Open Access Article
Open Access ArticleA series of Mn(I) photo-activated carbon monoxide-releasing molecules with benzimidazole coligands: synthesis, structural characterization, CO releasing properties and biological activity evaluation†
Mixia Hu abc,
YaLi Yand,
Baohua Zhu*ab,
Fei Changa,
Shiyong Yu
abc,
YaLi Yand,
Baohua Zhu*ab,
Fei Changa,
Shiyong Yu a and
Gaole Alatand
a and
Gaole Alatand
aCollege of Chemistry and Chemical Engineering, Inner Mongolia University, Hohhot 010021, China. E-mail: bhzhu@imu.edu.cn
bKey Lab of Fine Organic Synthesis Inner Mongolia Autonomous Region, Hohhot 010021, China
cCollege of Pharmacy, Inner Mongolia Medical University, Hohhot 010110, China
dSchool of Life Sciences, Inner Mongolia University, Hohhot 010021, China
First published on 2nd July 2019
Abstract
Five Mn(I) photo-activated carbon monoxide-releasing molecules (photo-CORMs) with benzimidazole coligands, namely [MnBr(CO)3L1] (1, L1 = 2-(2-pyridyl)benzimidazole), [Mn(CO)2L1(PPh3)2](ClO4) (2), [MnBr(CO)3L2] (3, L2 = 2,2′-bisbenzimidazole), [MnBr(CO)3L3]·CH3OH (4, L3 = 2,6-bis(benzimidazole-2′-yl)pyridine) and fac-[MnBr(CO)3L4] (5, L4 = 2,4-bis(benzimidazole-2′-yl) pyridine) were synthesized by reactions of MnBr(CO)5 with complexes L1–L4, respectively, and characterized via single crystal X-ray diffraction, elemental analysis, 1H-NMR, 13C-NMR, IR, UV-vis and fluorescence spectroscopy. The CO-release properties of 1–5 were investigated using the myoglobin assay and CO detection, and the results show that all of the complexes could release CO rapidly upon exposure to 365 nm UV light. Comparing their half-lives of CO release, we found that increasing the degree of unsaturation and conjugation of the ligand frame could be advantageous for prolonging the time of CO-release, and that the luminescence intensity of 1–5 could gradually be enhanced. The cellular fluorescence imaging tests demonstrate that these Mn(I) photo-CORMs can be taken up by human liver cells (HL-7702) and liver cancer cells (SK-Hep1), and exhibit good capabilities for bioimaging. A cell viability assay for SK-Hep1 shows that the anticancer activity of 3 is better than that of other complexes.
Introduction
Many studies have recently shown that carbon monoxide (CO), as one of the key signaling molecules in mammalian physiology and pathology,1 can induce aortic dilatation, prevent lung inflammation, inhibit cell proliferation, and help oxidatively damaged tissues to recover.2–6 However, it is also best known that CO is a highly toxic gas because of its high affinity toward haemoglobin, affecting oxygen transport,7,8 which restricts its practicability as an inhaled gas for therapeutic application. Therefore, CO-releasing molecules (CORMs) were designed and developed by R. Motterlini and co-workers in order to present an alternative way to deliver controlled amounts CO to cells and target tissues.3,7 The release of CO from CORMs can be triggered by light,9–13 ligand exchange,14–16 pH change,17 and solvent.18 In particular, photo-activated CORMs seem to be advantageous to deliver CO with well-defined control of the targeted sites, timing of activation and dosage of CO. Up to now, a number of different molecules have been reported as photo-CORMs, which can release CO under the control of light in the UV and visible range.9,19 Among these complexes, CORMs including manganese, as one of the essential elements in the life cycle of many life forms, were paid more attention due to their good biocompatibility compared to other transition metals.Recently, a few photo-CORMs based on the tripodal ligands tris(pyrazolyl)methane and tris(2-pyridylmethyl)amine κ3N-bound to fac-Mn(CO)3 moiety have been investigated by Schatzschneider and co-workers.20–22 Mascharak and co-workers have also reported a series of photoactive manganese carbonyl complexes, the apparent CO release rates of which increase steadily with varying degrees of conjugation in the ligand frame, thus generating photo-CORMs more sensitive to light in the more desirable visible range.23,24 The κ2N1, N2 Mn(I) tricarbonyl photo-CORMs with tridentate benzimidazole coligands were reported by Mansour group and the influence of the alkylation of benzimidazole ligand on the electronic structure was studied.25 As we know, in order to assess the location and extent of CO release in vivo, a crucial challenge is the appropriate design of photo-CORMs with suitable tracking of the CO donors within cellular matrixes. To date, close scrutiny of the literature reveals that there have been a very limited number of reports on luminescent Mn-based photo-CORMs,26,27 which can be tracked within the biological targets due to their inherent luminescent properties.
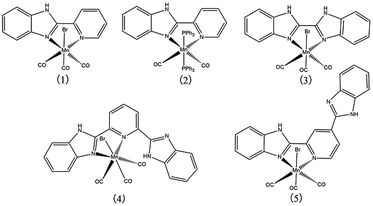
Furthermore, mebendazole (MBZ) which consists of a benzimidazole (BIM) ring in its structural framework, has been shown to possess anticancer properties in a broad range of preclinical studies across various cancer types.28–30 In these studies, MBZ's anticancer efficacy is thought to inhibit the polymerization of tubulins and normal spindle formation in cancer cells, resulting in mitotic arrest and apoptosis.31 With these in mind, we synthesized and characterized five photo-CORMs derived from Mn(I) and benzimidazole coligands, namely, [MnBr(CO)3L1] (1, L1 = 2-(2-pyridyl)benzimidazole), [Mn(CO)2L1(PPh3)2](ClO4) (2), [MnBr(CO)3L2] (3, L2 = 2,2′-bisbenzimidazole), [MnBr(CO)3L3]·CH3OH (4, L3 = 2,6-bis(benzimidazole-2′-yl)pyridine) and fac-[MnBr(CO)3L4] (5, L4 = 2,4-bis(benzimidazole-2′-yl)pyridine) (Fig. 1) for investigating the effect of varying ligands on the CO releasing properties, biological activities and the luminescence properties with the increasing of the rigidity and conjugate degree of ligands.
