Open Access Article
Open Access ArticleConstruction of self-signal DNA electrochemical biosensor employing WS2 nanosheets combined with PIn6COOH†
Jimin Yang‡
 ,
Lei Gao‡,
Cheng Peng and
Wei Zhang
,
Lei Gao‡,
Cheng Peng and
Wei Zhang *
*
School of Chemistry and Chemical Engineering, Linyi University, Linyi 276005, China. E-mail: zhangweiqust@126.com; Fax: +86-539-7258620; Tel: +86-539-7258620
First published on 26th March 2019
Abstract
In this work, a novel self-signal DNA electrochemical biosensor was constructed based on tungsten disulfide (WS2) nanosheets combined with poly(indole-6-carboxylic acid) (PIn6COOH) as the sensing interface. The WS2 nanosheets were synthesized via a simple solvent exfoliation method from bulk WS2, and then PIn6COOH was electropolymerized on the WS2 nanosheet-modified carbon paste electrode to obtain a unique nanocomposite. The electropolymerization efficiency was remarkably improved, ascribed to the physical adsorption between WS2 nanosheets and aromatic In6COOH monomers, resulting in an increase of the electrochemical response of PIn6COOH. Owing to the presence of π–π interactions between the conjugated PIn6COOH/WS2 nanocomposite and DNA bases, the probe ssDNA was noncovalently assembled on the nanocomposite substrate. After the hybridization of the probe ssDNA with the target DNA, the formation of the double-helix structure induced the resulting dsDNA to be released from the surface of the conjugated nanocomposite, accompanied with the self-signal regeneration of the nanocomposite (“signal-on”). The constructed PIn6COOH/WS2 nanocomposite was not only employed as an interface for DNA immobilization but also reflected the signal transduction stemming from DNA immobilization and hybridization without any external indicators or complex labeling processes. A detection limit of 2.3 × 10−18 mol L−1 has been estimated and a dynamic range of 1.0 × 10−17 mol L−1 to 1.0 × 10−11 mol L−1 has been shown for the detection of a PIK3CA gene related to lung cancer. Selectivity of the biosensor has been researched in the presence of noncomplementary and base mismatched DNA sequences.
1. Introduction
Detection of specific gene sequences through a simple and rapid strategy has emerged as an effective tool in lots of areas, such as drug screening, forensic investigation, environmental regulation, disease diagnosis and treatment, and other biomedical applications. Various methods have been utilized for DNA determination, involving piezoelectric, chemiluminescence, polymerase chain reaction, surface enhanced Raman scattering, fluorescence, and colorimetric and electrochemical techniques.1 Among these strategies, DNA electrochemical biosensors have received widespread concern as innovative assay platforms owing to their fast signal output, high specificity and sensitivity, portability, small dimensions, and inexpensive instrumentation.2,3 Nowadays, in order to meet the higher demand of DNA detection in different fields, DNA electrochemical biosensors have been combined with other techniques to improve their analytical capacity. Especially, various nanostructured materials have been employed to develop novel sensing interfaces to improve the recognition ability of the sensing layer and the signal output intensity of the sensing event.4–10The research of the biointeraction of the DNA–graphene and their bioanalysis has drawn much attention in recent years.11–16 With the development of graphene-like materials, two-dimensional (2D) transition-metal dichalcogenides (TMDs), another type of 2D materials, have aroused intensive interest recently owing to their unique structures, fine catalytic properties, attractive semiconducting characteristics and strong chemical stability.17–21 As a key member of the 2D TMDs, tungsten disulfide (WS2) with a structure composed of three stacked atom layers (S–W–S) bonded together by van der Waals forces has shown unparalleled performance in various fields, such as supercapacitors, hydrogen evolution, catalysts, lithium-ion batteries, sensors, and so on.22–26 Interestingly, compared to the intensive research reported on MoS2, the number of reports on the use of WS2 in electrochemical biosensing is smaller.
Poly(indole-6-carboxylic acid) (PIn6COOH), as a novel conjugated conducting polymer, has drawn much attention because of its several superiorities, such as excellent redox activity and wonderful stability, which has been exploited for determination of biomolecules.27,28 In this study, WS2 nanosheets supported PIn6COOH was employed as the platform for electrochemical sensing of the PIK3CA gene from lung cancer. The immobilization of the probe ssDNA was accomplished through the noncovalent π–π stacking between the rich-conjugated PIn6COOH/WS2 nanocomposite and the ring of nucleobases,29 accompanied with a decreased current response (“signal-off”), which might be due to that when the ssDNA probe film was coated on the electrode surface, the steric hindrance was introduced at the same time, which may influence the redox response of the PIn6COOH/WS2 composite.30 After the probe ssDNA hybridized with the target DNA, an increment of the redox current (“signal-on”) was obtained, which was caused by the releasing of the formed dsDNA helix from the surface of the nanocomposite,31,32 leading to the regeneration of the self-signals. The self-signal changes originated from DNA immobilization and hybridization could be sensitively identified by cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS).
