Open Access Article
Open Access ArticlePraseodymium promotes B–Z transition in self-assembled DNA nanostructures†
Madhabi M. Bhanjadeoab and
Umakanta Subudhi *ab
*ab
aDNA Nanotechnology & Application Laboratory, CSIR-Institute of Minerals & Materials Technology, Bhubaneswar 751 013, India. E-mail: usubudhi@immt.res.in; subudhisai@gmail.com
bAcademy of Scientific & Innovative Research (AcSIR), New Delhi 110025, India
First published on 6th February 2019
Abstract
Millimolar concentrations of PrCl3 can induce sequence-specific B–Z transition in various-self-assembled branched DNA (bDNA) nanostructures. Competitive dye binding and thermal kinetics suggest that the phosphate backbone and grooves of bDNA are wrapped with Pr3+ for stabilizing the Z-bDNA. Application of EDTA can convert Z-DNA back to the B-form.
The programmable self-assembly of branched DNA (bDNA) structures is the current trend for creating novel nanostructures with potential for diagnostic, therapeutic, and engineering applications.1 Formation of functional DNA devices will potentially involve dynamic structures that can endure controllable conformational transitions. Hitherto, most switchable DNA devices have been triggered by the addition of DNA single strands and toehold-mediated strand exchange, where transitions occur on time scales of minutes.2 In contrast, conformational transitions initiated by changes in ionic strength, pH, temperature, or light can be faster, and have been demonstrated to occur within seconds.3 Moreover, the B-to-Z DNA transition is one of the most exceptional conversions in the biological world where the transition from right-handed B-DNA to left-handed Z-DNA with opposite chirality happens in milliseconds.4 Recent studies on Z-DNA specific proteins that trigger the B-to-Z transition under physiological conditions have emphasized its role in various biological phenomena like transcription attenuation, gene regulation and participation in certain diseased conditions.5 After the discovery of Z-DNA structure, it is believed that B-to-Z transition is seen specifically on (CG)n or (GC)n sequences and requires extreme ionic conditions like 4 M NaCl.6 However, this vision has been changed gradually over a period of three decades. Now it is known that TA and AT repeat as well as some heterogenous-sequences can readily acquire Z-DNA in the presence of salts or metal complexes.7
Rare earth metals in this context stand as plausible candidates for inducing conformational modulations because of their strong affinity to bind protein and nucleic acids.8 Nevertheless, lanthanide complexes are of increasing importance in cancer diagnosis, therapy, bioimaging and sensing, owing to the versatile chemical and magnetic properties of the lanthanide-ion 4f electronic configuration.9 Recently, we have reported the LaCl3 and CeCl3 induced B–Z transition in bDNA nanostructures.10 Praseodymium is the third element in the lanthanide series and is well used in the medicine for its radioactive isotopes 59Pr141. Moreover, the stable form (59Pr140) is used in therapeutics as well as DNA binding probes for spectroscopic studies.9 However, reports regarding the interaction of PrCl3 with DNA, which involves B–Z transition, are yet to known. On this background, the PrCl3-induced B–Z transition was investigated in the self-assembled Y-shaped bDNA structures in order to understand how different sequences of bDNA affect and initiate the B–Z transition (Fig. 1). Particularly, the influence of loop length and overhang sequences on Pr-induced B–Z transition has been examined in the current communication. The details mechanism including the binding of various-dyes before and after the B–Z transition, and thermal kinetics was performed using Circular dichroism and fluorescence spectroscopy.