Experimental
Materials and reagents
In this work, all the starting analytical grade reagents and solvents were acquired from commercial sources and were used as received without further purification. The ligands 2,2′-bisbenzimidazole (L2),32 2,6-bis(benzimidazole-2′-yl)pyridine (L3)33 and 2,5-bis(benzimidazole-2′-yl)pyridine (L4)33 were synthesized according to published procedures. Human liver cell (HL-7702) and liver cancer cell (SK-Hep1) cells were purchased from ATCC, cultured in a 5% CO2, incubated at 37 °C, and grown in DMEM supplemented with 10% FBS and penicillin/streptomycin.Synthesis of [MnBr(CO)3L1] (1)
[MnBr(CO)5] (0.1 mmol) and 2-(2-pyridyl)benzimidazole (0.1 mmol) were dissolved in 25 mL dichloromethane under argon. The mixture was heated to reflux for 4 h in the absence of light, and an abundant yellow precipitate formed. The precipitate was filtered off, rinsed with a small amount of dichloromethane (1 mL each) (76% yield). Large block crystals suitable for X-ray analysis were readily formed by slow evaporation of a mixed CH2Cl2/CH3CH2OH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v
1 v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) solution of the complex. Anal. calcd for C15H9Br1Mn1N3O3: C, 43.51; H, 2.19; N, 10.15. Found: C, 44.23; H, 2.11, N, 10.47. FTIR (KBr pellets): νCO = 2028 and 1940 cm−1. 1H NMR (500 MHz in DMSO-d6) δ (ppm) = 7.56 (t, BIM-H, 2H), 7.77 (d, BIM-H,1H), 7.84 (d, BIM-H, 1H), 8.03 (t, Py-H, 1H), 8.31 (t, Py-H, 1H), 8.44 (d, Py-H, 1H), 9.26 (d, Py-H, 1H), 14.63 (s, N–H, 1H). 13C-NMR (DMSO-d6) δ (ppm) = 221.49 (C
v) solution of the complex. Anal. calcd for C15H9Br1Mn1N3O3: C, 43.51; H, 2.19; N, 10.15. Found: C, 44.23; H, 2.11, N, 10.47. FTIR (KBr pellets): νCO = 2028 and 1940 cm−1. 1H NMR (500 MHz in DMSO-d6) δ (ppm) = 7.56 (t, BIM-H, 2H), 7.77 (d, BIM-H,1H), 7.84 (d, BIM-H, 1H), 8.03 (t, Py-H, 1H), 8.31 (t, Py-H, 1H), 8.44 (d, Py-H, 1H), 9.26 (d, Py-H, 1H), 14.63 (s, N–H, 1H). 13C-NMR (DMSO-d6) δ (ppm) = 221.49 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), δ (ppm) = 221.98 (C
O), δ (ppm) = 221.98 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), δ (ppm) = 224.95 (C
O), δ (ppm) = 224.95 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O).
O).
Synthesis of [Mn(CO)2L1(PPh3)2](ClO4) (2)
This complex was synthesized by the reaction of complex 1 and AgClO4 in equimolar ratio in tetrahydrofuran (THF) and the resulting AgBr was filtered. Then, one equivalent of PPh3 in dichloromethane was added to the filtrate and the mixture in the absence of light was stirred at room temperature for 36 h.19 The solvent was removed under reduced pressure to give a viscous oil. Next, CH3CH2OH was added to the viscous oil and large block crystals suitable for X-ray analysis were readily formed by slow evaporation of CH3CH2OH solution (70% yield). Anal. calcd for C50H39Cl1Mn1N3O6P2: C, 64.56; H, 4.23; N, 4.52. Found: C, 64.92; H, 3.96, N, 5.11. FTIR (KBr pellets): νCO = 1926 and 1855 cm−1. 1H NMR (500 MHz in DMSO-d6) δ (ppm) = 7.03 (t, Ph-H, 14H), 7.61 (t, Ph-H, 16H, BIM-H, 5H), 8.08 (t, Py-H, 1H), 8.15 (d, BIM-H, 1H), 8.34 (d, Py-H, 1H), 8.87 (s, N–H, 1H). 13C-NMR (DMSO-d6) δ (ppm) = 222.10 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), δ (ppm) = 225.11 (C
O), δ (ppm) = 225.11 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O).
O).
