Open Access Article
Open Access ArticleOptimization of the extraction, preliminary characterization, and anti-inflammatory activity of crude polysaccharides from the stems of Trapa quadrispinosa
Feng Liac,
Xinhu Liub,
Xiaofeng Yuc,
Xiuquan Xu *c and
Huan Yang*c
*c and
Huan Yang*c
aAffiliated Hospital of Jiangsu University, Zhenjiang 212001, P. R. China
bTaizhou National Medical Hitech Development Zone, Taizhou 225300, P. R. China
cSchool of Pharmacy, Jiangsu University, Zhenjiang 212013, P. R. China. E-mail: xxq781026@ujs.edu.cn; Huanyang198109@163.com
First published on 22nd July 2019
Abstract
A novel method was developed using pressurized-assisted extraction (PAE) to efficiently extract crude Trapa quadrispinosa polysaccharides (TQCPS) from plant stems, and the extraction process was optimized using response surface methodology (RSM). At a water-to-material fixed ratio of 30 mL g−1, the highest yield of 3.72 ± 0.13% was obtained under the optimum conditions of extraction time of 32 min, extraction temperature at 47 °C, and extraction pressure at 1.87 Mpa, which were in agreement with the predicted value of 3.683%. Compared with conventional hot water extraction (HWE), the PAE method remarkably enhanced the extraction yield with the further advantages of short extraction time and low extraction temperature. The preliminary characteristics of TQCPS were analyzed through UV-vis, FT-IR, and chemical composition analysis. In subsequent anti-inflammatory studies, when RAW 264.7 mouse macrophage cells were treated with TQCPS, satisfactory anti-inflammatory activity was observed, and TQCPS significantly suppressed the release of nitric oxide (NO), tumor necrosis factor-α (TNF-α), and interleukin-6 (IL-6) and synchronously restrained the expression levels of inducible nitric oxide synthase (iNOS), TNF-α, and IL-6 mRNA induced by lipopolysaccharide (LPS) in a dose-dependent manner. These results indicate that PAE is a technology that can be used for efficient extraction of polysaccharides from medicinal plants, and TQCPS can be explored as a potential anti-inflammatory agent in medicine.
1. Introduction
Polysaccharides are a type of essential primary metabolite that exists in animals, plants, and fungi. In recent decades, there has been great interest in polysaccharides isolated from plants due to their various biological activities including anti-oxidant, anti-microbial, anti-tumor, anti-diabetic, and anti-inflammatory activities.1–5 Inflammation is a complicated biological response against injury, infection, and stress, and is mainly mediated by pro-inflammatory cells including macrophages.6 During the process of inflammation, activated macrophages can synthesize and release various inflammatory cytokines including tumor necrosis factor (TNF)-α, interleukin (IL)-1β, IL-6, and nitric oxide (NO) to promote inflammation or fight against pathogens.7 However, excess or inappropriate production of these inflammatory cytokines can also cause various inflammatory diseases such as rheumatoid arthritis, osteoarthritis, diabetes, Alzheimer's disease, and even cancer.8 Therefore, the inhibition of the synthesis or release of these inflammatory cytokines is an important target for the treatment of inflammatory diseases. It has been demonstrated that some natural polysaccharides exhibit obvious anti-inflammatory activities by inhibiting the production of inflammatory cytokines in cell or mouse models.9,10It is important to be able to efficiently extract these bioactive polysaccharides for their application or further research. Conventionally, polysaccharides are extracted from plant tissues by heating, boiling, or refluxing methods. However, these techniques have many disadvantages including requiring a great deal of time and energy, low yields, and degradation of polysaccharides with the loss of their pharmacological activities.11 It is necessary to find an efficient method for extracting polysaccharides in order to avoid these disadvantages of traditional extraction techniques. Recently, various advanced techniques have been developed to extract bioactive polysaccharides, such as ultrasonic-assisted extraction (UAE), microwave-assisted extraction (MAE), enzyme-assisted extraction (EAE), and pressurized-assisted extraction (PAE).12–15 Among these techniques, PAE is considered to be a relatively innovative and promising technique due to its higher efficiency and environmentally friendly characteristics of lower energy consumption and reduced emission of pollutant solvents.16,17 In the PAE process, the higher temperature and pressure increases the solubility and diffusion rate of target compounds, and reduces the viscosity and surface tension of the solvent, allowing increased penetration into the samples and thus achieving efficient extraction and remarkable activities over a wide range of polarities of compounds.18–21 PWE has been efficiently applied for extracting bioactive substances such as phenolic compounds and polysaccharide from different plants.22–24 Usually, extraction of bioactive substances from plant tissues by PAE is influenced by multiple independent variables such as extraction temperature, extraction time, extraction pressure, particle size, and liquid-to-sample ratio and their mutual interactions. Response surface methodology (RSM), an effective statistical technique, can be used to investigate and optimize complex processes when the independent variables have a combined effect on the response variables.25 The main advantage of RSM is to significantly reduce the experimental trials that are required for evaluating multiple parameters and their interactions.26
Trapa quadrispinosa, also named as Sijiaoling and belonging to the Trapaceae family, is regarded as a popular vegetable for its wonderful flavor and medicinal functions.27 The pericarps of T. quadrispinosa are used in allaying fever, diffusing dampness, dispelling pathogenic wind, and invigorating the spleen in traditional Chinese medicine.28 However, to the best of our knowledge, the composition and biological activities of the stem of T. quadrispinosa have rarely been examined.