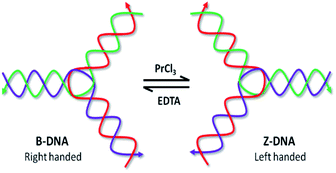 | ||
| Fig. 1 Schematic presentation of Pr-induced B–Z transition in bDNA structure and reversal transition by EDTA. | ||
To understand PrCl3-induced B–Z transition in bDNA, a series of eight Y-shaped bDNA structures (bDNA US-17 to US-24) have been designed as described previously.10 Four of the bDNA (US-17, 19, 21 and 23) bear 3T in the loop whereas the other set of four bDNAs (US-18, 20, 22 and 24) contain 5T in the loop without changing the complementary sequences. However, the overhang sequences of the bDNA structures are different from each other in each set of bDNA (ESI†). The interaction of bDNAs with PrCl3 is examined by employing both CD and absorbance spectroscopy. The B–Z transition in DNA has distinct optical signatures in the CD spectrum due to its helicity and base stacking.11 The CD spectra of bDNA US-17 is quite similar to that of bDNA US-2 with a positive peak at 280 nm and a negative peak at 250 nm.10 When bDNA US-17 is treated with 2.5 to 7.5 mM of PrCl3, a decreasing in spectral trend is observed showing the loosening of base stacking (Fig. 2a). On the contrary, with 10 mM of PrCl3 the CD spectra is dramatically changed showing a negative peak at 295 nm and a positive peak at 255 nm indicating a stable Z-DNA conformation. Since loop length plays an important role in the conformational flexibility,12 the effect of internal loop length on the B–Z transition is explored. The loop length is increased to 5T keeping the other sequences same and the assembled product bDNA US-18 is interacted with PrCl3. With increasing concentration of PrCl3, a clear condensation is noticed with gradual decrease in the positive and negative CD without any sign of transition (Fig. 2b). These results suggest that by addition of two more thymine bases in the loop, the conformational change in bDNA is resistant to B–Z transition. Possibly, the 5T-induced structural flexibility is different from the 3T-induced conformational flexibility which is sensitive for B–Z transition. To access the generality of the observations another set of bDNA US-19 (with 3T loop) and US-20 (with 5T loop) were interacted with PrCl3. The CD spectra of bDNA US-19 and US-20 is similar and no difference is noticed in the absorbance spectra of US-19 and US-20 (Fig. S1†). While addition of 5 mM PrCl3, a significant decrease in CD spectra is observed both in US-19 and US-20. However, with 7.5 and 10 mM of PrCl3 a typical Z-DNA signature is observed in bDNA US-19 unlike the progressive decrease in CD spectra of US-20. A remarkable hypochromism and bathochromic shift is also observed in the absorbance spectra of bDNA US-19 (Fig. S1†). Conversely, with 10 mM PrCl3 a partial B–Z transition is noticed in bDNA US-20 which is accompanied with a faint bathochromic shift (Fig. S1b and d†).
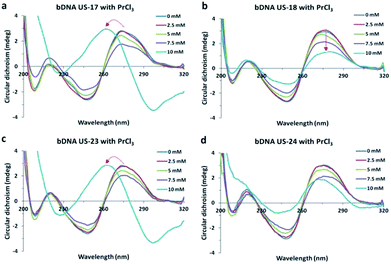 | ||
| Fig. 2 (a–d) Circular dichroism spectral changes of bDNA US-17, US-18, US-23 and US-24 induced by 2.5, 5.0, 7.5 and 10 mM PrCl3. | ||
To move further, another set of bDNA (US-21 with 3T and US-22 with 5T) is interacted with PrCl3. When bDNA US-21 is interacted with PrCl3 a small positive peak at around 260 nm is observed despite a clear negative peak as that of Z-DNA (Fig. S2a†). This phenomenon is indicative of incomplete transition where both the B-form and Z-form co-exists which is represented by a flattened peak at 265 nm. Moreover, the hypochromism and bathochromic shift of bDNA US-21 is clearly observed in Fig. S2c,† which is an indicator of B–Z transition. When PrCl3 is added to the bDNA US-22, a clear condensation is noticed similar to bDNA US-18 (Fig. S2b†). The progressive decreasing trend of both positive peak as well as negative peaks is the clear indication for the condensation of DNA. Similarly, when bDNA US-23 with 3T is interacted with 10 mM of PrCl3 the CD spectra is dramatically changed to Z-bDNA with a negative peak at 295 nm and a positive peak at 255 nm (Fig. 2c). When bDNA US-24 with 5T is subjected to CD, a similar spectrum is obtained like US-23. While addition of 5 and 7.5 mM PrCl3, a significant decrease in CD spectra is observed in US-24 alike to US-23, however, with 10 mM of PrCl3 a pseudo B–Z transition is noticed in bDNA US-24 (Fig. 2d). A clear bathochromic shift is also noticed in bDNA US-23 and bDNA US-24 in the presence of 10 mM PrCl3, which is comparable to the absorbance of bDNA US-17 (Fig. S3†).