Synthesis of [MnBr(CO)3L2] (3)
A mixture of 2,2′-bisbenzimidazole (0.1 mmol) and [MnBr(CO)5] (0.2 mmol) in a mixed CH2Cl2/CH3CH2OH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution was sealed in a 25 mL Teflon-lined stainless steel container and heated at 85 °C for 3 days before gently cooled to room temperature. The yellow flake crystals of 3 were readily formed by slow evaporation of the solution (70% yield). Anal. calcd for C17H10Br1Mn1N4O3: C, 45.06; H, 2.22; N, 12.36. Found: C, 45.62; H, 2.13; N, 13.01. FTIR (KBr pellets): νCO = 2024, 1932 and 1913 cm−1 (see the ESI† for details). NMR (500 MHz in DMSO-d6) δ (ppm) = 7.55–8.11 (BIM-H, 8H), 13.92 (s, N–H, 2H). 13C-NMR (DMSO-d6) δ (ppm) = 221.18 (C
1) solution was sealed in a 25 mL Teflon-lined stainless steel container and heated at 85 °C for 3 days before gently cooled to room temperature. The yellow flake crystals of 3 were readily formed by slow evaporation of the solution (70% yield). Anal. calcd for C17H10Br1Mn1N4O3: C, 45.06; H, 2.22; N, 12.36. Found: C, 45.62; H, 2.13; N, 13.01. FTIR (KBr pellets): νCO = 2024, 1932 and 1913 cm−1 (see the ESI† for details). NMR (500 MHz in DMSO-d6) δ (ppm) = 7.55–8.11 (BIM-H, 8H), 13.92 (s, N–H, 2H). 13C-NMR (DMSO-d6) δ (ppm) = 221.18 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), δ (ppm) = 221.88 (C
O), δ (ppm) = 221.88 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), δ (ppm) = 224.89 (C
O), δ (ppm) = 224.89 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O).
O).
Synthesis of [MnBr(CO)3L3]·CH3OH (4)
[MnBr(CO)5] (0.1 mmol) and 2,6-bis(benzimidazole-2′-yl)pyridine (0.1 mmol) were dissolved in 30 mL methanol under argon. The mixture was then heated to reflux for 3 h in the absence of light, and got the yellow solution. The yellow flake crystals of 4 were readily formed by slow evaporation of the solution (80% yield). Anal. calcd for C23H17Br1Mn1N5O4: C, 49.13; H, 3.05; N, 12.46. Found: C, 50.02; H, 2.21; N, 13.82. FTIR (KBr pellets): νCO = 2024 and 1920 cm−1. 1H NMR (500 MHz in DMSO-d6) δ (ppm) = 7.36 (t, BIM-H, 4H), 7.78 (d, BIM-H, 4H), 8.30 (t, Py-H, 3H), 13.1 (s, N–H, 2H). 13C-NMR (DMSO-d6) δ (ppm) = 220.50 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), δ (ppm) = 221.58 (C
O), δ (ppm) = 221.58 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), δ (ppm) = 226.32 (C
O), δ (ppm) = 226.32 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O).
O).
Synthesis of [MnBr(CO)3L4] (5)
[MnBr(CO)5] (0.1 mmol) and 2,4-bis(benzimidazole-2′-yl)pyridine (0.1 mmol) were dissolved in a mixed 30 mL methanol/acetone (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution under argon. The mixture was then heated to reflux for 3 h in the absence of light, and an abundant yellow precipitate of complex 5 was formed. Then, the precipitate was filtered off, rinsed with a small amount of methanol (1 mL each) (72% yield). Anal. calcd for C22H13Br1Mn1N5O3: C, 49.84; H, 2.47; N, 13.21. Found: C, 49.92; H, 2.33; N, 13.34. FTIR (KBr pellets): νCO = 2024 and 1932 cm−1. 1H NMR (500 MHz in DMSO-d6) δ (ppm) = 7.24 (t, BIM-H, 4H), 7.69 (d, BIM-H, 2H), 7.85 (d, BIM-H, 2H), 8.28 (t, Py-H, 1H), 9.05 (d, Py-H, 1H), 9.19 (d, Py-H, 1H), 13.70 (s, N–H, 1H). 13C-NMR (DMSO-d6) δ (ppm) = 218.76 (C
1) solution under argon. The mixture was then heated to reflux for 3 h in the absence of light, and an abundant yellow precipitate of complex 5 was formed. Then, the precipitate was filtered off, rinsed with a small amount of methanol (1 mL each) (72% yield). Anal. calcd for C22H13Br1Mn1N5O3: C, 49.84; H, 2.47; N, 13.21. Found: C, 49.92; H, 2.33; N, 13.34. FTIR (KBr pellets): νCO = 2024 and 1932 cm−1. 1H NMR (500 MHz in DMSO-d6) δ (ppm) = 7.24 (t, BIM-H, 4H), 7.69 (d, BIM-H, 2H), 7.85 (d, BIM-H, 2H), 8.28 (t, Py-H, 1H), 9.05 (d, Py-H, 1H), 9.19 (d, Py-H, 1H), 13.70 (s, N–H, 1H). 13C-NMR (DMSO-d6) δ (ppm) = 218.76 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), δ (ppm) = 222.43 (C
O), δ (ppm) = 222.43 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), δ (ppm) = 223.43 (C
O), δ (ppm) = 223.43 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O).
O).