2. Experimental section
2.1. Materials
The bulk WS2 was acquired from Shanghai Aladdin Biological Technology Corporation (China). Indole-6-carboxylic acid (In6COOH, 99%) was purchased from Sigma-Aldrich (USA). Graphite powder (spectral pure, diameter about 30 mm) was obtained from Shanghai Colloid Chemical Plant (China). N,N-Dimethylformamide (DMF), acetonitrile, phosphate-buffered saline (PBS) solution and sodium dodecyl sulfate (SDS) were provided by Shanghai Reagent Company (China). The mouse serum samples were provided by Linyi People's Hospital (China). All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Linyi People's Hospital and approved by the Animal Ethics Committee of Linyi People's Hospital.The 15-base synthetic oligonucleotides probe (ssDNA), its complementary DNA (cDNA, target DNA, namely an 15-base fragment of PIK3CA gene sequence), single-base mismatched DNA, three-base mismatched DNA and noncomplementary DNA (ncDNA) were synthesized by Shanghai Sangon Bioengineering Limited Company (China). Their base sequences were listed as follows:
Probe DNA (ssDNA): 5′-AGT GAT TTT AGA GAG-3′;
Complementary DNA (cDNA): 5′-CTC TCT AAA ATC ACT-3′;
Single-base mismatched DNA: 5′-CTC TGT AAA ATC ACT-3′;
Three-base mismatched DNA: 5′-CTC TGT AAA GTC TCT-3′;
Noncomplementary DNA (ncDNA): 5′-GTC TAC CAG CGA TGC-3′.
2.2. Instruments
The electrochemical measurements were conducted on a CHI660E electrochemical workstation (Shanghai Chenhua Instrument Company, China) by using a three-electrode system comprising a carbon paste electrode (CPE, 3 mm in diameter) as the working electrode, a saturated calomel electrode (SCE) as the reference electrode, and a platinum wire as the auxiliary electrode. Transmission electron microscopy (TEM) images were acquired using JEM-2100 (JEOL, Japan). FT-IR spectrum was conducted on a Tensor 27 FT-IR spectrophotometer (Bruker Company, Germany).2.3. Fabrication of the modified electrodes
The WS2 nanosheets were fabricated by a simple ultrasonic exfoliation method. Briefly, bulk WS2 was dispersed in DMF at a concentration of 1.0 mg mL−1 by sonication for 6 h. The bare CPE was prepared as reported previously.33 In order to obtain a smooth surface, the surface of bare CPE was pre-treated by polishing with a weighing paper. 10 μL of the above dispersion was drop casted over the pre-treated surface of bare CPE carefully and then allowed to dry for 24 h to achieve the WS2/CPE.The prepared electrode (WS2/CPE) was undergoes the electropolymerization in a acetonitrile solution containing 0.05 mol L−1 In6COOH monomers at a constant potential of 1.4 V for 600 s. The modified electrode was denoted as PIn6COOH/WS2/CPE. For comparison, the PIn6COOH/CPE was also prepared under the similar procedure just without the WS2 nanosheets.
2.4. Immobilization and hybridization of DNA
10 μL of PBS solution (pH 7.0) containing 1.0 × 10−11 mol L−1 probe ssDNA was drop-cast to cover the PIn6COOH/WS2/CPE and air-dried. After that, the modified electrode was immersed in 0.1% SDS solution for 5 min to remove the unbound capture probe. Subsequently, the electrode modified with probe DNA was obtained and denoted as ssDNA/PIn6COOH/WS2/CPE.The hybridization capacity of the fabricated biosensor was investigated by dripping 10 μL of hybridization solution (PBS, pH 7.0) containing target DNA with desired concentrations on the probe-captured electrode. After naturally dried in the air, the electrode was kept at −0.7 V for 300 s to release the formed dsDNA from the electrode surface, followed by rinsed with SDS to remove the non-specifically attached DNA, and the hybridized electrode was achieved. The procedure was also applied for the hybridization of the biosensor with the other base-mismatched and noncomplementary DNA fragments.