In our previous study, polysaccharides were detected in the stem of T. quadrispinosa, and they engaged in a variety of free radical scavenging activities.29 It is well known that reactive oxygen species (ROS) are closely related to the occurrence and development of inflammation and tumors.30 Trapa quadrispinosa polysaccharides (TQCPS) possess excellent antioxidant abilities, and may exhibit satisfactory anti-inflammatory activity. However, there is very little data on the extraction and anti-inflammatory activity of TQCPS. Thus, the objective of the present study was to explore the potential use of pressure technology on the extraction of crude polysaccharides from the stem of T. quadrispinosa and evaluate their anti-inflammatory activity.
2. Experimental
2.1. Reagents and materials
The stems of T. quadrispinosa were collected from Weishan Lake, Weishan County, Shandong province, China. The dried materials were pulverized and screened through a 60-mesh sieve. The powder was extracted with petroleum ether (60–90 °C) and 95% ethanol for 6 h in a Soxhlet apparatus to remove some colored materials and other micromolecular substances. Thereafter, the samples were obtained by filtration and dried for further use.The RAW 264.7 murine macrophage cell line was obtained from the American Type Culture Collection (Rockville, MD, USA). Lipopolysaccharide (LPS) from Escherichia coli (0111:B4), 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), and Griess reagent were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), streptomycin, penicillin, and TRIzol reagent were all purchased from Gibco Laboratories (Grand Island, NY, USA). Mouse IL-6 and TNF-α enzyme-linked immunosorbent assay (ELISA) kits were purchased from Invitrogen (Carlsbad CA, USA). All other chemical reagents used in experiments were of analytical grade, and double-distilled water was used throughout the experiments.
2.2. Pressure-assisted extraction (PAE)
The PAE was carried out in a high pressure micro-reactor (WHF-0.25L, automatically controlled reaction kettle, Weihai Co. Ltd., Shandong, China). Samples (2.0 g) were extracted with distilled water at different temperatures (30–70 °C) and different water-to-material ratios (20–40 mL g−1) under extraction pressure (1.2–2.4 Mpa) for various periods (10–50 min). After extraction, the mixtures were centrifuged at 4000 rpm for 10 min, and the collected supernatants were concentrated to a certain volume by a rotary evaporator. Then, the crude polysaccharides were precipitated by the addition of absolute ethanol to a final concentration of 80% (v/v) at 4 °C for 24 h. The obtained crude polysaccharides were re-dissolved in double-distilled water for polysaccharide content and anti-inflammatory activity determination.The percentage yield of polysaccharide from T. quadrispinosa stems was determined with the phenol-sulfuric method.31 Glucose was chosen as a standard, and the polysaccharide yield (%) was calculated as follows:
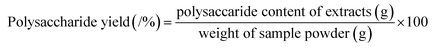 | (1) |
For comparison, in hot water extraction (HWE), a 5.0 g sample was extracted using 150 mL of distilled water at 80 °C for 4 h.
Based on the preliminary single factor results, RSM was employed to further optimize the PAE conditions for TQCPS from the stems of T. quadrispinosa. Extraction time (X1, min), extraction temperature (X2, °C), and extraction pressure (X3, Mpa) were the preferred independent variables. A Box-Behnken Design (BBD) with three variables and three levels was constructed. The yield of TQCPS was designated as the response-dependent value. Based on the experimental data from the BBD, regression analysis was carried out, and a second-order polynomial model was obtained as follows:
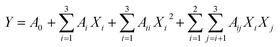 | (2) |
Design-Expert version 8.0.5b software (Stat-Ease Inc., Minneapolis, MN, USA) was used to analyze the experimental data. Analysis of variance (ANOVA) was used to evaluate the significant terms in the model. The various statistical analysis parameters including lack of fit tests, P value, F value, determination coefficient (R2), adjusted determination coefficient (Radj2), and coefficient of variation (C.V.) were chosen to evaluate the adequacy of the models.
2.3. Scanning electron microscopy analysis
In order to understand the mechanism of PAE, the microstructure of an untreated sample as well as residue after extraction were collected and observed by scanning electron microscopy (SEM). The dried sample powders were fixed on a specific carbon film support and covered with gold using a sputter coater. The shape and the surface characteristics were observed by a SEM system (Hitachi S4800, Japan) under high vacuum conditions.2.4. Analysis of TQCPS
2.5. Anti-inflammatory activity assay in vitro
The anti-inflammatory effects of TQCPS were evaluated using the LPS-induced RAW 264.7 cell assay as previously reported.7For the cytokine assay, after stimulation by LPS, the cell culture supernatants were collected, and the concentrations of IL-6 and TNF-α were carefully determined using ELISA kits according to the manufacturer's instructions.