Physical measurements
FT-IR spectra were recorded in the range of 400–4000 cm−1 on a Bruker Vector 22 FT-IR spectrophotometer using KBr pellet. UV-vis spectra were obtained with a Varian Cary 50 UV-vis spectrophotometer. The 1H NMR and 13C NMR spectra were recorded a Bruker Avance DPX-400 MHz spectrometer. Elemental analyses (C, H, and N) were determined with a PerkinElmer 2400 Series II elemental analyzer. CO measurement in ppm was recorded using a single gas detector (HD5S+) with an error less than 3%. The luminescence spectra were measured on an Aminco Bowman Series 2 Spectrofluorometer with a xenon arc lamp as the light source. A laser scanning confocal microscope (OLYMPUS, IX81) was used to confirm the cell targeting process and cell imaging capacity.Single crystal X-ray diffraction
Single crystal X-ray data on complexes 1–4 were collected at 293 K on a Bruker Smart Apex CCD area-detector diffractometer with graphite-monochromated Mo Kα radiation (λ = 0.71073 Å). The structures were solved by direct methods using SHELXTL and non-hydrogen atoms were refined on F2 with SHELX97. Details of the data collection and refinement parameters of complexes 1–4 are given in Table S1,† and selected distances and bond angles were listed in Table S2 in the ESI.†Myoglobin assay
The release of CO from the metal carbonyl complexes was studied spectrophotometrically by measuring the conversion of deoxy-myoglobin (deoxy-Mb) to carbonyl myoglobin (Mb-CO). First, 10.2 mg horse skeletal muscle myoglobin was dissolved in 10 mL phosphate buffer (PBS, 0.1 M, pH 7.4) and reduced by adding sodium dithionite (0.1%). 40 μM complexes 1–5 dissolved in DMSO were added to 3 mL deoxy-Mb in a 1 cm × 1 cm quartz cuvette. They were overlaid with 500 μL light mineral oil to prevent CO from escaping or the myoglobin from being oxygenated, and were irradiated with a 365 nm UV lamp in 2 min intervals. The extent of conversion of Mb to carboxymyoglobin (MbCO) was monitored at regular intervals.CO measurement by using a CO detector
Complexes 1–5 dissolved in DMSO at a concentration of 2.0 mg mL−1 were incubated in a sealed vessel with 1 mL gas space. The liberated CO gas of 1–5 in ppm was measured using a CO gas detector with different times irradiated with a 365 nm UV lamp. The number of mg was calculated from the ppm number based on 1 mL volume.Cellular uptake study
The in vitro targeting ability of the complexes 1–5 were evaluated by luminescence imaging in human liver cell (HL-7702) and liver cancer cell (SK-Hep1). The cells were cultured onto a 6-well plate for 24 h and afterwards treated with 25 μM of complexes 1–5 allowing to incubate at 37 °C with 5% CO2 (g) for 1 h. Before being observed using a laser confocal microscope (OLYMPUS, IX81) under an excitation wavelength of 473 nm, the HL-7702 and SK-Hep1 cells were washed several times with phosphate-buffered saline (PBS) buffer.MTT assay
Liver cancer cells (SK-Hep1) were used to evaluate the in vitro cytotoxicities and antitumor efficiencies of complexes 1–5 by MTT assay. First of all, SK-Hep1 were plated in 96-well plates (104 cells per well) and incubated for 24 h. The DMEM (growth medium) were removed from the wells and replaced with fresh DMEM containing the complexes 1–5 with different concentrations (15, 30, 45, 60, 75, and 90 μmol L−1), respectively. Next, the plates were incubated completely under a low power LED-light source for about 40 min and subsequently incubated for an additional 20 min. Next, all DMEM were removed from each well before fresh DMEM were added, and the cells were allowed to recover for 4 h. At last, the cell viability was assessed as a percentage of the absorbance value of the sample well compared to that of the control cell. All experiments were executed in triplicate, and the results were averaged.Results and discussion
Molecular structures
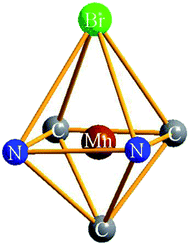
The molecular structures for complexes 1–4 have been authenticated by single-crystal X-ray crystallography (Fig. 2). Unfortunately, crystals suitable for X-ray analysis of complex 5 have not been obtained. The Mn(I) center in complexes 1–4 resides in a distorted octahedral coordination sphere, three CO ligands of which resides in a facial disposition except for complex 2 (Fig. 3). The two N atoms of the ligands and two C atoms of CO constitute the equatorial planes, while one CO and the Br atom occupy the two axial positions. In complex 2, the two N atoms of the L1 and two C atoms from CO constitute the equatorial plane while two PPh3 occupy the axial positions. There is a distinct difference between the two Mn–N bond lengths: the Mn(1)–N(1) (pyridyl) bond length (2.093(3) Å) is much longer than that of Mn(1)–N(3) (imidazolyl, 2.032(2) Å) in complex 1, indicating that the imidazolyl nitrogen atom is a stronger electron donor than that of the pyridyl ring. The same trend was also observed for the structures of complexes 2 and 4 (see Table S2†). The Mn–C distance in complex 1 (1.790(4), 1.801(4) and 1.808(4) Å) is noticeably longer than the corresponding Mn–C distances in complex 2 (1.7775(16) and 1.7895(16) Å), which maybe resulted in rapid CO release as evident by reduced myoglobin (Mb) assay. In the synthesis of complex 4, when methanol mixed with a little water and the heating temperature with oil bath is higher than 100 °C, CO was released from MnBr(CO)5 to form the complex composed of the Mn and L3 (Fig. S5†). The octahedral coordination sphere of Mn center is more distorted than others (Fig. S6†).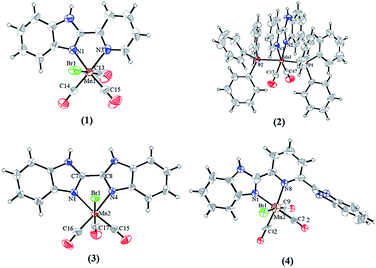 | ||
| Fig. 2 Molecular structures of photo-CORMs 1–4, shown with displacement ellipsoids at 50% probability level. The ClO4− of 2 and the free CH3OH molecules of 4 are omitted for clarity. | ||
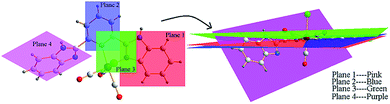
In complexes 1–2 and 4, the benzimidazole, pyridyl ring and the five-membered ring formed by Mn and ligand (MnNNCC) are almost in the same plane. The dihedral angle of plane 1 (the benzimidazole) and plane 2 (pyridyl ring) is 1.13° for complex 1, 1.81° for complex 2, 1.54° for complex 4, and the dihedral angle of plane 1 and plane 3 (the five-membered ring MnNNCC) is 1.78° for complex 1, 0.77° for complex 2, 8.55° for complex 4 (Fig. 4, and S7†). Due to the influence of steric hindrance, the dihedral angle of plane 4 (uncoordinated benzimidazole) and plane 1 (55.92°) is larger than others in complex 4. For complex 3, the dihedral angles of two benzimidazole rings, the benzimidazole ring and the MnNNCC ring are 5.69° and 4.99°, respectively. These differences in degree of planar conjugation may be the one of main reasons to cause the changes in luminescence intensity of complexes 1–5.