The schematic illustration of the construction process of the self-signal electrochemical sensing platform was displayed in Scheme 1.
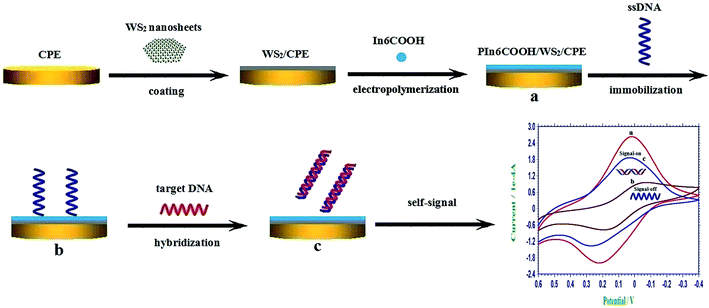 | ||
| Scheme 1 The schematic illustration of the construction process of the self-signal electrochemical sensing platform. | ||
2.5. Electrochemical measurements
The CV experiments were recorded in 0.3 mol L−1 PBS (pH 7.0) at a scan rate of 100 mV s−1 from 0.6 V to −0.4 V. The EIS experiments were also made in 0.3 mol L−1 PBS (pH 7.0). The AC voltage amplitude was 5 mV, and the voltage frequencies were ranged from 105 Hz to 1 Hz.During the entire experiments, the solution was continually bubbled with nitrogen for removing the oxygen. Unless specifically indicated, all the experiments were carried out at room temperature.
3. Results and discussion
3.1. Structural characterizations
The TEM has been adopted to characterize the morphologies of the as-prepared samples, as displayed in Fig. 1. The TEM image in Fig. 1A revealed the layered WS2 sheets, illustrating the flake-like shape of WS2. As shown in Fig. 1B, the formed PIn6COOH/WS2 nanocomposite indicated a large amount of prominences compared with Fig. 1A, revealing that the WS2 sheets served as the substrate for the nucleation and growth of PIn6COOH and turned to a relatively coarse structures. The obtained PIn6COOH film electropolymerized without the WS2 sheets (Fig. 1C) displayed a rough surface with many corrugations.The FT-IR spectrum of the PIn6COOH/WS2 composite was shown in Fig. S1 (ESI†). The intense band at 1684 cm−1 was related to the stretching vibration of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group. The broad band at 2549 cm−1 corresponded to the –OH stretching vibration. And then, the band at 1422 cm−1 was pertinent to the C–O stretching vibration. The bands at 756 cm−1 was assigned to the three adjacent C–H deformation vibration of ring hydrogens on benzene ring. In addition, the broad peak of N–H bond at 3430 cm−1 was the characteristic absorption of the N–H bond. This can provide more evidence of the successful modification of PIn6COOH.
O group. The broad band at 2549 cm−1 corresponded to the –OH stretching vibration. And then, the band at 1422 cm−1 was pertinent to the C–O stretching vibration. The bands at 756 cm−1 was assigned to the three adjacent C–H deformation vibration of ring hydrogens on benzene ring. In addition, the broad peak of N–H bond at 3430 cm−1 was the characteristic absorption of the N–H bond. This can provide more evidence of the successful modification of PIn6COOH.
3.2. Electrochemical characterization of different modified electrodes
As observed in Fig. 2, the electrochemical properties of different modified electrodes were contrasted by monitoring the self-redox response of PIn6COOH in 0.3 mol L−1 PBS (pH 7.0). There were no obvious CV peaks on the bare CPE (curve a). The CV response of the PIn6COOH/CPE (curve b) consisted of a distinct couple of redox peaks, demonstrating that an electroactive polymer film had been formed on the electrode surface. The CV peak current of the PIn6COOH/WS2/CPE (curve c) displayed the highest response compared with the bare CPE (curve a) and PIn6COOH/CPE (curve b), which could be owing to the physical adsorption between WS2 nanosheets and aromatic rings of In6COOH monomers, which could supply more growth sites and significantly enhance the electropolymerization efficiency, followed by a promoted electrochemical self-redox response of PIn6COOH.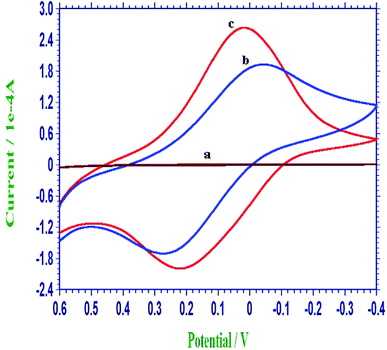 | ||
| Fig. 2 Representative cyclic voltammograms of the bare CPE (a), PIn6COOH/CPE (b) and PIn6COOH/WS2/CPE (c) in 0.3 mol L−1 PBS (pH 7.0). | ||
3.3. Optimization of the experimental conditions