From the above observations it is confirmed that the B–Z transition is clearly dependent on the loop length. The 3T variant of bDNA (US-17, US-19 and US-23) is readily exhibiting B–Z transition in presence of 10 mM of PrCl3, whereas with 5T internal loop bDNA is showing either clear condensation (US-18 and US-22) or pseudo transition (US-20 and US-24). The differential response of 5T bearing bDNA structures to B–Z transition might be due to the different overhang sequences. Another interesting observation was that when one of the overhangs AGCT was changed to AATT in bDNA US-19, the B–Z transition was observed with 7.5 mM PrCl3, whereas with three GATC overhang in bDNA US-21, a partial transition was observed. Thus, the overhangs as well as the loop length control the degree of B–Z transition. In general, 10 mM of PrCl3 is sufficient to induce B–Z transition in bDNA structure. Nevertheless, among the transitions, bDNA US-23 is showing a clear B–Z transition, and its 5T variant also showing a pseudo-transition with clear bathchromic shift. Therefore, bDNA US-23 is chosen for thermal kinetics and dye binding assay.
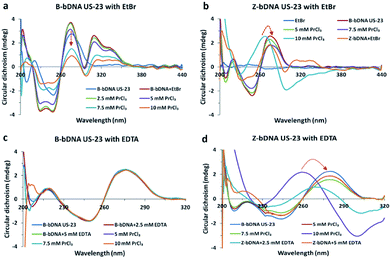
Among the differences between B-DNA and Z-DNA, the absence of major groove in Z-DNA is an attribute which is used to differentiate the Z-DNA from the B-form of DNA.4 In this regard, methyl green (MG) has been chosen that binds selectively to the major grooves of B-DNA, but unable to interact with Z-DNA. Fig. 3 showed CD spectral changes of B-bDNA and Z-bDNA in the presence of MG. When MG is bound to B-bDNA US-23, four induced CD signals (one positive peak at 650 nm and three negative peaks at 320, 425 and 620 nm) are observed which are typical characteristic of bound MG.13 With incremental addition of PrCl3, the induced CD signal is gradually decreased. Interestingly, with 10 mM of PrCl3 the spectrum changed to Z-bDNA with its characteristic negative peak at 298 nm and a positive peak at 262 nm. This finding supports that Pr3+ could bind to the bDNA in a manner that excludes MG from the major groove. To examine the effect of MG binding to Pr-induced Z-bDNA, initially B-bDNA US-23 is interacted with 10 mM PrCl3 to induce B–Z transition and then MG is applied to the Z-bDNA US-23. It is observed that there is no induced negative peak at 320, 425 and 620 nm unlike Fig. 3a except a rudimentary positive peak at 650 nm. Moreover, the PrCl3-induced Z-bDNA conformation is unaltered in presence of MG (Fig. 3b). This result clearly supports the fact that Z-bDNA is devoid of major grooves and therefore, MG is unable to bind to the lost major groove in Pr3+ induced Z-bDNA US-23. Thus, 10 mM PrCl3 is sufficient to induce the B–Z transition in bDNA nanostructures. Possibly, the PrCl3-induced change in helicity leads to the drastic decrease in π–π stacking between the bases and hence losing of major grooves in bDNA.