Spectroscopic properties
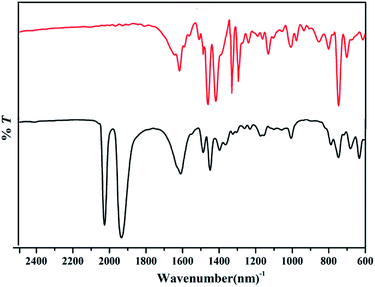
The elemental analysis for complex 4 (calcd for C23H17Br1Mn1N5O4: C, 49.13; H, 3.05; N, 12.46. Found: C, 50.02; H, 2.21; N, 13.82) was probably done on a desolvated sample, so the result is almost identical to complex 4 without CH3OH. The IR spectra of complexes 1–5 exhibit two strong bands in the carbonyl region (Fig. 7 and Fig. S1†). The CO stretching vibrations are observed at 2028, 1940 cm−1 for 1, at 1926, 1855 cm−1 for 2, at 2024, 1932, 1913 cm−1 for 3, at 2024, 1920 cm−1 for 4, as well as at 2024, 1932 cm−1 for 5. A minor peak splitting of a broad νCO stretch has been observed in complex 3, which suggests the distinction between the two CO ligands bound in the equatorial plane. The higher symmetry of the 1, 4 and 5 is reflected in two sharp νCO bands.24 For complex 2, a strong stretch around 1100 cm−1 was assigned to ClO4− vibration, and the 1102 cm−1, 1049 cm−1, 1009 cm−1 were assigned to PPh3 vibration. UV-vis absorption spectra of 1–5 in DMSO at 298 K show similar spectral features. Two absorption bands with λmax ≈ 260 and 330 nm were observed for 1–5, which are assigned to π–π* intra-ligand (IL) transitions. The less intense and lower-energy absorption at about 420 nm is assigned to a metal-to-ligand charge-transfer (MLCT) dπ(Mn)–π*(ligand) (Fig. S2†).34 In the 1H NMR spectra of complexes 1–5 in DMSO-d6 (Fig. S3†), the chemical shifts of the imino protons were observed at 13.1–14.7 ppm as a singlet peak. The pyridine ring H signals appeared at 8.03–9.26 ppm and the BIM proton signals appeared at 7.24–8.15 ppm, which are corresponding with those of the complexes reported in literatures.35,36 All complexes are diamagnetic as evidenced by clean 1H NMR spectra in solvents, which ensures that only Mn(I) is present. The signals observed for 1–5 in the 13C-NMR (DMSO-d6) spectra as follow: δ (ppm) = 221.49, 221.98, 224.95 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O) for 1; δ (ppm) = 222.10, 225.11 (C
O) for 1; δ (ppm) = 222.10, 225.11 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O) for 2; δ (ppm) = 221.18, 221.88, 224.89 (C
O) for 2; δ (ppm) = 221.18, 221.88, 224.89 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O) for 3; δ (ppm) = 220.50, 221.58, 226.32 (C
O) for 3; δ (ppm) = 220.50, 221.58, 226.32 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O) for 4, δ (ppm) = 218.76, 222.43, 223.43 (C
O) for 4, δ (ppm) = 218.76, 222.43, 223.43 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O) for 5 suggest the CO ligands in a fac-arrangement (Fig. S4†).
O) for 5 suggest the CO ligands in a fac-arrangement (Fig. S4†).