The electrochemical response of the PIn6COOH/WS2/CPE could be dramatically affected by the electrochemical polymerization parameters, such as the polymerization time of PIn6COOH. Here, PIn6COOH was electropolymerized by keeping a fixed potential of 1.4 V, and different polymerization time was investigated. The results showed that the electrochemical response of PIn6COOH/WS2/CPE increased with the prolonging of the polymerization time and achieved a maximum at 600 s, and then decreased with further electrochemical polymerization. Therefore, 600 s was utilized as the polymerization time in our experiments.The amount of WS2 nanosheets also had a prominent effect on the performance of the obtained PIn6COOH/WS2 nanocomposite. EIS was adopted to optimize the influence of the amount of WS2 on the formation of the nanocomposite. With the increase of the amount of WS2, the impedance response of the PIn6COOH/WS2 nanocomposite decreased, and the minimum was reached at 10 μL of 1.0 mg mL−1 WS2. However, when the amount of WS2 was higher than 10 μL of 1.0 mg mL−1, the impedance increased. This might be ascribed to the coverage of the electrode surface by a excessively thick WS2, which may block the effective adsorption and electropolymerization of In6COOH, followed by the depressed conductivity of the formed nanocomposite. Hence, 10 μL of 1.0 mg mL−1 WS2 nanosheets was chosen for the electrode fabrication in experiments.
The polymer had fine electrochemical redox behavior when the pH value was ≤7. And the redox activity of the polymer became poor in alkaline conditions. And in order to maintain the fine bioactivity of DNA molecules, pH 7.0 was used in experiments.
3.4. Electrochemical self-signal monitoring of DNA immobilization and hybridization
The changes of the self-redox signals of PIn6COOH was employed to probe the immobilization and hybridization of DNA on the PIn6COOH/WS2 nanocomposite. As displayed in curve a of Fig. 3, the PIn6COOH/WS2/CPE exhibited favorable electrochemical activities in a neutral environment. When the ssDNA probes were immobilized on the substrate of the PIn6COOH/WS2 nanocomposite by noncovalent conjunction between the rich-conjugated nanocomposite and the ring of nucleobases,29 the self-redox signals of the ssDNA/PIn6COOH/WS2/CPE reduced notably (curve b). When the ssDNA probe film was coated on the electrode surface, the steric hindrance was introduced at the same time, which may influence the redox response of the PIn6COOH/WS2 composite.30 Thereby, the signals of the ssDNA/PIn6COOH/WS2 layer decreased (“signal-off”), illustrating the successful attachment of ssDNA probes on the PIn6COOH/WS2 substrate. However, after the hybridization of the ssDNA probes with the target DNA, an evident increment of the self-signal response (curve c) was observed compared with curve b as a result of the release of the formed dsDNA from the nanocomposite substrate, accompanied with the self-signal regeneration of the nanocomposite (“signal-on”). As a matter of fact, the formation of stable hydrogen bonds among the nucleobases and their shielding inside the phosphate backbone after hybridization is the propulsive effort for the above release.31,32 And the reason due to that the dsDNA has much higher negative charge density than that of ssDNA is also an important influence factor of the desorption. The above results proved that the self-signal changes of the PIn6COOH layer could be exploited as a facile and competent implement for direct DNA determination without needing any external indicators or adding any labels to the biomolecules.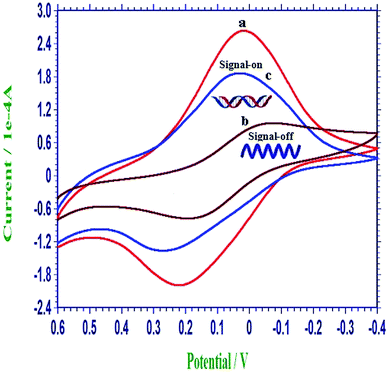 | ||
| Fig. 3 Representative cyclic voltammograms of the PIn6COOH/WS2/CPE (a), ssDNA/PIn6COOH/WS2/CPE (b) and dsDNA/PIn6COOH/WS2/CPE (c) in 0.3 mol L−1 PBS (pH 7.0). | ||
3.5. Selectivity of the constructed DNA biosensor