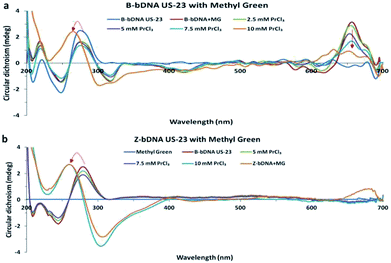 | ||
| Fig. 3 (a and b) Circular dichroism spectra of B-bDNA US-23 with MG followed by PrCl3 and (b) CD spectra of Z-bDNA US-23 with MG. | ||
To understand the molecular mechanism of interaction of bDNA US-23 with PrCl3, we employed a well-established dye binding assay.6 Since nucleic acid grooves are known to interact with metal ions, two well-known minor groove binders such as DAPI and Hoechst 33342 are chosen to bind competitively with PrCl3 bound bDNA US-23. DAPI alone did not show any CD signal, however, after interaction with bDNA US-23, an induced peak is observed at 370 nm (Fig. S4†). Addition of PrCl3 to the bDNA-DAPI complex led to decrease in the induced CD as well as the absorbance. With increasing concentration of PrCl3 the induced CD at 370 nm and absorbance at 260 nm is further decreased and interestingly, with ≥10 mM PrCl3, B-to-Z transition is observed suggesting Pr3+ possibly replaces DAPI from bDNA US-23. In another set of experiment, when DAPI is interacted with Pr-induced Z-bDNA a minor induced peak is observed at 370 nm (Fig. S4c†). Similarly, Hoechst 33342 is interacted with bDNA US-23 and an induced positive peak is observed at 340 nm and the absorbance of bDNA is also increased at 260 nm (Fig. S5†). Addition of PrCl3 up to 5 mM did not alter the induced peak at 340 nm, however 7.5 mM led to decrease in the induced CD and with ≥10 mM PrCl3, B-to-Z transition is observed suggesting Pr3+ possibly replaces Hoechst 33342. The increased absorbance is also decreased after addition of PrCl3. On the contrary, when Hoechst 33342 is interacted with Pr-induced Z-bDNA, an intense peak is observed at 330 nm and the usual DNA peak at 262 nm is enhanced. Collectively our results suggest that Pr-induced Z-conformation of bDNA US-23 is stable in presence of Hoechst and DAPI. It appears that binding affinity of Pr3+ towards bDNA US-23 is higher than minor groove binders. Nevertheless, it is noticed that PrCl3 easily displaced DAPI than Hoechst 33342.
To check whether the mode of binding of PrCl3 is through intercalation we repeated the same competitive dye binding assay with ethidium bromide (EtBr). When bDNA US-23 is interacted with EtBr, two positive peaks are observed at 270 and 300 nm and also two negative peaks are observed at 240 and 250 nm (Fig. 4a). Addition of ≥10 mM PrCl3 led to decrease in the induced CD at respective peaks without the B-to-Z transition. Possibly Pr3+does not intercalate with the B-bDNA US-23 or PrCl3 unable to displace the EtBr from bDNA US-23. May be the EtBr bound bDNA are in minimized energy, as a result the bases do not flip to acquire left-handed conformation even after addition of PrCl3. In another set of reaction, when Pr-induced Z-bDNA US-23 is interacted with EtBr, it is observed that the negative peaks at 295 nm got decreased and the positive peak at 263 nm shifted to 278 nm which correspond to the B-bDNA (Fig. 4b). Possibly, the intercalation of EtBr to Z-bDNA is disturbed and hence, resulting in a ruminant peak at 300 and 270 nm. Z-bDNA–EtBr complex had a decreased absorbance at 290 nm as compared to B-bDNA–EtBr complex which can be easily corroborated with the CD data (Fig. S6†).