CO release measurement via myoglobin assay
All the complexes are stable in air as long as they are stored in the dark and soluble in common organic solvents, DMSO, MeCN, THF, and so on, but they are almost insoluble in water. Stability of 1–5 in DMSO was checked by recording the absorption spectrum periodically for a 24 h period. When the solution was kept in the dark, no significant change was observed (Fig. S2†). Such solutions are capable of releasing CO, when they are exposed to low power UV light. The CO release from complexes 1–5 was studied using the standard myoglobin assay, which is based on the UV/vis spectroscopic detection of the conversion of deoxy-Mb to Mb–CO (Mb = myoglobin). In the range of 500–600 nm, the maximal absorption peak of deoxy-Mb is observed at 560 nm while the two maximal absorption peaks of Mb–CO appear at 540 and 578 nm. The neat DMSO solutions of all five complexes were transferred into a cuvette containing 60 μM deoxy-Mb in phosphate buffer (pH 7.4). In the absence of light, no spectral changes were observed over 1 h, indicating that complexes 1–5 do not release CO when kept in the dark. Exposure to 365 nm UV light at regular intervals, the intensity of two new bands at 540 and 577 nm increases, while the intensity of the Mb Q-band at 560 nm slowly decreases (Fig. 5). The half-lives (t1/2, min) were listed in Table 1 and Fig. S8.†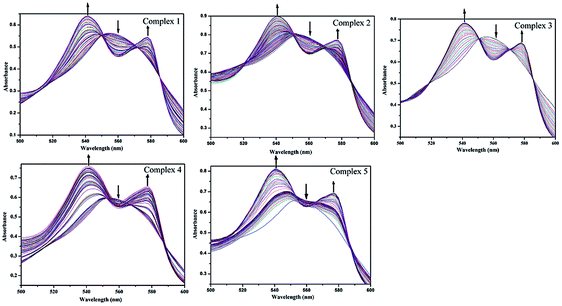 | ||
| Fig. 5 Change of absorption of myoglobin with the irradiation at 365 nm UV light for a solution of complexes 1–5 (40 μM in 0.1 PBS at pH 7.4) with sodium dithionite (10 mM) by UV/vis spectroscopy. | ||
| Complex | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| t1/2 of CO released (min) | 5 | 13 | 6 | 15 | 10 |
| Quantum yield values | 0.36 | 0.10 | 0.28 | 0.12 | 0.16 |
The half-lives of CO release were 5 min for 1, and 13 min for 2. It is noteworthy that complex 2 through substitution of the bromide and a CO ligand of 1 with a good π-acceptor ligand PPh3 was synthesized, as a result, complex 2 exhibits a relatively slower CO release rate than that of complex 1.37 Comparison CO release rates of complexes 1, 3 and 4, it can be found that the half-lives of CO release increases from 5 min of complex 1 to 15 min of complex 4, with the increasing of the degree of unsaturation and conjugation in their ligand frame. The difference in rates between 4 and 5 may be attributed to differences in the steric profile of the benzimidazolyl unit to the resulting impact upon the substitution of CO by solvent molecules under UV light irradiation. Therefore, we can adjust the rate of CO-release with a proper control of the structural parameters and functionalization of the ligands.
The half-lives of CO release for complexes 1–5 is slower than that observed in the three Mn(I) carbonyl complexes [Mn(tpa)(CO)3]ClO4, [Mn(dpa)(CO)3]Br and [Mn(pqa)(CO)3]ClO4, as reported by Mascharak and co-workers (t1/2 in the range of 1.4–2.2 min).24 Therefore, complexes 1–5 show an excellent capacity of CO delivery as photo-CORMs for biological studies under illumination in PBS. The quantum yield values measured with potassium ferrioxalate as the actinometer for 1–5 are shown in Table 1.23
CO release measurement by using a CO detector
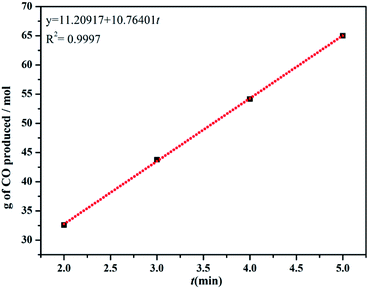
Although the myoglobin assay confirm the properties of releasing CO for complexes 1–5, the dithionite used in the assay for deoxygenating myoglobin may interferes with the CO releasing.26 In order to overcome this problem from interferences, commercial CO detector was used to follow the CO release.During the test time, the production of carbon monoxide is relatively linear, and the rates of CO produced (kCO) for 1–5 are shown in Table 2. The present results suggest that the rates of CO produced for complex 1 is the fastest and for complex 4 is the slowest, which is consistent with the myoglobin assay (Fig. 6 and S9†).
| Complex | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Apparent rate g (mol−1 min−1) | 10.8 | 0.8 | 8.0 | 1.9 | 3.4 |
The MeCN solution of complexes 1–5 were taken in a glass Petri dish and exposed to low power UV light for 40 min. Then, the photolyzed solutions were dried under vacuum. The results of IR spectra for the solid clearly indicated the loss of all CO molecules from 1–5 upon UV light illumination (Fig. 7 and S1†).28,38
The luminescence properties
At ambient temperature, the luminescence spectra of ligands L1–L4 show emission bands in the range of 350–600 nm in DMSO solution (λmax values: 365 nm (L1), 404 nm (L2), 378 nm (L3), 480 nm (L4)), which are assigned to intraligand π*–π transition39 (Fig. S10†). The luminescence spectra of the solutions of complexes 1–5 (5 μmol) in DMSO (3 mL) were measured at room temperature and green luminescence were observed for all complexes. As shown in Fig. 8, the fluorescent emissions bands of complexes 1–5 are red-shifted to around 565–575 nm by comparison with the emission spectra of free ligands L1–L4, because of the metal-to-ligand charge-transfer transition (MLCT) in these complexes.40 As a result, complex 4 exhibits significantly higher emission intensity compared to the other complexes, which makes it possible to serve as attractive candidates for optical imaging.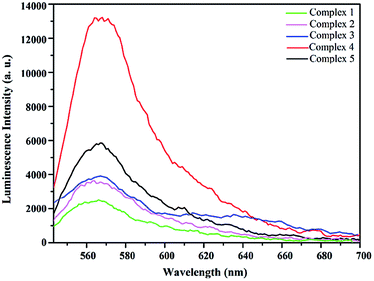 | ||
| Fig. 8 The luminescence emission spectra of complexes 1–5 in DMSO at room temperature (λex ≈ 450 nm). | ||
Live cell imaging and cell viability assay
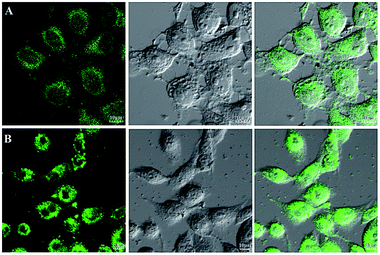
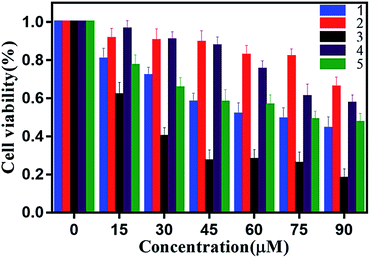
The emission bands of complexes 1–5 extend to the visible-light region, suggesting that the luminescent properties can already fulfill the requirements of bioimaging studies. In order to demonstrate the capability of the complexes 1–5 for bioimaging, we carried out fluorescence imaging tests using HL-7702 cells and SK-Hep1 cells as shown in Fig. 9, and S11.† When HL-7702 cells and SK-Hep1 cells are incubated with all five complexes, bright green fluorescence is observed in the cell. All the images indicated considerable extent of cellular internalization and verified the luminescence imaging capability of all complexes in living cell systems. Complex 4 with significantly higher emission intensity displays great potential for bio-applications.Because of the facile cellular uptake of these complexes by the cancer cells prompted us to evaluate the potential of CO induced eradication of SK-Hep1 cells. The viability of the cells was finally assessed by MTT assay. As shown in Fig. 10, with the increase of complex concentration, anticancer activity against SK-Hep1 cells intensifies. Complex 3 shows excellent anticancer activity possibly due to high liposolubility and good biocompatibility of benzimidazolyl unit and the suitable molecular size than complexes 1, 4 and 5. Because of the low solubility, complex 2 exhibits weaker anticancer activity than other complexes.
Conclusions
In summary, five luminescent Mn(I) carbonyl complexes with benzimidazole coligands have been synthesized and characterized. These complexes have a good stability in the dark and exhibit moderate CO release upon illumination with 365 nm UV light after being dissolved in a minimum amount of DMSO. The rate of CO-release can be adjusted by extending the degree of unsaturation and conjugation of the ligand frame. Complexes 1–5 show interesting luminescence properties, with the increasing of the rigidity and conjugate degree of ligands, the luminescence intensity gradually enhance and the maximum emission wavelength shift to the green region (565–575 nm). Fluorescence imaging tests show that the complexes display efficient cellular uptake in HL-7702 cells and SK-Hep1 cells and thus verify the fluorescence imaging capability of the complex in living cell systems. The viability cell assay shows that complex 3 possesses excellent anticancer activity.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We acknowledge the financial support from the National Nature Science Foundation of China (No. 20561002, 21261012), Natural Science Foundation of Inner Mongolia Autonomous Region (No. 2017MS0207) and Grassland Talent Project of Inner Mongolia Autonomous Region (No. CYYC20130042).References
- H. P. Kim, S. W. Ryter and A. M. Choi, Annu. Rev. Pharmacol. Toxicol., 2006, 46, 411s–419 CrossRef PubMed.
- M. L. Wu, Y. C. Ho and S. F. Yet, Antioxid. Redox Signaling, 2011, 15, 1835–1846 CrossRef CAS PubMed.
- A. B. Stein, Y. Guo, W. Tan, W. J. Wu, X. Zhu, Q. Li, C. Luo, B. Dawn, T. R. Johnson, R. Motterlini and R. Bolli, J. Mol. Cell. Cardiol., 2005, 38, 127–134 CrossRef CAS PubMed.
- S. Faller and A. Hoetzel, Curr. Pharm. Biotechnol., 2012, 13, 777–786 CAS.
- F. Gullotta, A. Di Masi and P. Ascenzi, IUBMB Life, 2012, 64(5), 378–386 CrossRef CAS PubMed.
- L. Rochette, Y. Cottin, M. Zeller and C. Vergely, Pharmacol. Ther., 2013, 137, 133–152 CrossRef CAS PubMed.
- T. R. Johnson, B. E. Mann, J. E. Clark, R. Foresti, C. J. Green and R. Motterlini, Angew. Chem., Int. Ed., 2003, 42, 3722–3729 CrossRef CAS PubMed.
- S. W. Ryter and L. E. Otterbein, BioEssays, 2004, 26, 270–280 CrossRef CAS PubMed.
- J. Niesel, A. Pinto, H. W. Peindy, K. N′Dongo, I. Merz, I. Ott, R. Gust and U. Schatzschneider, Chem. Commun., 2008, 1798 RSC.
- A. E. Pierri, A. Pallaoro, G. Wu and P. C. Ford, J. Am. Chem. Soc., 2012, 134, 18197–18200 CrossRef CAS PubMed.
- S. J. Carrington, I. Chakraborty, J. M. L. Bernard and P. K. Mascharak, ACS Med. Chem. Lett., 2014, 5, 1324–1328 CrossRef CAS PubMed.
- U. R. Gandra, J. Axthelm, P. Hoffmann and N. Taye, J. Am. Chem. Soc., 2017, 139, 4991–4994 CrossRef PubMed.
- A. M. Mansour and A. Friedrich, Polyhedron, 2017, 131, 13–21 CrossRef CAS.
- L. Long, X. J. Jiang, X. Wang, Z. Y. Xiao and X. M. Liu, Dalton Trans., 2013, 42, 15663–15669 RSC.