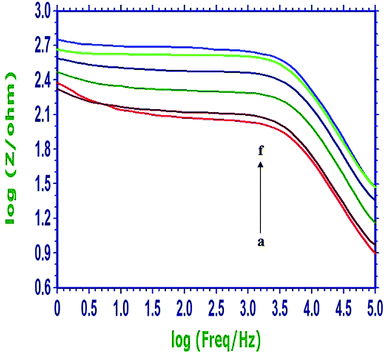
The specificity of the constructed sensing protocol was examined by testing the system with the complementary, noncomplementary and mismatched DNA fragments related to the PIK3CA gene, and the obtained Bode plots in 0.3 mol L−1 PBS (pH 7.0) were shown in Fig. 4. The curve a (red) was the Bode plot of the PIn6COOH/WS2/CPE. When the probe ssDNA was noncovalently attached on the PIn6COOH/WS2 nanocomposite, an evident increase of the resistance response was observed (curve f, light blue). After the biosensor was hybridized with the complementary sequence, the resistance response decreased apparently (curve b, brown), which suggested that the hybridization reaction had been occurred, and the release of the formed duplex from the substrate of the nanocomposite caused “signal-on”. However, when the biosensor was hybridized with the single-base mismatched sequence (curve c, dark green) or three-base mismatched sequence (curve d, dark blue), the resistance response was higher than that obtained from the hybridization with the complementary DNA, but was still smaller than the probe electrode, illustrating that complete hybridization was not attained because of the base mismatch. As expected, noncomplementary oligonucleotides displayed a negligible change in the response (curve e, light green) compared with the curve f (light blue), indicating that the hybridization reaction did not happen. These results demonstrated that the developed DNA biosensor possessed splendid selectivity for the hybridization detection.3.6. Quantitative detection of PIK3CA gene target sequences
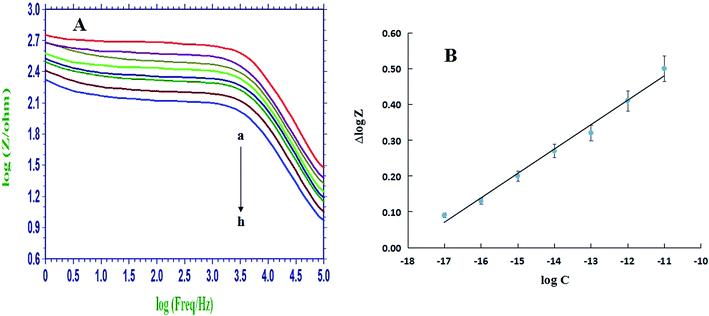
The sensitivity of the DNA biosensor was further investigated by using different concentrations of the complementary sequences for hybridization. Fig. 5A showed the Bode plots recorded on ssDNA/PIn6COOH/WS2/CPE (curve a) and that after hybridization with various concentrations of complementary sequences, and the impedance decreased with the gradual enhancement of the cDNA concentrations from 1.0 × 10−17 mol L−1 to 1.0 × 10−11 mol L−1. The difference (namely Δlog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Z) between the log
Z) between the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Z values of the probe-captured electrode and that after hybridization with the cDNA was employed as the measurement signal. Fig. 5B showed a linear dependence between the Δlog
Z values of the probe-captured electrode and that after hybridization with the cDNA was employed as the measurement signal. Fig. 5B showed a linear dependence between the Δlog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Z value and the logarithm of complementary target sequence concentrations with a regression equation of Δlog
Z value and the logarithm of complementary target sequence concentrations with a regression equation of Δlog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Z = 0.0682
Z = 0.0682![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C + 1.2293 (r = 0.9960). The detection limit was estimated to be 2.3 × 10−18 mol L−1 based on 3σ, where σ was the relative standard deviation (RSD) of a blank solution with seven parallel measurements. The analytical performance of this proposed biosensor was also compared with some other DNA electrochemical biosensors based on the poly(indole-6-carboxylic acid), and the results were listed in Table 1. It is apparent that the proposed DNA biosensor had the lower detection limit and the wider linear range for the target DNA assay, which might be mainly ascribed to the large specific surface area of the WS2 nanosheets that improved the immobilization amount of ssDNA probes and the fine electronic conduction ability of PIn6COOH that improved the detection signals.