3. Results and discussion
3.1. Single-factor optimization for extraction
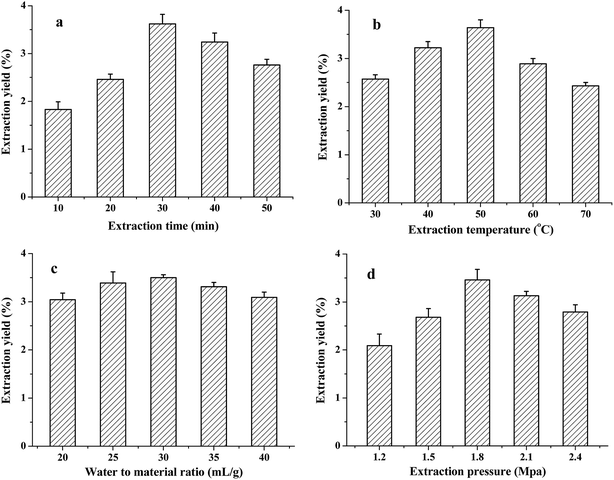
In this study, the PAE method was applied to extract TQCPS from the stems of T. quadrispinosa. The extraction time, water-to-material ratio, extraction temperature, and extraction pressure were chosen for investigation, and the results are shown in Fig. 1.According to the single factor results, extraction time, extraction temperature, and extraction pressure more significantly affected the yield of TQCPS. Therefore, these three parameters were preferred for further experimentation.
3.2. Optimization of TQCPS extraction by BBD
| Run | Independent variables | Response | |||
|---|---|---|---|---|---|
| X1 (min) | X2 (°C) | X3 (Mpa) | Experimental | Predicted | |
| 1 | −1(20) | −1(40) | 0(1.8) | 2.50 | 2.55 |
| 2 | −1(20) | 1(60) | 0(1.8) | 2.76 | 2.79 |
| 3 | −1(20) | 0(50) | −1(1.5) | 2.54 | 2.52 |
| 4 | −1(20) | 0(50) | 1(2.1) | 3.20 | 3.22 |
| 5 | 1(40) | −1(40) | 0(1.8) | 3.36 | 3.33 |
| 6 | 1(40) | 1(60) | 0(1.8) | 2.42 | 2.38 |
| 7 | 1(40) | 0(50) | −1(1.5) | 2.92 | 2.98 |
| 8 | 1(40) | 0(50) | 1(2.1) | 3.02 | 3.04 |
| 9 | 0(30) | −1(40) | −1(1.5) | 2.73 | 2.70 |
| 10 | 0(30) | −1(40) | 1(2.1) | 3.20 | 3.14 |
| 11 | 0(30) | 1(60) | −1(1.5) | 2.55 | 2.53 |
| 12 | 0(30) | 1(60) | 1(2.1) | 2.66 | 2.69 |
| 13 | 0(30) | 0(50) | 0(1.8) | 3.62 | 3.63 |
| 14 | 0(30) | 0(50) | 0(1.8) | 3.68 | 3.63 |
| 15 | 0(30) | 0(50) | 0(1.8) | 3.60 | 3.63 |
Based on multiple regression analysis of the experimental data, the model is expressed by a second-order polynomial equation as follows:
| Yyield = 3.63 + 0.09X1 − 0.18X2 + 0.17X3 − 0.30X1X2 − 0.14X1X3 − 0.09X2X3 − 0.37X12 − 0.50X22 − 0.34X32 | (3) |
The analysis of variance (ANOVA) for the second-order polynomial model response is given in Table 2. According to the results, the high F value (71.63) and the low P value (P < 0.0001) suggested that the regression models are more significant. The goodness-of-fit of regression model was evaluated by determination coefficient (R2) and adjusted determination coefficient (Radj2). The high values of R2 (0.9923) and Radj2 (0.9785) indicated a satisfactory correlation between the experimental and predicted values.38 Furthermore, the low value of coefficient of variation (C.V.) at 2.15% and high value of adeq. precision at 24.059 demonstrated a very high degree of precision and a good deal of reliability of the experimental values.39 The low F value (3.28) and the high P value (P = 0.2425 > 0.05) for lack of fit suggested that it was not significant relative to the pure error, and a 24.25% chance could occur due to noise.
