res., 2017, 5, 483–496 Search PubMed.
- H. Dvir, I. Silman, M. Harel, T. L. Rosenberry and J. L. Sussman, Chem.-Biol. Interact., 2010, 187, 10–22 CrossRef CAS PubMed.
- M. Harel, G. Kryger, T. L. Rosenberry, W. D. Mallender, T. Lewis, R. J. Fletcher, J. M. Guss, I. Silman and J. L. Sussman, Protein Sci., 2000, 9, 1063–1072 CrossRef CAS PubMed.
- G. Amitai, A. Shemesh, E. Sitbon, M. Shklar, D. Netanely, I. Venger and S. Pietrokovski, J. Mol. Biol., 2004, 344, 1135–1146 CrossRef CAS PubMed.
- Q. Wang, X. Ma, J. He, Y. Li and H. Li, RSC Adv., 2015, 5, 44696–44704 RSC.
- D. P. Yeggoni, M. Gokara, D. M. Manidhar, A. Rachamallu, S. Nakka, C. S. Reddy and R. Subramanyam, Mol. Pharmaceutics, 2014, 11, 1117–1131 CrossRef CAS PubMed.
- M. Raza, Y. Wei, Y. Jiang, A. Ahmad, S. Raza, S. Ullah, Y. Han, Q. U. Khan and Q. Yuan, New J. Chem., 2017, 41, 8203–8213 RSC.
- P. Mitra, U. Pal, N. Chandra Maiti, A. Ghosh, A. Bhunia and S. Basu, RSC Adv., 2016, 6, 53454–53468 RSC.
- H. Yang, Y. Huang, J. Liu, P. Tang, Q. Sun, X. Xiong, B. Tang, J. He and H. Li, Sci. Rep., 2017, 7, 11126 CrossRef.
- S. J. Wang, Y. L. Peng, C. G. Zhang, Q. P. Ma, X. X. Peng and L. L. Ren, Bull. Korean Chem. Soc., 2017, 38, 735–743 CrossRef.
- E. Kaspchak, L. I. Mafra and M. R. Mafra, Food Chem., 2018, 252, 1–8 CrossRef CAS PubMed.
- G. L. Ellman, K. D. Courtney, V. Andres and R. M. Featherstone, Biochem. Pharmacol., 1961, 7, 88–95 CrossRef CAS.
- S. Prasanth and C. Sudarsanakumar, New J. Chem., 2017, 41, 9521–9530 RSC.
- Y. Zhan, J. Yang, L. Guo, F. Luo, B. Qiu, G. Hong and Z. Lin, Sens. Actuators, B, 2019, 279, 61–68 CrossRef CAS.
- P. Barai, N. Raval, S. Acharya, A. Borisa, H. Bhatt and N. Acharya, Behav. Brain Res., 2019, 356, 18–40 CrossRef CAS.
- M. M. Yin, P. Dong, W. Q. Chen, S. P. Xu, L. Y. Yang, F. L. Jiang and Y. Liu, Langmuir, 2017, 33, 5108–5116 CrossRef CAS PubMed.
- S. Zhuang, H. Wang, K. Ding, J. Wang, L. Pan, Y. Lu, Q. Liu and C. Zhang, Chemosphere, 2016, 144, 1050–1059 CrossRef CAS PubMed.
- Q. Wang, Q. Sun, P. Tang, B. Tang, J. He, X. Ma and H. Li, RSC Adv., 2015, 5, 81696–81706 RSC.
- V. Sinisi, C. Forzato, N. Cefarin, L. Navarini and F. Berti, Food Chem., 2015, 168, 332–340 CrossRef CAS PubMed.
- N. Gan, Q. Sun, M. Zhang, P. Tang, L. Zhao, T. Xie, Y. Zhang and H. Li, J. Biomol. Struct. Dyn., 2018 DOI:10.1080/07391102.2018.1502686.
| This journal is © The Royal Society of Chemistry 2019 |