C + 1.2293 (r = 0.9960). The detection limit was estimated to be 2.3 × 10−18 mol L−1 based on 3σ, where σ was the relative standard deviation (RSD) of a blank solution with seven parallel measurements. The analytical performance of this proposed biosensor was also compared with some other DNA electrochemical biosensors based on the poly(indole-6-carboxylic acid), and the results were listed in Table 1. It is apparent that the proposed DNA biosensor had the lower detection limit and the wider linear range for the target DNA assay, which might be mainly ascribed to the large specific surface area of the WS2 nanosheets that improved the immobilization amount of ssDNA probes and the fine electronic conduction ability of PIn6COOH that improved the detection signals.
| This work | Ref. 34 | Ref. 35 | |
| Electrodes | Poly(indole-6-carboxylic acid)/WS2/CPE | Poly(indole-6-carboxylic acid)/GCE | Poly(indole-6-carboxylic acid)-MWNT/GCE |
| Detection methods | EIS (PBS) | CV (NaAc–HAc) | CV (NaAc–HAc) |
| Detection limit (mol L−1) | 2.3 × 10−18 | 5.79 × 10−12 | 2.0 × 10−15 |
| Detection range (mol L−1) | 1.0 × 10−17 to 1.0 × 10−11 | 3.5 × 10−10 to 2.0 × 10−8 | 1.0 × 10−14 to 5.0 × 10−12 |
3.7. Stability, repeatability and precision of the DNA biosensor
The stability experiments showed that after a fabricated biosensor was stored at 4 °C in a dry environment for 10 days, the obtained hybridization response remained at 93.6% compared with that before storage, exhibiting that the constructed biosensor had a favorable stability. In addition, the repeatability of the biosensor was monitored by five parallel-made probe DNA biosensors to detect 1.0 × 10−17 mol L−1 target DNA sequences, and the results showed that a RSD of 5.26% was estimated, displaying a fine repeatability of the developed DNA biosensor.In order to investigate the precision of the developed DNA biosensor, 1.0 fM pure complementary DNA was added into PBS buffer, and then analyzed by the developed biosensor. The recovery was calculated to be 103.8%, suggesting the fine precision of the biosensor.
3.8. Detection of target DNA in serum samples by the biosensor
The proposed biosensor was finally utilized to evaluate the applicability for real serum samples. According to the standard addition method, three different concentrations of target DNA were spiked into diluted serum samples. As shown in Table S1 (ESI†), the recoveries were in the range of 97.5–101%, and the RSDs were 3.86–5.19%. These results indicate that the proposed strategy for DNA detection can be applied to detect the target in complex samples with satisfactory results.4. Conclusions
In summary, a new approach has been proposed for the self-signal electrochemical detection of PIK3CA gene related to lung cancer with a novel nanocomposite film based on WS2 nanosheets combined with PIn6COOH. The morphological and surface properties of this nanocomposite deposited on the CPE surface have been studied, where it has been confirmed that this film is well suited for grafting the biolayer. WS2 nanosheets incorporated with polymer enhances the performance of biosensors in terms of high selectivity, low detection limit, wide dynamic range, fine stability and repeatability integrated with the competence to distinguish mismatched sequences from complementary target. These superiorities could make this platform as a new paradigm in biosensor design and pave the way for the building of other innovative analytical systems.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Natural Science Foundation of Shandong Province (No. ZR2017QB013), the Undergraduate Training Program for Innovation and Entrepreneurship of Linyi University (No. 201810452170), and the students study evaluation reform course (Applied Electrochemistry) of Linyi University.References
- A. Sassolas, B. D. Leca-Bouvier and L. J. Blum, DNA Biosensors and Microarrays, Chem. Rev., 2008, 108, 109–139 CrossRef CAS PubMed.
- A. L. Liu, K. Wang, S. H. Weng, Y. Lei, L. Q. Lin, W. Chen, X. H. Lin and Y. Z. Chen, Development of electrochemical DNA biosensors, Trends Anal. Chem., 2012, 37, 101–111 CrossRef CAS.
- G. Evtugyn and T. Hianik, Electrochemical DNA sensors and aptasensors based on electropolymerized materials and polyelectrolyte complexes, Trends Anal. Chem., 2016, 79, 168–178 CrossRef CAS.
- C. L. Zhang, J. L. He, Y. C. Zhang, J. Chen, Y. L. Zhao, Y. Z. Niu and C. Yu, Cerium dioxide-doped carboxyl fullerene as novel nanoprobe and catalyst in electrochemical biosensor for amperometric detection of the CYP2C19*2 allele in human serum, Biosens. Bioelectron., 2018, 102, 94–100 CrossRef CAS PubMed.
- H. Imran, P. N. Manikandan and V. Dharuman, Graphene oxide supported liposomes for efficient label free electrochemical DNA biosensing, Sens. Actuators, B, 2018, 260, 841–851 CrossRef CAS.