| Source | Coefficient estimate | Sum of squares | DF | Mean square | F value | P value | Significance |
|---|---|---|---|---|---|---|---|
| a Notes: * significant at 0.05 level; ** significant at 0.01 level; *** significant at 0.001 level. | |||||||
| Model | 3.63 | 2.65 | 9 | 0.29 | 71.63 | <0.0001 | ***<0.001 |
| X1 | 0.09 | 0.065 | 1 | 0.065 | 15.79 | 0.0106 | *<0.05 |
| X2 | −0.18 | 0.25 | 1 | 0.25 | 59.71 | 0.0006 | ***<0.001 |
| X3 | 0.17 | 0.22 | 1 | 0.22 | 54.70 | 0.0007 | ***<0.001 |
| X1X2 | −0.30 | 0.36 | 1 | 0.36 | 87.73 | 0.0002 | ***<0.001 |
| X1X3 | −0.14 | 0.078 | 1 | 0.078 | 19.11 | 0.0072 | **<0.01 |
| X2X3 | −0.09 | 0.032 | 1 | 0.032 | 7.90 | 0.0375 | *<0.05 |
| X12 | −0.37 | 0.50 | 1 | 0.50 | 122.63 | 0.0001 | ***<0.001 |
| X22 | −0.50 | 0.94 | 1 | 0.94 | 228.72 | <0.0001 | ***<0.001 |
| X32 | −0.34 | 0.44 | 1 | 0.44 | 106.59 | 0.0001 | ***<0.001 |
| Residual | 0.021 | 5 | 4.10 × 10–3 | ||||
| Lack of fit | 0.017 | 3 | 5.68 × 10−3 | 3.28 | 0.2424 | >0.05 | |
| Pure error | 3.47 × 10−3 | 2 | 1.73 × 10−3 | ||||
| Col total | 2.67 | 14 | |||||
| R2 = 0.9923 Radj2 = 0.9785 C.V. = 2.15 adeq. precision = 24.059 | |||||||
Usually, the P value and F value are used as a tool to check the significance of each coefficient.40 The smaller the P value and the larger the F value, the more significant the corresponding coefficient. According to Table 2, the yield of TQCPS was significantly influenced by the linear terms of extraction time (X1, P < 0.05), extraction temperature (X2, P < 0.001), extraction pressure (X3, P < 0.001), and all three quadratic terms of X12, X22, and X32 (P < 0.001). Among the interaction terms, X1X1, X1X3, and X2X3 were significant at P < 0.001, 0.01, and 0.05 levels, respectively. The F value showed that the sequence of factors influencing the extraction yield was extraction temperature > extraction pressure > extraction time, and the order of interaction effect was X1X2 > X1X3 > X2X3.
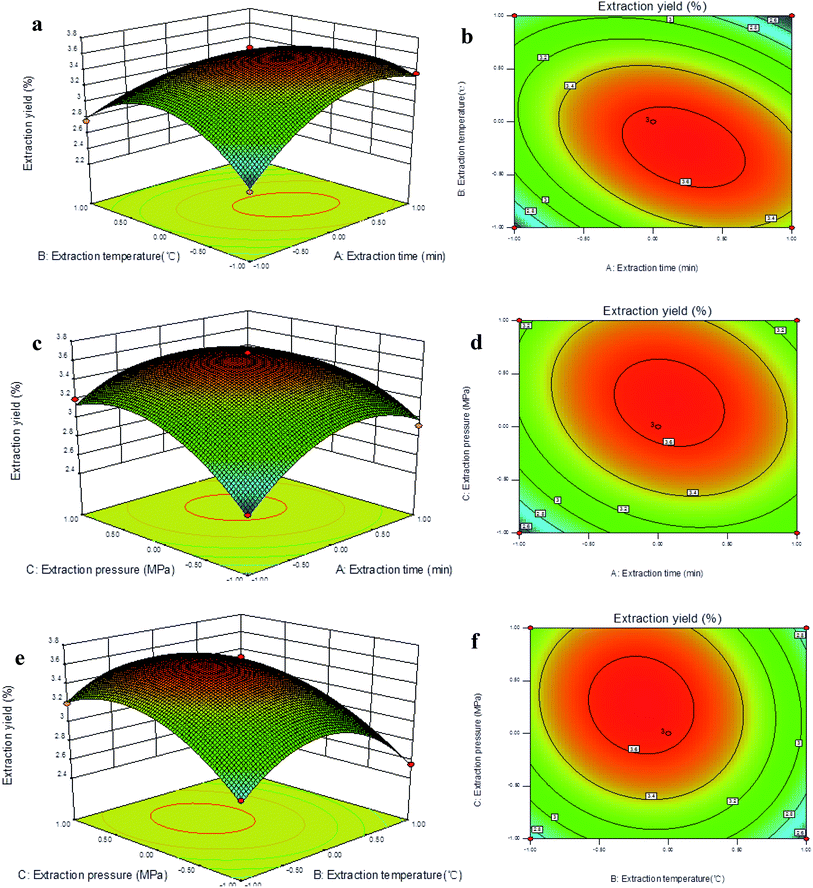
In the present study, the 3D response surface and 2D contour plots illustrate the relationship between extraction time (X1), extraction temperature (X2), extraction pressure (X3), and extraction yield of TQCPS (Fig. 2). Fig. 2a and b illustrate the effect of extraction time and extraction temperature on the yield when fixing the extraction pressure at the zero level (1.8 Mpa). The extraction temperature exhibited a quadratic effect on the yield. When the extraction time was maintained at a lower level, the yield initially increased and then decreased with increasing extraction temperature. At the same time, the result indicated that the curved surface of the extraction temperature was steeper than that of extraction time. This signified that the influences of extraction temperature on the extraction yield were more significant than that of extraction time. Furthermore, the yield was significantly influenced by the interaction between extraction temperature and extraction time because of the most elliptical contour shape among all of them.
Fig. 2c and d exhibit the effects of extraction time and extraction pressure on the yield when fixing the extraction temperature at the zero level (50 °C), and show that both the extraction time and extraction pressure had a positive impact on the yield. There was an obvious increase in the yield when increasing extraction time and extraction pressure, and the yield slightly decreased when the extraction time and pressure increased up to a threshold level. Additionally, the extraction time and pressure exhibited the lowest interaction effects because of the smallest degree of an elliptical shape on 2D contour plots. Fig. 2e and f show the mutual interactions between extraction temperature and extraction pressure on the yield of TQCPS when fixing the extraction time at 30 min. When the extraction temperature was maintained at a lower level, the yield increased with increasing extraction pressure. However, the yield increased and then slightly decreased with increasing extraction temperature when the extraction pressure was fixed. It was indicated that a greater yield could be obtained when a higher extraction pressure and a lower extraction temperature was selected, and the TQCPS was more likely to be degraded under the condition of high temperature.
3.3. Analysis of microscopic changes
To study the structural alteration of samples treated by PAE, the T. quadrispinosa stem samples were observed by SEM. As shown in Fig. 3a, it was obvious that there was a complete parenchyma and no destruction of cell walls for the untreated samples. However, samples subjected to PAE exhibited a lack of cellular integrity, and there were many huge perforations on the external surfaces (Fig. 3b). The damaged cells easily released their intracellular polysaccharides into the extraction solvent, which contributed to the higher extraction efficiency.43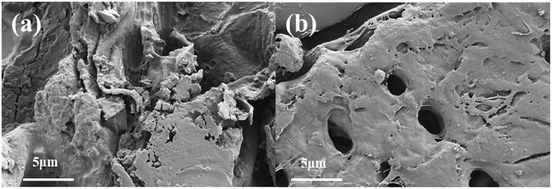 | ||
| Fig. 3 Scanning electron microscopy images of T. quadrispinosa stem samples: (a) untreated raw material and (b) material treated by PAE under optimum conditions. | ||
3.4. Preliminary characterization of TQCPS
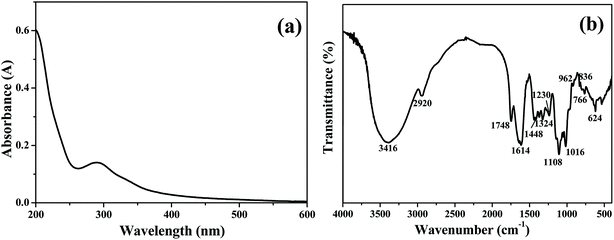
The chemical composition of TQCPS was determined, and the amounts of carbohydrate, uronic acid, protein, and total polyphenols in TQCPS were 62.42%, 17.84%, 2.44%, and 0.53%, respectively. The UV-vis spectrum of TQCPS was scanned at the range of 200–600 nm. As shown in Fig. 4a, there was weak absorbance at 280 nm, indicating the presence of proteins, which was consistent with the results of the chemical composition analysis of TQCPS. Additionally, there was no absorbance at 260 or 520 nm in the UV spectrum, indicating the absence of nucleic acid and pigment, respectively. The FT-IR spectrum of TQCPS was recorded in the range of 4000–400 cm−1 (Fig. 4b). A strong absorbance band at 3416 cm−1 represents O–H stretching vibration in the constituent sugar residues. The weak band at 2920 cm−1 indicates C–H stretching vibration in the sugar ring.44 The bands at 1748 and 1614 cm−1 denote the unique absorption of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and COO− belonging to uranic acid.45 The bands at 1448, 1324, and 1230 cm−1 were assigned to C–O stretching vibrations and O–H deformation vibrations.46 The strong absorption bands in the 1060 cm−1 region (1108, 1016 cm−1) were identified to be the vibrations of the C–O–C bond.47 Furthermore, the bands at 962 and 836 cm−1 relating to α- and β-configurations were also characteristic of glycosidic structures.48
O and COO− belonging to uranic acid.45 The bands at 1448, 1324, and 1230 cm−1 were assigned to C–O stretching vibrations and O–H deformation vibrations.46 The strong absorption bands in the 1060 cm−1 region (1108, 1016 cm−1) were identified to be the vibrations of the C–O–C bond.47 Furthermore, the bands at 962 and 836 cm−1 relating to α- and β-configurations were also characteristic of glycosidic structures.48
3.5. Anti-inflammatory activity of TQCPS
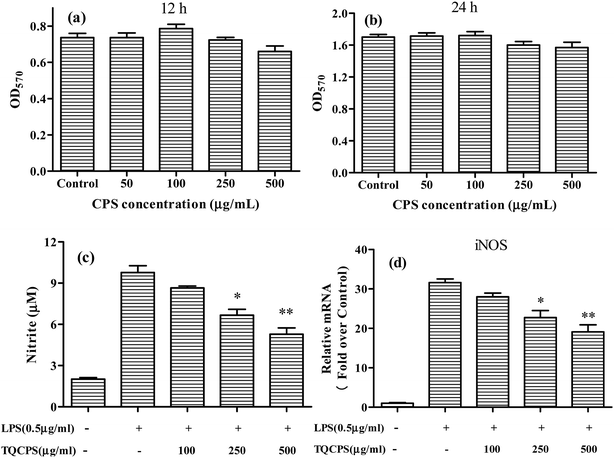
LPS is a prototypical bacterial endotoxin that can initiate a variety of inflammatory responses by directly activating macrophages. The activated macrophages produce increasing amounts of NO and cytokines such as IL-1, IL-6, and TNF-α, all of which are involved in the pathogenesis of many diseases. Therefore, macrophages in the presence of LPS have been widely used for evaluating the anti-inflammatory potential of samples in vitro.49 In this study, RAW 264.7, the most commonly used mouse macrophage cell line, was selected to evaluate the anti-inflammatory activity of TQCPS.The results are shown in Fig. 5c and d. As shown in Fig. 5c, LPS alone markedly increased NO production up to 9.77 μM compared to 2.01 μM NO from untreated RAW 264.7 cells. The level of NO decreased in a dose-dependent manner when RAW 264.7 cells were co-incubated with TQCPS and LPS. When TQCPS was added at 500 μg mL−1, the production level of NO was determined to be 5.28 μM, a significant 46% decrease compared to cells treated with LPS alone (p < 0.01). Correspondingly, the stimulation of RAW 264.7 cells with LPS was noteworthy and increased mRNA expression of iNOS against untreated cells by 31.6 fold. However, the mRNA level of iNOS decreased from 28.0 fold to 19.1 fold after treatment with TQCPS at concentrations ranging from 100 to 500 μg mL−1 in a dose-dependent manner (Fig. 5d). These results suggest that TQCPS could decrease NO production by the downregulation of iNOS expression at a transcriptional level in RAW 264.7 cells stimulated by LPS.
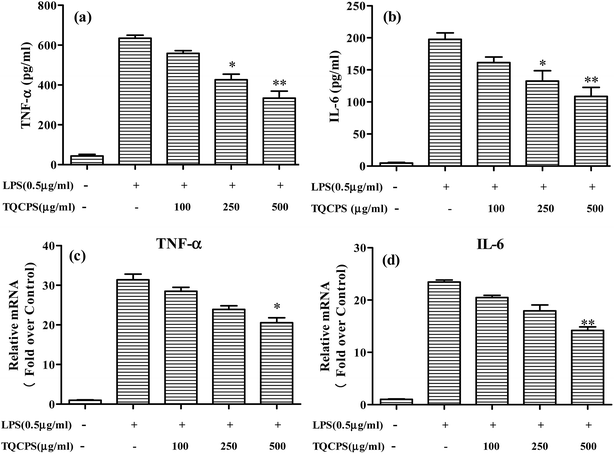
The results revealed that the secretion of TNF-α and IL-6 significantly increased from 43.6 and 4.7 pg mL−1 to 634.3 and 197.7 pg mL−1, respectively, when cells were stimulated by LPS. The secretion levels of TNF-α and IL-6 were decreased to 331.3 and 108.7 pg mL−1, respectively, when 500 μg mL−1 TQCPS was added to the LPS-stimulated RAW 264.7 cells (Fig. 6a and b). Furthermore, the effect of TQCPS on the level of mRNA expression was also determined by RT-PCR. As shown in Fig. 6c and d, there was a dose-dependent decrease in the mRNA expression of TNF-α and IL-6 (20.57 and 14.18 fold, respectively, at 500 μg mL−1 TQCPS) compared to the stimulation group (31.38 and 23.44 fold, respectively). These results strongly indicate that TQCPS could suppress the expression and secretion of inflammatory cytokines in LPS-induced RAW 264.7 cells.
It is well known that the bioactivities of polysaccharides are mostly dependent on their physicochemical characteristics including composition, distribution of average molecular weight, monosaccharide component, backbone, connection location, repeating unit, and advanced structures.52,53 Thus, it is necessary to carry out a further study on the purification and physical characterization analysis of purified polysaccharide and its detailed mechanism of anti-inflammatory activity.
4. Conclusions
In this study, pressurized-assisted extraction was successfully used to extract polysaccharides from the stem of T. quadrispinosa. Response surface methodology was employed to optimize extraction conditions. Fixing the ratio of water to material at 30 mL g−1, the optimal extraction conditions were as follows: extraction time of 32 min, extraction temperature at 47 °C, and extraction pressure at 1.87 Mpa, with the highest yield of polysaccharides at 3.72% ± 0.13%. The application of pressurized-assisted extraction significantly decreased extraction time, and improved the extraction efficiency compared with the conventional hot water extraction method. Investigation of the anti-inflammatory activity of TQCPS on murine macrophage cell line RAW 264.7 revealed that TQCPS significantly downregulates the production of NO and the secretion of inflammatory cytokines, including IL-6 and TNF-α. This indicates that TQCPS can be explored as a potential anti-inflammatory agent for use in medicine.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
The financial support for this investigation was given by the National Natural Science Foundation of China (81402840), Natural Science Foundation of Jiangsu Province, China (BK20130495), College of Natural Science Research Program of Jiangsu Province, China (13KJB350001), and the Natural Science Foundation of Jiangsu Province, Administration of Traditional Chinese Medicine, China (YB2017101).References
- B. T. Zhao, J. Liu, X. Chen, J. Zhang and J. L. Wang, RSC Adv., 2018, 8, 11731–11743 RSC.
- I. Ghazala, A. Sila, F. Frikha, D. Driss, S. Ellouz-Chaabouni and A. Haddar, J. Food Sci. Technol., 2015, 52, 6953–6965 CrossRef CAS.
- C. L. Lu, W. Zhu, M. Wang, M. M. Hu, W. L. Chen, X. J. Xu and C. J. Lu, Carbohydr. Polym., 2015, 122, 428–436 CrossRef CAS PubMed.
- Q. Y. Wu, H. S. Qu, J. Q. Jia, C. Kuang, Y. Wen, H. Yan and Z. Z. Gui, Carbohydr. Polym., 2015, 132, 31–40 CrossRef CAS PubMed.
- C. Chen, L. J. You, A. M. Abbasi, X. Fu and R. H. Liu, Carbohydr. Polym., 2015, 130, 122–132 CrossRef CAS PubMed.
- C. H. Liao and J. Y. Lin, Food Chem., 2012, 135, 1818–1827 CrossRef CAS PubMed.
- H. L. Liu, R. Xu, L. L. Feng, W. J. Guo, N. Cao, C. Qian, P. Teng, L. Wang, X. F. Wu, Y. Sun, J. X. Li, Y. Shen and Q. Xu, PLoS One, 2012, 7, 37168 CrossRef PubMed.
- T. Luo, J. Qin, M. Liu, J. Luo, F. Ding, M. L. Wang and L. M. Zheng, Inflammation Res., 2015, 64, 205–212 CrossRef CAS PubMed.
- Y. Diao, Y. Q. Xin, Y. Zhou, N. Li, X. L. Pan, S. M. Qi, Z. L. Qi, Y. M. Xu, L. Luo, H. G. Wan, L. Lan and Z. M. Yin, Int. Immunopharmacol., 2014, 18, 12–19 CrossRef CAS PubMed.
- J. J. Cheng, C. H. Chao, P. C. Chang and M. K. Lu, Food Hydrocolloids, 2016, 53, 37–45 CrossRef CAS.
- R. Z. Chen, S. Z. Li, C. M. Liu, S. M. Yang and X. L. Li, Process Biochem., 2012, 47, 2040–2050 CrossRef CAS.
- J. B. Pu, B. H. Xia, Y. J. Hu, H. J. Zhang, J. Chen, J. Zhou, W. Q. Liang and P. Xu, Molecules, 2015, 20, 22220–22235 CrossRef CAS PubMed.
- C. L. Cao, Q. Huang, B. Zhang, C. Li and X. Fu, Int. J. Biol. Macromol., 2018, 109, 357–368 CrossRef CAS PubMed.
- S. Q. Qian, X. H. Fang, D. M. Dan, E. J. Diao and Z. X. Lu, RSC Adv., 2018, 8, 42145–42152 RSC.
- Y. Q. Xu, F. Cai, Z. Y. Yu, L. Zhang, X. G. Li, Y. Yang and G. J. Liu, Food Chem., 2016, 194, 650–658 CrossRef CAS PubMed.
- M. Plaza and C. Turner, TrAC, Trends Anal. Chem., 2015, 71, 39–54 CrossRef CAS.
- H. N. Zhang, W. Tchabo and Y. K. Ma, Emir. J. Food Agric., 2017, 29, 815–819 CrossRef.
- H. Kamali, T. A. Sani, A. Mohammadi, P. Alesheikh, E. Khodaverdi and F. Hadizadeh, J. Supercrit. Fluids, 2018, 133, 535–541 CrossRef CAS.
- W. Tchabo, Y. K. Ma, E. Kwaw, H. N. Zhang, X. Li and N. A. Afoakwah, Food Bioprocess Technol., 2017, 10, 1210–1223 CrossRef CAS.
- F. J. Leyva-Jimenez, J. Lozano-Sanchez, I. Borras-Linares, D. Arraez-Roman and A. Segura-Carretero, Food Res. Int., 2018, 109, 213–222 CrossRef CAS PubMed.
- Q. Yuan, S. Lin, Y. Fu, X. R. Nie, W. Liu, Y. Su, Q. H. Han, L. Zhao, Q. Zhang, D. R. Lin, W. Qin and D. T. Wu, Int. J. Biol. Macromol., 2019, 127, 178–186 CrossRef CAS PubMed.
- J. Gao, L. Z. Lin, B. G. Sun and M. M. Zhao, Food Funct., 2017, 8, 3043–3052 RSC.
- H. Kamali, E. Khodaverdi, F. Hadizadeh and S. H. Ghaziaskar, J. Supercrit. Fluids, 2016, 107, 307–314 CrossRef CAS.
- H. N. Zhang and Y. K. Ma, Czech J. Food Sci., 2017, 35, 180–187 CrossRef CAS.
- M. L. Zhong, S. Y. Huang, H. H. Wang, Y. L. Huang, J. R. Xu and L. J. Zhang, RSC Adv., 2019, 9, 1576–1585 RSC.
- X. L. Luo, J. M. Cui, H. H. Zhang, Y. Q. Duan, D. Zhang, M. H. Cai and G. Y. Chen, Ind. Crops Prod., 2018, 112, 296–304 CrossRef CAS.
- P. Y. Chiang and J. Y. Ciou, LWT--Food Sci. Technol., 2010, 43, 361–365 CrossRef CAS.
- H. N. Yu and S. R. Shen, LWT--Food Sci. Technol., 2015, 61, 238–243 CrossRef CAS.
- A. Raza, F. Li, X. Q. Xu and J. Tang, Int. J. Biol. Macromol., 2017, 94, 335–344 CrossRef CAS PubMed.
- J. Fang, T. Seki and H. Maeda, Adv. Drug Delivery Rev., 2009, 61, 290–302 CrossRef CAS PubMed.
- M. Dubois, K. A. Gilles, J. K. Hamilton, P. Rebers and F. Smith, Anal. Chem., 1956, 28, 350–356 CrossRef CAS.
- N. Blumenkrantz and G. Asboe-Hansen, Anal. Biochem., 1973, 54, 484–489 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- M. F. Vázquez, L. R. Comini, J. M. Milanesio, S. N. Montoya, J. L. Cabrera, S. Bottini and R. E. Martini, J. Supercrit. Fluids, 2015, 101, 170–175 CrossRef.
- A. Valdés, L. Vidal, A. Beltrán, A. Canals and M. C. Garrigós, J. Agric. Food Chem., 2015, 63, 5395–5402 CrossRef PubMed.
- Ó. Benito-Román, E. Alonso and M. Cocero, J. Supercrit. Fluids, 2013, 73, 120–125 CrossRef.
- J. L. Liu, S. L. Zheng, Q. J. Fan, J. C. Yuan, S. M. Yang and F. L. Kong, Int. J. Biol. Macromol., 2015, 76, 80–85 CrossRef CAS PubMed.
- X. J. Hou and W. Chen, Carbohydr. Polym., 2008, 72, 67–74 CrossRef CAS.
- X. F. Wang, H. Zhang, Z. Y. Wang and H. N. Bai, RSC Adv., 2015, 5, 106800–106808 RSC.
- C. P. Zhu and X. L. Liu, Carbohydr. Polym., 2013, 92, 1197–1202 CrossRef CAS PubMed.
- S. A. Shen, D. J. Chen, X. Li, T. Li, M. Yuan, Y. H. Zhou and C. B. Ding, Carbohydr. Polym., 2014, 104, 80–86 CrossRef CAS PubMed.
- J. Zhang, S. Y. Jia, Y. Liu, S. H. Wu and J. Y. Ran, Carbohydr. Polym., 2011, 86, 1089–1092 CrossRef CAS.
- J. Xi and L. G. Yan, Sep. Purif. Technol., 2017, 175, 170–176 CrossRef CAS.
- C. X. Yang, N. He, X. P. Ling, M. L. Ye, C. X. Zhang, W. Y. Shao, C. Y. Yao, Z. Y. Wang and Q. B. Li, Sep. Purif. Technol., 2008, 63, 226–230 CrossRef CAS.
- W. K. Hsu, T. H. Hsu, F. Y. Lin, Y. K. Cheng and J. P. W. Yang, Carbohydr. Polym., 2013, 92, 297–306 CrossRef CAS PubMed.
- X. Jin, Q. L. Wang, X. Yang, M. Guo, W. J. Li, J. X. Shi, M. Adu-Frimpong, X. M. Xu, W. W. Deng and J. N. Yu, Int. J. Food Sci. Technol., 2018, 53(10), 2298–2307 CrossRef CAS.
- X. J. Jia, C. Zhang, J. F. Qiu, L. L. Wang, J. L. Bao, K. Wang, Y. L. Zhang, M. W. Chen, J. B. Wan, H. X. Su, J. P. Han and C. W. He, Carbohydr. Polym., 2015, 132, 67–71 CrossRef CAS PubMed.
- Q. X. Yuan, L. Y. Zhao, Q. Q. Cha, Y. Sun, H. Ye and X. X. Zeng, J. Agric. Food Chem., 2015, 63, 7986–7994 CrossRef CAS PubMed.
- Y. Diao, Y. Q. Xin, Y. Zhou, N. Li, X. L. Pan, S. M. Qi, Z. L. Qi, Y. M. Xu, L. Luo, H. G. Wan, L. Lan and Z. M. Yin, Int. Immunopharmacol., 2014, 18, 12–19 CrossRef CAS PubMed.
- W. W. Lee, G. Ahn, J. P. W. Arachchillage, Y. M. Kim, S. K. Kim, B. J. Lee and Y. J. Jeon, J. Med. Food, 2011, 14, 1546–1553 CrossRef CAS PubMed.
- T. T. Zhang, C. L. Lu, J. G. Jiang, M. Wang, D. M. Wang and W. Zhu, Carbohydr. Polym., 2015, 130, 307–315 CrossRef CAS PubMed.
- X. Y. Li and L. Wang, Int. J. Biol. Macromol., 2016, 83, 270–276 CrossRef CAS PubMed.
- Z. F. Zhang, G. Y. Lv, H. J. Pan, L. G. Shi and L. F. Fan, Food Sci. Technol. Res., 2011, 17, 461–470 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2019 |