EDTA, a known metal chelator is used earlier to study the reversibility of the metal ion induced B-to-Z DNA transition.10 To check the Pr3+ chelation by EDTA, the bDNA US-23 is preincubated with EDTA and then PrCl3 is added. As expected in presence of EDTA no change in B-conformation is observed suggesting that before PrCl3 being interacting with bDNA all Pr3+ are trapped by EDTA, hence no B–Z transition or any other change is noticed (Fig. 4c). The absorbance spectra are also unaltered after addition of PrCl3 to the preincubated bDNA–EDTA solution (Fig. S6c†). On the contrary, when Pr-induced Z-bDNA is titrated with increasing concentration of EDTA the Z-conformation of bDNA slowly moves back to the B-conformation (Fig. 4d). This suggests that EDTA-chelates the PrCl3 and Z-bDNA are back to its native right-handed conformation. The absorbance spectra also support this observation (Fig. S6d†).
To compare these observations and understand how different fluorophores are released from B-conformation of bDNA, the fluorescence competitive binding assay is conducted after binding to PrCl3. The fluorescence quenching experiment is widely used to establish DNA binding mode.10,14 In this assay DAPI, Hoechst and EtBr dyes are interacted with B-bDNA US-23. When Hoechst 33342 (excitation wavelength, 361 nm) and DAPI (excitation wavelength, 358 nm) are interacted with bDNA US-23, an enhanced fluorescence is observed (Fig. S7†). Now, if Pr3+ competitively binds to the same site of bDNA where DAPI or Hoechst are bound, the fluorescence of DAPI and Hoechst would significantly decrease, because the strong binding of Pr3+ with bDNA should exclude these minor groove binders. As expected when PrCl3 is incrementally added to this enhanced fluorescence, invariably the emission intensity decreased than the DAPI or Hoechst bound to bDNA (Fig. S7†). With addition of 10 mM PrCl3, fluorescence intensity of DNA declined to 30% and 60% for Hoechst 33342 and DAPI, respectively. This is suggesting that DAPI is easily released and replaced by PrCl3 similar to the spectroscopic observations. On the other hand, intercalating dye EtBr (excitation wavelength 480 nm) is widely used as a suitable fluorescence probe for DNA-binding with complexes in solution. When PrCl3 is added to the bDNA–EtBr complex, the fluorescence intensity of bDNA–EtBr complex has gradually weakened up to 50% at about 590 nm. As shown in Fig. S7,† when PrCl3 is added to US-23-dye complex, the decrease in emission is highest with DAPI and lowest with Hoechst suggesting that Pr3+ has DAPI like binding to the bDNA US-23. It is indicated that EtBr, DAPI or Hoechst could still bind to DNA even in the presence of Pr3+. In combination with CD, competitive binding results of major and minor groove binders and fluorescence experiments further supported that Pr3+could bind to major and minor grooves of bDNA. Nevertheless, the binding of Pr3+ is very strong towards DNA; once it is bound neither the minor groove nor major groove binders could replace and reverse the transition. The Z-conformation of the bDNA is retained even in the presence of minor and major groove binders.
Another property of left-handed Z-DNA is its thermal instability. From the thermal melting curve, it is observed that the Tm of B-bDNA US-23 is ∼75 °C whereas the Tm of Z-DNA is found to be lesser than 33 °C in both absorbance and CD spectroscopy (Fig. S8–S11†). The reason for the reduced Tm of Z-DNA can be explained as follows. The conversion of B-DNA to Z-DNA is associated with a flipping over of the base pairs so that they had an upside-down orientation relative to that of B-DNA. This flipping over resulted both in the production of a syn-conformation in purine bases and a change in the deoxyribose-ring pucker in the respective bases. Moreover, in Z-DNA, the base pairs occupy a position at the periphery instead of the centre as in B-DNA.15 This may be the cause of lower Tm of Z-DNA since base stacking and base pairing are not in proper order like B-DNA. Interestingly, the Tm value of the B-bDNA is also unaltered during the process of annealing. Thus, both the CD and absorbance spectroscopy results advocate the reversible nature of B-bDNA US-23 during the process of denaturation and renaturation unlike the Pr3+ bound Z-bDNA. We speculate a thorough wrapping of Pr3+ on the backbone of Z-DNA which hinders the formation of hydrogen bonding during the process renaturation. However, when EDTA is applied to Pr-induced Z-bDNA, the Pr3+ is chelated and a clear Z–B transition was observed as mentioned earlier.