- S. Bizid, S. Blili, R. Mlika, A. H. Said and H. Korri-Youssoufi, Direct E-DNA sensor of Mycobacterium tuberculosis mutant strain based on new nanocomposite transducer (Fc-ac-OMPA/MWCNTs), Talanta, 2018, 184, 475–483 CrossRef CAS PubMed.
- S. Shahrokhian and R. Salimian, Ultrasensitive detection of cancer biomarkers using conducting polymer/electrochemically reduced graphene oxide-based biosensor: Application toward BRCA1 sensing, Sens. Actuators, B, 2018, 266, 160–169 CrossRef CAS.
- W. Zhang, Z. C. Dai, X. Liu and J. M. Yang, High-performance electrochemical sensing of circulating tumor DNA in peripheral blood based on poly-xanthurenic acid functionalized MoS2 nanosheets, Biosens. Bioelectron., 2018, 105, 116–120 CrossRef CAS PubMed.
- Y. K. Ye, Y. Q. Liu, S. D. He, X. Xu, X. D. Cao, Y. W. Ye and H. S. Zheng, Ultrasensitive electrochemical DNA sensor for virulence invA gene of Salmonella using silver nanoclusters as signal probe, Sens. Actuators, B, 2018, 272, 53–59 CrossRef CAS.
- M. Chen, Y. Y. Wang, H. L. Su, L. Mao, X. N. Jiang, T. Zhang and X. Z. Dai, Three-dimensional electrochemical DNA biosensor based on 3D graphene-Ag nanoparticles for sensitive detection of CYFRA21-1 in non-small cell lung cancer, Sens. Actuators, B, 2018, 255, 2910–2918 CrossRef CAS.
- Y. Wang, Z. H. Li, T. J. Weber, D. H. Hu, C. T. Lin, J. H. Li and Y. H. Lin, In situ live cell sensing of multiple nucleotides exploiting DNA/RNA aptamers and graphene oxide nanosheets, Anal. Chem., 2013, 85, 6775–6782 CrossRef CAS PubMed.
- Y. Wang, L. H. Tang, Z. H. Li, Y. H. Lin and J. H. Li, In situ simultaneous monitoring of ATP and GTP using a graphene oxide nanosheet–based sensing platform in living cells, Nat. Protoc., 2014, 9, 1944–1955 CrossRef CAS PubMed.
- Q. Zhang, Y. Qiao, F. Hao, L. Zhang, S. Y. Wu, Y. Li, J. H. Li and X. M. Song, Fabrication of a biocompatible and conductive platform based on a single-stranded DNA/graphene nanocomposite for direct electrochemistry and electrocatalysis, Chem.–Eur. J., 2010, 16, 8133–8139 CrossRef CAS PubMed.
- L. H. Tang, H. X. Chang, Y. Liu and J. H. Li, Duplex DNA/graphene oxide biointerface: from fundamental understanding to specific enzymatic effects, Adv. Funct. Mater., 2012, 24, 3083–3088 CrossRef.
- Y. Wang, Z. H. Li, D. H. Hu, C. T. Lin, J. H. Li and Y. H. Lin, Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells, J. Am. Chem. Soc., 2010, 132, 9274–9276 CrossRef CAS PubMed.
- L. H. Tang, Y. Wang and J. H. Li, The graphene/nucleic acid nanobiointerface, Chem. Soc. Rev., 2015, 44, 6954–6980 RSC.
- H. Wang, H. B. Feng and J. H. Li, Graphene and graphene-like layered transition metal dichalcogenides in energy conversion and storage, Small, 2014, 10, 2165–2181 CrossRef CAS PubMed.
- G. Zhang, H. J. Liu, J. H. Qu and J. H. Li, Two-dimensional layered MoS2: rational design, properties and electrochemical applications, Energy Environ. Sci., 2016, 9, 1190–1209 RSC.
- G. Zhang and Y. W. Zhang, Thermoelectric properties of two-dimensional transition metal dichalcogenides, J. Mater. Chem. C, 2017, 5, 7684–7698 RSC.
- Z. H. Hu, Z. T. Wu, C. Han, J. He, Z. H. Ni and W. Chen, Two-dimensional transition metal dichalcogenides: interface and defect engineering, Chem. Soc. Rev., 2018, 47, 3100–3128 RSC.
- A. Sajedi-Moghaddam, E. Saievar-Iranizad and M. Pumera, Two-dimensional transition metal dichalcogenide/conducting polymer composites: synthesis and applications, Nanoscale, 2017, 9, 8052–8065 RSC.