- L. M. Chen, X. J. Jiang, X. Wang, L. Long, J. Y. Zhang and X. M. Liu, New J. Chem., 2014, 38, 5957–5963 RSC.
- C. J. Gao, X. H. Liang, Z. X. Guo, B. P. Jiang, X. M. Liu and X. C. Shen, ACS Omega, 2018, 3, 2683–2689 CrossRef CAS PubMed.
- F. Zobi, A. Degonda, M. C. Schaub and A. Y. Bogdanova, Inorg. Chem., 2010, 49, 7313–7322 CrossRef CAS PubMed.
- V. Yempally, S. J. Kyran, R. K. Raju, W. Y. Fan, E. N. Brothers, D. J. Darensbourg and A. A. Bengali, Inorg. Chem., 2014, 53, 4081–4088 CrossRef CAS PubMed.
- S. J. Carrington, I. Chakraborty and P. K. Mascharak, Chem. Commun., 2013, 49, 11254–11256 RSC.
- H. Pfeiffer, A. Rojas, J. Niesel and U. Schatzschneider, Dalton Trans., 2009, 4292–4298 RSC.
- C. Nagel, S. McLean, R. K. Poole, H. Braunschweig, T. Kramera and U. Schatzschneider, Dalton Trans., 2014, 43, 9986–9997 RSC.
- M. T. Trejo, N. Rana, C. Nagel, H. E. Jesse, T. W. Smith, L. K. Wareham, M. Hippler, U. Schatzschneider and R. K. Poole, Antioxid. Redox Signaling, 2016, 24, 765–780 CrossRef PubMed.
- M. A. Gonzalez, S. J. Carrington, N. L. Fry, J. L. Martinez and P. K. Mascharak, Inorg. Chem., 2012, 51, 11930–11940 CrossRef CAS PubMed.
- M. A. Gonzalez, M. A. Yim, S. Cheng, A. Moyes, A. J. Hobbs and P. K. Mascharak, Inorg. Chem., 2012, 51, 601–608 CrossRef CAS PubMed.
- A. M. Mansour and A. Friedrich, Inorg. Chem. Front., 2017, 4, 1517–1524 RSC.
- S. J. Carrington, I. Chakraborty, J. M. L. Bernard and P. K. Mascharak, ACS Med. Chem. Lett., 2014, 5, 1324–1328 CrossRef CAS PubMed.
- J. Jimenez, I. Chakraborty, A. Dominguez, J. Martinez-Gonzalez, W. M. Chamil Sameera and P. K. Mascharak, Inorg. Chem., 2018, 57, 1766–1773 CrossRef CAS PubMed.
- T. Mukhopadhyay, J. Sasaki, R. Ramesh and J. A. Roth, Mebendazole elicits a potent antitumor effect on human cancer cell lines both in vitro and in vivo, Clin. Cancer Res., 2002, 8, 2963–2969 CAS.
- D. Martarelli, P. Pompei, C. Baldi and G. Mazzoni, Cancer Chemother. Pharmacol., 2008, 61, 809–817 CrossRef CAS PubMed.
- P. Nygren, M. Fryknas, B. Ågerup and R. Larsson, J. Cancer Res. Clin. Oncol., 2013, 139, 2133–2140 CrossRef CAS PubMed.
- K. Gull, P. J. Dawson, C. Davis and E. H. Byard, Biochem. Soc. Trans., 1987, 15, 59–60 CrossRef CAS PubMed.
- J. Liu, Q. Lin, H. Yao, M. Wang, Y. M. Zhang and T. B. Wei, Chin. Chem. Lett., 2014, 25, 35–38 CrossRef CAS.
- A. W. Addison and P. J. Burke, J. Heterocycl. Chem., 1981, 18, 803 CrossRef CAS.
- S. M. Kuang, D. G. Cuttell, D. R. McMillin, P. E. Fanwick and R. A. Walton, Inorg. Chem., 2002, 41, 3313–3322 CrossRef CAS PubMed.
- D. F. Zhang and L. M. Yan, J. Phys. Chem. B, 2010, 114, 12234–12241 CrossRef CAS PubMed.
- S. C. Zimmermann, T. Tichý, J. Vávra, R. P. Dash, C. E. Slusher, A. J. Gadiano, Y. Wu, A. Jančařík, L. Tenora, L. Monincová, E. Prchalová, G. J. Riggins, P. Majer, B. S. Slusher and R. Rais, J. Med. Chem., 2018, 61, 3918–3929 CrossRef CAS PubMed.
- I. Chakraborty, S. J. Carrington and P. K. Mascharak, Acc. Chem. Res., 2014, 47, 2603–2611 CrossRef CAS PubMed.
- J. Jorge, C. Indranil, D. Annmarie, M. G. Jorge, W. M. S. Chamil and P. K. Mascharak, Inorg. Chem., 2018, 57, 1766–1773 CrossRef PubMed.
- W. L. Jia, T. McCormick, Y. Tao, J. P. Lu and S. Wang, Inorg. Chem., 2005, 44, 5706–5712 CrossRef CAS PubMed.
- J. L. Chen, X. F. Cao, W. Gu, H. R. Wen, L. X. Shi, G. Rong and P. Luo, Inorg. Chem. Commun., 2011, 14, 1894–1897 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Additional figures and characterization data. CCDC 1548082 for 1, 1548083 for 2, 1851697 for 3 and 1548084 for 4. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ra01370a |
| This journal is © The Royal Society of Chemistry 2019 |