In summary, for the first time our results indicate that low concentration of PrCl3 can induce a left-handed helical structure in self-assembled Y-shaped bDNA with sequence and structure selectivity. Interestingly, the bDNA having 3T in the loop are more sensitive for B–Z transition than 5T bearing bDNA. One of the most important findings in our study is that bDNA US-17, US-19, and US-23 structures with 3T are found to be more specific affinity towards PrCl3 and have considerable conformational flexibility for switching from right-handed to left-handed Z-DNA. However, the interaction between the bDNA structure and PrCl3 is different than other lanthanides discussed in earlier reports. While 5 and 7.5 mM of LaCl3 and CeCl3 respectively, are enough to induce a left-handed Z-conformation, 10 mM of PrCl3 is required for a stable Z-bDNA conformation. Thermal kinetics and dye binding experiments suggest that the B–Z transition is associated with an array of Pr3+ loaded on the phosphate backbone of bDNA structure as well as to the grooves of DNA and when Pr3+ is chelated by EDTA the bDNA transit to the original B-conformation.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Council of Scientific and Industrial Research, Government of India under National Laboratory Scheme EMPOWER OLP-19 and YSP-05 to U.S. and 12th Five year plan project (ESC-401).Notes and references
- (a) N. C. Seeman, Annu. Rev. Biochem., 2010, 79, 65 CrossRef CAS PubMed; (b) A. K. Nayak and U. Subudhi, RSC Adv., 2014, 4, 54506 RSC; (c) S. Saha, V. Prakash, S. Halder, K. Chakraborty and Y. Krishnan, Nat. Nanotechnol., 2015, 10, 645 CrossRef CAS PubMed; (d) P. D. Halley, C. R. Lucas, E. M. McWilliams, M. J. Webber, R. A. Patton, C. Kural, D. M. Lucas, J. C. Byrd and C. E. Castro, Small, 2016, 12, 308 CrossRef CAS PubMed; (e) S. Nahar, A. K. Nayak, A. Ghosh, U. Subudhi and S. Maiti, Nanoscale, 2018, 10, 195 RSC.
- (a) Y. Li, C. Zhang, C. Tian and C. Mao, Org. Biomol. Chem., 2014, 12, 2543 RSC; (b) X. Shen, A. Asenjo-Garcia, Q. Liu, Q. Jiang, F. Javier Garcia de Abajo, N. Liu and B. Ding, Nano Lett., 2013, 13, 2128 CrossRef CAS PubMed.
- (a) C. Mao, W. Sun, Z. Shen and N. C. Seeman, Nature, 1999, 397, 144 CrossRef CAS PubMed; (b) A. Rajendran, M. Endo and H. Sugiyama, Angew. Chem., Int. Ed., 2012, 51, 874 CrossRef CAS PubMed; (c) S. Modi, C. Nizak, S. Surana, S. Halder and Y. Krishnan, Nat. Nanotechnol., 2013, 8, 859 CrossRef PubMed.
- (a) A. Rich, A. Nordheim and A. H. J. Wang, Annu. Rev. Biochem., 1984, 53, 791 CrossRef CAS PubMed; (b) A. Rich and S. Zhang, Nature, 2003, 4, 566 CAS.
- (a) S. C. Ha, N. K. Lokanath, D. Van Quyen, C. A. Wu, K. Lowenhaupt, A. Rich, Y. G. Kim and K. K. Kim, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 14367 CrossRef CAS PubMed; (b) S. C. Ha, K. Lowenhaupt, A. Rich, Y. G. Kim and K. K. Kim, Nature, 2005, 437, 1183 CrossRef CAS PubMed; (c) S. C. Ha, J. Choi, H. Y. Hwang, A. Rich, Y. G. Kim and K. K. Kim, Nucleic Acids Res., 2008, 37, 629 CrossRef PubMed; (d) S. Bae, D. Kim, K. K. Kim, Y. G. Kim and S. J. Hohng, J. Am. Chem. Soc., 2010, 133, 668 CrossRef PubMed; (e) J. Geng, C. Zhao, J. Ren and X. Qu, Chem. Commun., 2010, 46, 7187 RSC; (f) Y. M. Lee, H. E. Kim, C. J. Park, A. R. Lee, H. C. Ahn, S. J. Cho, K. H. Choi, B. S. Choi and J. H. Lee, J. Am. Chem. Soc., 2012, 134, 5276 CrossRef CAS PubMed.
- M. J. Ellison, R. J. Kelleher, A. H. Wang, J. F. Habener and A. Rich, Proc. Natl. Acad. Sci. U. S. A., 1985, 82, 8320 CrossRef CAS.
- (a) J. Kim, C. Yang and S. DasSarma, J. Biol. Chem., 1996, 271, 9340 CrossRef CAS PubMed; (b) J. R. Bothe, K. Lowenhaupt and H. M. Al-Hashimi, J. Am. Chem. Soc., 2011, 133, 2016 CrossRef CAS PubMed; (c) Z. Wu, T. Tian, J. Yu, X. Weng, Y. Liu and X. Zhou, Angew. Chem., Int. Ed., 2011, 50, 11962 CrossRef CAS PubMed; (d) S. Wang, Y. Long, J. Wang, Y. Ge, P. Guo, Y. Liu, T. Tian and X. Zhou, J. Am. Chem. Soc., 2014, 136, 56 CrossRef CAS PubMed; (e) Y. J. Jang, C. Lee and S. K. Kim, Sci. Rep., 2015, 5, 9943 CrossRef CAS PubMed.
- (a) C. Bouzigues, T. Gacoin and A. Alexandrou, ACS Nano, 2011, 5, 8488 CrossRef CAS PubMed; (b) C. Liu, Y. Hou and M. Gao, Adv. Mater., 2014, 26, 6922 CrossRef CAS PubMed; (c) S. Sharma, R. R. Samal, U. Subudhi and G. B. N. Chainy, Int. J. Biol. Macromol., 2018, 115, 853 CrossRef CAS PubMed.
- (a) M. Khorasani-Motlagh, M. Noroozifar and S. Khmmarnia, Spectrochim. Acta, Part A, 2011, 78, 389 CrossRef PubMed; (b) R. D. Teo, J. Termini and H. B. Gray, J. Med. Chem., 2016, 59, 6012 CrossRef CAS PubMed.
- (a) A. K. Nayak, A. Mishra, B. S. Jena, B. K. Mishra and U. Subudhi, Sci. Rep., 2016, 6, 26855 CrossRef CAS PubMed; (b) M. M. Bhanjadeo, A. K. Nayak and U. Subudhi, Biochem. Biophys. Res. Commun., 2017, 482, 916 CrossRef CAS PubMed.
- J. Kypr, I. Kejnovska, D. Renciuk and M. Vorlickova, Nucleic Acids Res., 2009, 37, 1713 CrossRef CAS PubMed.
- Y. He and C. Mao, Chem. Commun., 2006, 9, 968 RSC.
- L. Feng, A. Zhao, J. Ren and X. Qu, Nucleic Acids Res., 2013, 41, 7987 CrossRef CAS PubMed.
- S. A. Tysoe, R. J. Morgan, A. D. Baker and T. C. Strekas, J. Phys. Chem., 1993, 97, 1707 CrossRef CAS.
- Y. Kim, H. Li, Y. He, X. Chen, X. Ma and M. Lee, Nat. Nanotechnol., 2017, 12, 551 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra10164g |
| This journal is © The Royal Society of Chemistry 2019 |