- A. Q. Liang, D. Q. Li, W. Q. Zhou, Y. L. Wu, G. Ye, J. Wu, Y. N. Chang, R. Wang, J. K. Xu, G. M. Nie, J. Hou and Y. K. Du, Robust flexible WS2/PEDOT:PSS film for use in high-performance miniature supercapacitors, J. Electroanal. Chem., 2018, 824, 136–146 CrossRef CAS.
- Y. J. Chen, C. H. Kang, R. H. Wang, Z. Y. Ren, H. Y. Fu, Y. T. Xiao and G. H. Tian, CoP/WS2 nanoflake heterostructures as efficient electrocatalysts for significant improvement in hydrogen evolution activity, Appl. Surf. Sci., 2018, 442, 352–360 CrossRef CAS.
- A. Khataee, P. Eghbali, M. H. Irani-Nezhad and A. Hassani, Sonochemical synthesis of WS2 nanosheets and its application in sonocatalytic removal of organic dyes from water solution, Ultrason. Sonochem., 2018, 48, 329–339 CrossRef CAS PubMed.
- X. Yu, C. G. Pei, W. S. Chen and L. G. Feng, 2 dimensional WS2 tailored nitrogen-doped carbon nanofiber as a highly pseudocapacitive anode material for lithium-ion battery, Electrochim. Acta, 2018, 272, 119–126 CrossRef CAS.
- T. T. Xu, Y. Y. Liu, Y. Y. Pei, Y. P. Chen, Z. M. Jiang, Z. F. Shi, J. M. Xu, D. Wu, Y. T. Tian and X. J. Li, The ultra-high NO2 response of ultra-thin WS2 nanosheets synthesized by hydrothermal and calcination processes, Sens. Actuators, B, 2018, 259, 789–796 CrossRef CAS.
- P. C. Pandey, D. S. Chauhan and V. Singh, Poly(indole-6-carboxylic acid) and tetracyanoquinodimethane-modified electrode for selective oxidation of dopamine, Electrochim. Acta, 2009, 54, 2266–2270 CrossRef CAS.
- G. M. Nie, Z. M. Bai, W. Y. Yu and L. Zhang, Electrochemiluminescence biosensor for Ramos cells based on a nanostructured conducting polymer composite material (PICA-MWNTs), J. Polym. Sci., 2013, 51, 2385–2392 CrossRef CAS.
- S. Liu, L. Wang, Y. L. Luo, J. Q. Tian, H. L. Li and X. P. Sun, Polyaniline nanofibres for fluorescent nucleic acid detection, Nanoscale, 2011, 3, 967–969 RSC.
- T. Yang, Q. Guan, X. H. Guo, L. Meng, M. Du and K. Jiao, Direct and freely switchable detection of target genes engineered by reduced graphene oxide-poly(m-aminobenzenesulfonic acid) nanocomposite via synchronous pulse electrosynthesis, Anal. Chem., 2013, 85, 1358–1366 CrossRef CAS PubMed.
- X. X. Wang, F. X. Nan, J. L. Zhao, T. Yang, T. Ge and K. Jiao, A label-free ultrasensitive electrochemical DNA sensor based on thin-layer MoS2 nanosheets with high electrochemical activity, Biosens. Bioelectron., 2015, 64, 386–391 CrossRef CAS PubMed.
- Y. W. Hu, T. Yang, X. X. Wang and K. Jiao, Highly sensitive indicator-free impedance sensing of DNA hybridization based on poly(m-aminobenzenesulfonic acid)/TiO2 nanosheet membranes with pulse potentiostatic method preparation, Chem.–Eur. J., 2010, 16, 1992–1999 CrossRef CAS PubMed.
- H. Momeneh and M. B. Gholivand, Mycophenolate mofetil sensor based on molecularly imprinted polymer/multi-walled carbon nanotubes modified carbon paste electrode, Anal. Biochem., 2018, 557, 97–103 CrossRef CAS PubMed.
- G. M. Nie, Y. Zhang, Q. F. Guo and S. S. Zhang, Label-free DNA detection based on a novel nanostructured conducting poly(indole-6-carboxylic acid) films, Sens. Actuators, B, 2009, 139, 592–597 CrossRef CAS.
- G. M. Nie, Z. M. Bai, J. Chen and W. Y. Yu, Simple label-free femtomolar DNA detection based on a nanostructure composite material: MWNT-doped poly(indole-6-carboxylic acid), ACS Macro Lett., 2012, 1, 1304–1307 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra10266j |
| ‡ These two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |