Open Access Article
Open Access ArticleXiao Qing Long Tang essential oil exhibits inhibitory effects on the release of pro-inflammatory mediators by suppressing NF-κB, AP-1, and IRF3 signalling in the lipopolysaccharide-stimulated RAW264.7 cells
Gan Luo ab,
Jing Konga,
Brian Chi-Yan Chengcd,
Hui Zhaoe,
Xiu-Qiong Fuf,
Li-Shan Yana,
Yu Dinga,
Yan-Ling Liua,
Si-Yuan Pana,
Shuo-Feng Zhanga and
Yi Zhang
ab,
Jing Konga,
Brian Chi-Yan Chengcd,
Hui Zhaoe,
Xiu-Qiong Fuf,
Li-Shan Yana,
Yu Dinga,
Yan-Ling Liua,
Si-Yuan Pana,
Shuo-Feng Zhanga and
Yi Zhang *a
*a
aSchool of Chinese Materia Medica, Beijing University of Chinese Medicine, Beijing, China. E-mail: yizhang714@163.com
bBeijing Key Laboratory for Production Process Control and Quality Evaluation of Traditional Chinese Medicine, Beijing Municipal Science & Technology Commission, Beijing 102400, China
cCollege of Professional and Continuing Education, Hong Kong Polytechnic University, Hung Hom, Hong Kong
dQuality Healthcare Medical Services, Kowloon Bay, Hong Kong
eSchool of Traditional Chinese Medicine, Capital Medical University, Beijing, China
fCentre for Cancer and Inflammation Research, School of Chinese Medicine, Hong Kong Baptist University, Kowloon Tong, Hong Kong
First published on 26th April 2019
Abstract
Xiao Qing Long Tang (literally “Minor blue dragon decoction” in Chinese), a traditional Chinese formula, is prescribed to treat respiratory diseases. However, only few studies have been reported on its anti-inflammatory mechanisms. In this study, we investigated the inhibitory effects of Xiao Qing Long Tang essential oil on inflammatory mediators and explored the mechanisms of action of XQEO in the lipopolysaccharide (LPS)-stimulated RAW264.7 cells. XQEO was prepared via steam distillation and characterized by GC-MS analysis. MTT and Griess assays were used to measure cell viability and NO production, respectively. The mRNA expression and the production of LPS-induced pro-inflammatory cytokines (IL-1β, IL-6, TNF-α, and IL-10) and chemokines (MCP-1, Rantes, and MIP-1α) were determined by real-time PCR and enzyme-linked immunosorbent assay, respectively. Furthermore, we determined the protein levels of the components of NF-κB, AP-1 and IRF3 signalling by Western blotting. Immunofluorescence assay was used to estimate the nuclear translocation of NF-κB, AP-1 and IRF3. The results showed that XQEO inhibited the secretion of NO and PGE2 and down-regulated the mRNA and protein levels of iNOS and COX-2. We also found that XQEO suppressed the LPS-induced overproduction of pro-inflammatory mediators. Moreover, XQEO inhibited the phosphorylation of NF-κB/p65, AP-1/c-Jun, and IRF3 by suppressing their upstream kinases, such as MAPKs, TBK1, Akt, IKKα/β, and IκB, reducing the LPS-induced NF-κB, AP-1 and IRF3 translocation to the nucleus. These findings suggest that XQEO effectively suppresses the production of pro-inflammatory mediators possibly through the inhibition of NF-κB, AP-1, and IRF3 signalling in the LPS-stimulated RAW264.7 cells.
Introduction
In response to infection or tissue damage, immune cells initiate signalling cascades that trigger an inflammatory response and promote the release of cytokines and chemokines against invading pathogens;1 one of the main triggers of inflammation is the recognition of microbes by receptors, such as Toll-like receptor 4 (TLR4), which is apparent in innate immunity and responsible for presenting antigens or allergens such as lipopolysaccharides (LPSs), of the innate immune system.2 After an LPS binds to the TLR4 in the outer membrane of macrophages, various intracellular signalling pathways will be activated; this will lead to the activation of transcription factors such as nuclear factor-κB (NF-κB), activator protein-1 (AP-1), and interferon regulatory factor 3 (IRF3).3–5 Subsequently, these transcription factors will enter the nucleus and promote the secretion of pro-inflammatory cytokines and chemokines.6 Then, these inflammatory mediators will trigger inflammatory responses, initiating cellular damage and tissue injury.7 Thus, the inhibition of TLR4 signalling transduction is one of the focuses in the development of effective anti-inflammatory agents.Recent attention has focused on the control of pathogenesis for inflammatory diseases using NSAID and glucocorticoid.8 However, the therapeutic effect of these drugs is not clinically satisfactory due to their adverse effects and toxicity.9 Therefore, effective and safe therapeutic agents are urgently needed to ameliorate inflammatory disorders. Traditional Chinese medicine (TCM), as a type of complementary and alternative medicine, is a sophisticated and time-honored form of healthcare in China.10 Many TCMs are widely used to treat inflammatory diseases due to their safety and efficacy.11,12 Xiao Qing Long Tang ( , literally: Minor blue dragon decoction) is a traditional Chinese formula consisting of Herba Ephedrae, Rhizoma Zingiberis, Ramulus Cinnamomi, Radix et Rhizoma Asari, Rhizoma Pinellia, Fructus Schisandrae Chinensis, Radix et Rhizoma Glycyrrhizae, and Radix paeoniae alba at the ratio of 1
, literally: Minor blue dragon decoction) is a traditional Chinese formula consisting of Herba Ephedrae, Rhizoma Zingiberis, Ramulus Cinnamomi, Radix et Rhizoma Asari, Rhizoma Pinellia, Fructus Schisandrae Chinensis, Radix et Rhizoma Glycyrrhizae, and Radix paeoniae alba at the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1:1
1:1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1:1
1:1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1:1
1:1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.13 It was first reported in the Shang Han Lun and has been commonly used for the treatment of cold syndrome, bronchitis, and nasal allergies for thousands of years in China.14 Previous studies have shown that Xiao Qing Long Tang has potential anti-inflammatory effects in the airway of asthmatic animals.15 Moreover, it exerted preventive effects against asthma in an allergic asthma mouse model via neurotrophin regulation.16 Recently, some studies have reported that many essential oils extracted from the major constituents of Xiao Qing Long Tang, such as Rhizoma Zingiberis17,18 and Ramulus Cinnamomi,19 exhibit anti-inflammatory properties in vitro and in vivo. However, no pharmacological studies have been reported on the investigation of the anti-inflammatory activity of essential oils extracted from Xiao Qing Long Tang. Therefore, in this study, we employed the RAW264.7 cell line as an inflammatory cell model, which is commonly used in medical research.20 We also examined the inhibitory effects of the Xiao Qing Long Tang essential oil (XQEO) on the secretion of inflammatory mediators and explored the underlying mechanisms of action of XQEO.
1.13 It was first reported in the Shang Han Lun and has been commonly used for the treatment of cold syndrome, bronchitis, and nasal allergies for thousands of years in China.14 Previous studies have shown that Xiao Qing Long Tang has potential anti-inflammatory effects in the airway of asthmatic animals.15 Moreover, it exerted preventive effects against asthma in an allergic asthma mouse model via neurotrophin regulation.16 Recently, some studies have reported that many essential oils extracted from the major constituents of Xiao Qing Long Tang, such as Rhizoma Zingiberis17,18 and Ramulus Cinnamomi,19 exhibit anti-inflammatory properties in vitro and in vivo. However, no pharmacological studies have been reported on the investigation of the anti-inflammatory activity of essential oils extracted from Xiao Qing Long Tang. Therefore, in this study, we employed the RAW264.7 cell line as an inflammatory cell model, which is commonly used in medical research.20 We also examined the inhibitory effects of the Xiao Qing Long Tang essential oil (XQEO) on the secretion of inflammatory mediators and explored the underlying mechanisms of action of XQEO.
Materials and methods
Herbal preparation
Herba Ephedrae, Rhizoma Zingiberis, Ramulus Cinnamomi, Radix et Rhizoma Asari, Rhizoma Pinellia, Fructus Schisandrae Chinensis, Radix et Rhizoma Glycyrrhizae, and Radix paeoniae alba were purchased from Beijing Tong Ren Tang (Tong Ren Tang Co. Ltd., Beijing, China). All of them were authenticated by Lecturer Guang Xi Ren (School of Chinese Materia Medica, Beijing University of Chinese Medicine). Voucher specimens (Herba Ephedrae, Rhizoma Zingiberis, Ramulus Cinnamomi etc.) have been deposited at the Department of Pharmacology, School of Chinese Medicine, Beijing University of Chinese Medicine.Air-dried whole plants composed of Xiao Qing Long Tang were mechanically ground, and then, 114 g of the powder was used for extracting the essential oil by hydro-distillation using a Clevenger-type apparatus with the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 of material to water. XQEO was obtained continuously within 8 h and dried by anhydrous sodium sulfate. After separating anhydrous sodium sulfate by filtration, 500 μL XQEO was obtained and then kept at 4 °C in dark vials until testing. A solution of XQEO was prepared by dissolving the extract in dimethyl sulfoxide (DMSO) and then used in the following experiments.
5 of material to water. XQEO was obtained continuously within 8 h and dried by anhydrous sodium sulfate. After separating anhydrous sodium sulfate by filtration, 500 μL XQEO was obtained and then kept at 4 °C in dark vials until testing. A solution of XQEO was prepared by dissolving the extract in dimethyl sulfoxide (DMSO) and then used in the following experiments.
GC-MS analysis of XQEO
The volatile constituents in XQEO were separated using the Agilent 7890B/5975A gas chromatograph (Agilent Technologies, Palo Alto, CA, United States) with the HP-5MS 5% phenylmethylsiloxane capillary column (30.00 m × 0.25 mm, 0.25 μm film thickness). Helium (99.999%) was used as the carrier gas at the flow rate of 2 mL min−1, and a 1 μL sample (10 mg of XQEO in 1 mL of methanol) was injected in the split mode distributed at 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The oven temperature program was setted as follows: the temperature was maintained at 60 °C for 1 min, then consecutively ramped to 140 °C at 4 °C min−1, from 140 °C to 170 °C at 2.5 °C min−1 and then from 170 °C to 240 °C at 8 °C min−1. The injector and detector temperatures were set at 240 and 280 °C, respectively. The mass spectrometer was operated at a 70 eV electron impact, with the ion source temperature of 230 °C, and the data were obtained in the full scan mode over the mass scan range m/z of 30–500. The identification of oil components was accomplished by comparison of their mass spectral fragmentation patterns (NIST 14 mass spec library and search programs). The peak area percent was used for obtaining quantitative data.
1. The oven temperature program was setted as follows: the temperature was maintained at 60 °C for 1 min, then consecutively ramped to 140 °C at 4 °C min−1, from 140 °C to 170 °C at 2.5 °C min−1 and then from 170 °C to 240 °C at 8 °C min−1. The injector and detector temperatures were set at 240 and 280 °C, respectively. The mass spectrometer was operated at a 70 eV electron impact, with the ion source temperature of 230 °C, and the data were obtained in the full scan mode over the mass scan range m/z of 30–500. The identification of oil components was accomplished by comparison of their mass spectral fragmentation patterns (NIST 14 mass spec library and search programs). The peak area percent was used for obtaining quantitative data.
Reagents
Lipopolysaccharide (LPS, Escherichia coli 0111:B4), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl thiazolium bromide (MTT), bovine serum albumin, and Griess reagent (modified) were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Fetal bovine serum (FBS) was obtained from Biological Industries (Beth-Haemek, Israel), and the Dulbecco's Modified Eagle Medium (DMEM) was obtained from Corning Cellgro (Manassas, VA, USA). Penicillin–streptomycin solution was bought from Caisson labs (Smithfield, UT, USA). The RAW264.7 cells, a BALB/c-derived murine macrophage cell line (ATCC no. TIB-71), was purchased from ATCC (Rockville, MD, USA). The prostaglandin E2 (PGE2) ELISA kit was obtained from Enzo life science (Exeter, UK). Interleukin 1β (IL-1β), IL-6, IL-10, tumor necrosis factor α (TNF-α), monocyte chemoattractant protein (MCP)-1, macrophage inflammatory protein (MIP)-1α and regulated on activation normal T-cell expressed and secreted (Rantes) ELISA kits were obtained from Thermo fisher scientific (San Diego, CA, USA). p65 (catalog no. 8242), phospho-p65 (Ser536, catalog no. 3033), Akt (catalog no. 9272), phospho-Akt (Ser473, catalog no. 4060), c-Jun (catalog no. 9165), phospho-c-Jun (Ser73, catalog no. 9164), cyclooxygenase-2 (COX-2, catalog no. 12282), inducible nitric oxide synthase (iNOS, catalog no. 13120), phospho-IRF3 (Ser396, catalog no. 4947), IκBα (catalog no. 4814), phospho-IκBα (Ser32, catalog no. 2859), phospho-IKKα/β (Ser176/180, catalog no. 2697), extracellular signal-regulated kinase (ERK, catalog no. 4695), phospho-ERK (Thr202/Tyr204, catalog no. 4370), c-Jun N-terminal kinase (JNK, catalog no. 9252), phospho-JNK (Thr183/Tyr185, catalog no. 4668), p38 mitogen-activated protein kinase (p38, catalog no. 8690), phospho-p38 (Thr180/Tyr182, catalog no. 4511), TANK-binding kinase 1 (TBK1, catalog no. 3013), phospho-TBK1 (Ser172, catalog no. 5483), anti-rabbit IgG HRP-linked antibody (catalog no. 7074) and alexa fluor 488-conjugated secondary antibodies (catalog no. 4412) were obtained from Cell signaling technology (Boston, MA, USA). Interferon regulatory factor 3 (IRF3, catalog no. ab68481) was purchased from Abcam (Cambridge, UK). IκB kinase α/β (IKKα/β, catalog no. sc-7607) and sp1 (catalog no. sc-420) monoclonal antibodies and anti-mouse IgG HRP-linked antibody (catalog no. sc-516102) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).Cell culture
The murine macrophage cell line RAW267.4 was maintained in the DMEM medium (4.5 g L−1 glucose, L-glutamine & sodium pyruvate) containing 10% heat-inactivated fetal bovine serum (FBS) and 1% antibiotics (penicillin/streptomycin) at 37 °C under 5% CO2.Cell viability assay
The effect of XQEO on the viability of the RAW264.7 cells was determined using the MTT assay. The cells were seeded at 5000 cells per well in 96-well plates and allowed to grow for 24 h. Before treatment, XQEO was dissolved in DMSO as a stock solution and diluted with DMEM to various concentrations (3.125, 6.25, 12.5, 25, and 50 μg mL−1). 6 replicate wells were setted for each concentration. After 24 h incubation, the XQEO was added to the cells followed by incubation for 1 h, and then, the cells were treated with or without LPS (1 μg mL−1) for 24 h at 37 °C. Subsequently, 10 μL of MTT solution (5 mg mL−1) was added to each well, the cells were incubated for 3 h at 37 °C, and 100 μL DMSO was added to dissolve the crystals. The absorbance in each well was measured at 570 nm by a microplate reader (BMG SPECTROstar Nano, Germany). The cell viability in the control group (cells were not treated with XQEO or LPS) was set as 100% for the MTT assays and calculated by the GraphPad Prism 6.0 Software.RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Cells were seeded in 6-well plates (4 × 105 cells per mL) for 24 h, treated with or without XQEO for 1 h, and then stimulated with LPS (1 μg mL−1) for 6 h. Total ribonucleic acid (RNA) was extracted from the RAW264.7 cells using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) and reverse-transcripted with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Lithuania) according to the manufacturer's protocol. After this, the cDNA samples were subjected to real-time PCR analyses by monitoring the increase in the fluorescence of SYBR green (Thermo Fisher Scientific, Woolston, UK) using the Step One Plus Real Time PCR system (Applied Biosystems, CA, USA). The expression of the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene in each sample was evaluated as an internal control. Table 1 shows the primer sequences of various genes (iNOS, COX-2, mPGES1, IL-1β, IL-6, IL-10, TNF-α, MCP-1, MIP-1α and Rantes), which were determined via the real-time PCR analysis. Each sample was amplified in triplicate for quantification. The data were analyzed by relative quantitation using the ΔΔCt method and normalized to GAPDH.| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) | |
|---|---|---|---|
| iNOS | AGCAACTACTGCTGGTGGTG | TCTTCAGAGTCTGCCCATTG | |
| COX-2 | CTGGAACATGGACTCACTCAGTTTG | AGGCCTTTGCCACTGCTTGT | |
| mPGES1 | ATGAGGCTGCGGAAGAAGG | GCCGAGGAAGAGGAAAGGATAG | |
| IL-1β | GAAGAAGAGCCCATCCTCTG | TCATCTCGGAGCCTGTAGTG | |
| IL-6 | AGTCCGGAGAGGAGACTTCA | ATTTCCACGATTTCCCAGAG | |
| IL-10 | TTGAATTCCCTGGGTGAGAAG | TCCACTGCCTTGCTCTTATTT | |
| TNF-α | ATGAGAAGTTCCCAAATGGC | CTCCACTTGGTGGTTTGCTA | |
| MCP-1 | AATGCTAACGCCACCGAGAG | CCTTGTTCTGCTCCTCATAGTCC | |
| MIP-1α | CCCAGCCAGGTGTCATTTTCC | GCATTCAGTTCCAGGTCAGTG | |
| Rantes | CATATGGCTCGGACACCA | ACACACTTGGCGGTTCCT | |
| GAPDH | GGCCTTCCGTGTTCCTACC | TGCCTGCTTCACCACCTTC | |
Determination of the nitric oxide (NO) content, PGE2, cytokines and chemokines
The RAW264.7 cells were seeded at 2 × 105 cells per well in 24-well culture plates and allowed to grow for 24 h. The cells were incubated with 1 mL of XQEO (0, 6.25, 12.5, 25 and 50 μg mL−1) for 1 h and then in the presence or absence of LPS (1 μg mL−1) for another 24 h. The cell-free supernatants were obtained for the determination of the NO content using the typical commercial Griess reagent (Sigma Aldrich). The levels of PGE2, IL-1β, IL-6, IL-10, TNF-α, MCP-1, MIP-1α, and Rantes in the culture medium were also quantified by the ELISA kits following the manufacturer's instructions. Four replicate wells were used.Western blot analysis
The cells were seeded in 60 mm-diameter culture dishes (4 × 105 cells) and pre-treated with XQEO at 12.5–50 μg mL−1 for 1 h; then, LPS (1 μg mL−1) was added, and the cells were incubated at 37 °C for 30 min or 60 min. The cells were washed twice with PBS and re-suspended in the RIPA protein extraction buffer (Beyotime biotechnology, Beijing, China) followed by incubation for 30 min at 4 °C. Cell debris was removed by a microcentrifuge (4 °C, 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min), and then, the protein concentration of the supernatants was ascertained by the Bio-rad protein assay reagent according to the manufacturer's instructions. The cytoplasmic and nuclear proteins were extracted using a nuclear extraction kit (Solarbio, Beijing, China) according to the manufacturer's instructions. Then, 20 μg of protein was fractionated by 10% SDS PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (0.45 μm). After incubation for 1 h with 5% skim milk in Tris-buffered saline/Tween 20 (TBS/T) at room temperature, the membranes were incubated with indicated primary antibodies (1
000 rpm, 10 min), and then, the protein concentration of the supernatants was ascertained by the Bio-rad protein assay reagent according to the manufacturer's instructions. The cytoplasmic and nuclear proteins were extracted using a nuclear extraction kit (Solarbio, Beijing, China) according to the manufacturer's instructions. Then, 20 μg of protein was fractionated by 10% SDS PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (0.45 μm). After incubation for 1 h with 5% skim milk in Tris-buffered saline/Tween 20 (TBS/T) at room temperature, the membranes were incubated with indicated primary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) at 4 °C for 24 h. The membranes were rinsed thrice with TBS/T and incubated with secondary antibodies (1
1000) at 4 °C for 24 h. The membranes were rinsed thrice with TBS/T and incubated with secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) for 1 h at room temperature followed by washing thrice with TBS/T. The blots were developed using enhanced chemiluminescence detection agents (Tanon, Shanghai, China) and analysed using the ImageJ software (National Institutes of Health [NIH], Bethesda, MD, USA). The protein levels were normalized to the matching densitometric value of the internal control β-actin. Three replicates were used.
2000) for 1 h at room temperature followed by washing thrice with TBS/T. The blots were developed using enhanced chemiluminescence detection agents (Tanon, Shanghai, China) and analysed using the ImageJ software (National Institutes of Health [NIH], Bethesda, MD, USA). The protein levels were normalized to the matching densitometric value of the internal control β-actin. Three replicates were used.
Immunofluorescence staining
After the RAW264.7 cells (1 × 104 cells per well) were seeded in chamber slides for 24 h, the cells were washed twice with ice-cold PBS and then fixed with 4% formaldehyde for 15 min at RT. After washing with PBS for three times, the cells were permeabilized by incubation with 400 μL of a 0.25% Triton X-100 solution for 30 min at 37 °C and followed by incubated for 10 min at room temperature. After blocking the cells with 3% BSA for 1 h, the cells were incubated with the polyclonal antibodies for NF-κB/p65, AP-1/c-Jun, and IRF3 diluted at a 2% BSA solution followed by incubation overnight at 4 °C. After being washed 3 times with PBS, the cells were incubated with the Alexa Fluor 488-conjugated secondary antibody for 1 h at RT. The nuclei were stained with DAPI (YESEN, Shanghai, China), and the fluorescence was visualized using the Nikon A1R Eclipse Ti confocal microscope (Nikon Corp., Tokyo, Japan).Statistical analysis
All the statistical analyses were performed using the Microsoft Excel and GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). The data are presented as mean ± standard error of mean (SEM). Moreover, one-way analysis of variance (ANOVA) followed by the Dunnett's multiple-comparison test was used to assess the differences between the experimental groups. Differences were considered significant at p < 0.05.Results
Chemical profiling of the XQEO by GC-MS
The GC-MS analysis shows that there are 24 different compounds in XQEO, representing 91.41% of the total oil (Fig. 1 and Table 2). The XQEO was predominantly composed of safrole (19.705%), methyl eugenol (14.168%), 3,5-dimethoxytoluene (11.269%), (Z)-3-phenylacrylaldehyde (8.002%), and 2-allyl-1,4-dimethoxy-3-methyl-benzene (5.829%). Among other compounds, a variety of compounds were found at significant amounts including 1,3-benzodioxole,4-methoxy-6-(2-propenyl)-(3.508%), (1R)-2,6,6-trimethylbicyclo [3.1.1] hept-2-ene (3.224%), β-pinene (2.923%), camphene (2.466%), 3-carene (2.520%), 2,4-cycloheptadien-1-one,2,6,6-trimethyl-(1.995%), eucalyptol (1.975%), endo-borneol (1.726%), and α-terpineol (1.595%).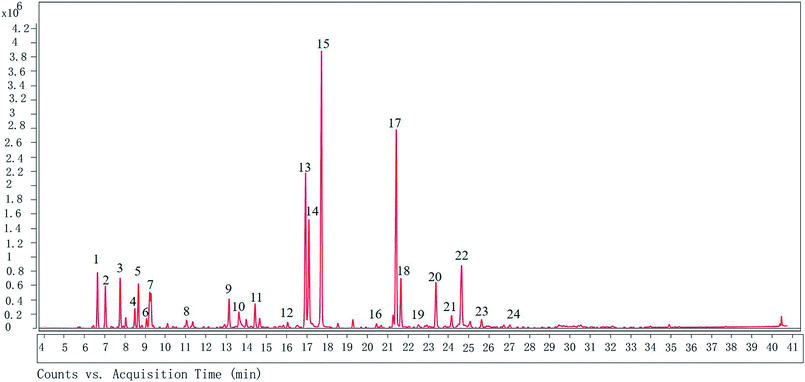 | ||
| Fig. 1 The GC chromatogram of XQEO. The volatile constituents in the essential oils were separated using GC chromatogram. | ||
| Peak number | Compounds | Ret. time (min) | Formula | Relative abundance (%) |
|---|---|---|---|---|
| 1 | (1R)-2,6,6-Trimethylbicyclo [3.1.1] hept-2-ene | 6.643 | C10H16 | 3.224 |
| 2 | Camphene | 7.033 | C10H16 | 2.466 |
| 3 | β-Pinene | 7.763 | C10H16 | 2.923 |
| 4 | α-Phellandrene | 8.489 | C10H16 | 1.124 |
| 5 | 3-Carene | 8.664 | C10H16 | 2.520 |
| 6 | Cyclohexane,1-methylene-4-(1-methylethenyl)- | 9.226 | C10H16 | 2.548 |
| 7 | Eucalyptol | 9.289 | C10H18O | 1.975 |
| 8 | Cyclohexene,1-methyl-4-(1-methylethylidene)- | 11.049 | C10H16 | 0.579 |
| 9 | 2,4-Cycloheptadien-1-one,2,6,6-trimethyl- | 13.149 | C10H14O | 1.995 |
| 10 | Endo-borneol | 13.636 | C10H18O | 1.726 |
| 11 | α-Terpineol | 14.433 | C10H18O | 1.595 |
| 12 | 2,6-Octadienal, 3,7-dimethyl-, (Z)- | 16.048 | C10H16O | 0.391 |
| 13 | 3,5-Dimethoxytoluene | 16.93 | C9H12O2 | 11.269 |
| 14 | (Z)-3-Phenylacrylaldehyde | 17.097 | C9H8O | 8.002 |
| 15 | Safrole | 17.716 | C10H12O2 | 19.705 |
| 16 | Ylangene | 20.437 | C15H24 | 0.306 |
| 17 | Methyl eugenol | 21.413 | C11H14O2 | 14.168 |
| 18 | Benzene, 1,2,3-trimethoxy-5-methyl- | 21.644 | C10H14O3 | 3.348 |
| 19 | 1H-cyclopropa[a]naphthalene, 1a,2,3,5,6,7,7a,7b-octahydro-1,1,7,7a-tetramethyl-, [1aR-(1aα,7α,7aα,7bα)]- | 22.512 | C15H24 | 0.310 |
| 20 | 1,3-Benzodioxole, 4-methoxy-6-(2-propenyl)- | 23.377 | C11H12O3 | 3.508 |
| 21 | Benzene,1-(1,5-dimethyl-4-hexenyl)-4-methyl- | 24.152 | C15H22 | 1.001 |
| 22 | 2-Allyl-1,4-dimethoxy-3-methyl-benzene | 24.641 | C11H12O3 | 5.829 |
| 23 | Cyclohexene, 3-(1,5-dimethyl-4-hexenyl)-6-methylene-, [S-(R*,S*)]- | 25.629 | C15H24 | 0.641 |
| 24 | 1,6,10-Dodecatrien-3-ol,3,7,11-trimethyl-, (E)- | 27.028 | C15H26O | 0.253 |
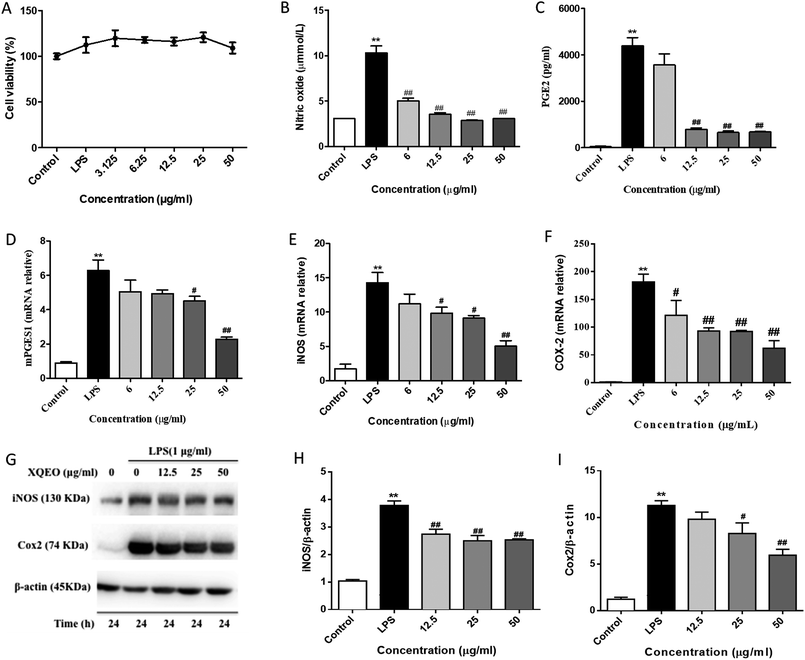
XQEO inhibited the LPS-induced overproduction of NO and PGE2 by suppressing the expression of iNOS and COX-2 in the RAW264.7 macrophages
NO and PGE2 are the key mediators in the inflammatory process.21 The effects of XQEO on the production of inflammatory mediators, including NO and PGE2, were firstly determined. Our results showed that treatment of the LPS-stimulated RAW264.7 cells with XQEO (6–50 μg mL−1) significantly inhibited the secretion of NO in these cells (p < 0.01). Moreover, the production of PGE2 was markedly suppressed by XQEO (12.5–50 μg mL−1) in a concentration-dependent manner (p < 0.01). These inhibitory effects of XQEO were not caused by its nonspecific cytotoxicity because based on the MTT assay results, XQEO at the concentration of up to 50 μg mL−1 showed no effect on cell viability. As NO and PGE2 are generated by iNOS and COX-2,22 we determined the protein and mRNA expression levels of these enzymes by Western blotting and PCR, respectively. The results showed that the protein and mRNA expression levels of iNOS and COX-2 were obviously down-regulated by the XQEO treatment in the LPS-stimulated RAW264.7 cells (p < 0.05 or p < 0.01). We also found that the mRNA expression of another key enzyme for the synthesis of PGE2 (mPGES1) was suppressed in a concentration-dependent manner after pre-treatment with XQEO in the LPS-stimulated RAW264.7 macrophages (p < 0.05 or p < 0.01) (Fig. 2).Effects of XQEO on the LPS-induced mRNA overexpression of pro-inflammatory cytokines and chemokines in the RAW264.7 macrophages
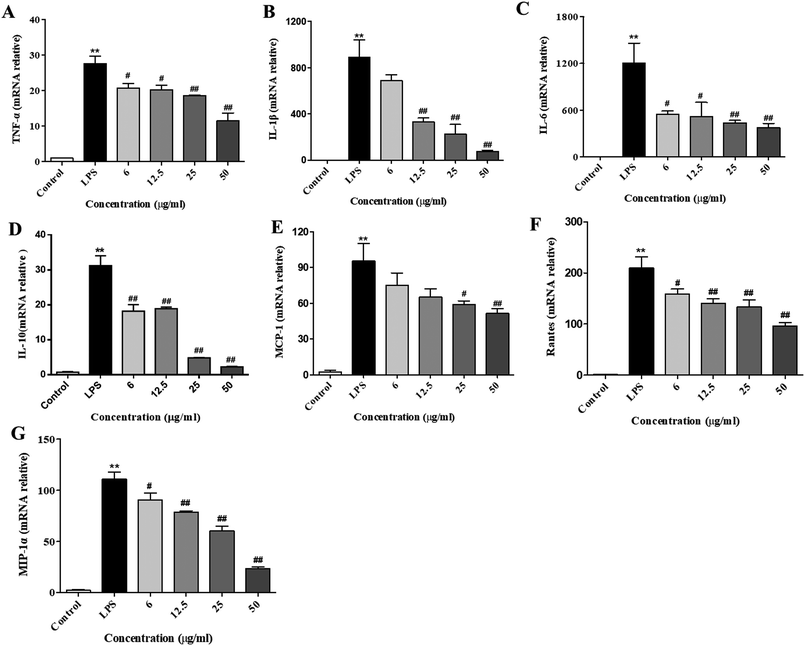
After stimulation of the macrophages with LPS, the gene expressions of cytokines (e.g., IL-10, IL-1β, IL-6, and TNF-α) and chemokines (MCP-1, Rantes, and MIP-1) were remarkably elevated. Subsequently, the secretion of these cytokines and chemokines were increased, which was necessary to activate the inflammatory response.23 To examine the inhibitory effect of XQEO on the gene expression of pro-inflammatory cytokines and chemokines, we investigated the mRNA expression levels of TNF-α, IL-1β, IL-6, IL-10, MCP-1, Rantes, and MIP-1α in the LPS-induced RAW264.7 macrophages. The results indicate that the mRNA expression levels of cytokines (IL-6, IL-1β, IL-10, and TNF-α, as shown in Fig. 3A–D) and chemokines (MCP-1, Rantes, and MIP-1, as shown in Fig. 3E–G) were obviously up-regulated after the LPS treatment (p < 0.01). However, the XQEO decreased the expression levels of cytokines (IL-6, IL-1β, IL-10, and TNF-α, as shown in Fig. 3A–D) and chemokines (MCP-1, Rantes, and MIP-1α, as shown in Fig. 3 E–G) in the LPS-stimulated RAW264.7 cells (p < 0.01 or p < 0.05) in a concentration-dependent manner. These results indicate that XQEO can suppress the expression of these inflammatory cytokines and chemokines at the transcriptional level.Effects of the XQEO on the LPS-induced overproduction of pro-inflammatory cytokines and chemokines in the RAW264.7 macrophages
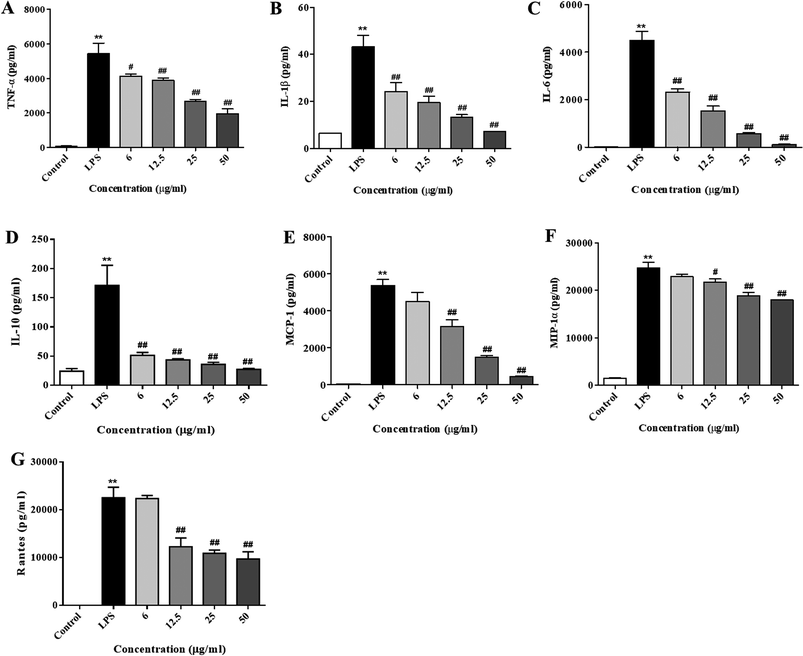
Next, we further investigated the production of cytokines and chemokines in the LPS-stimulated RAW264.7 cells. Consistent with the RT-PCR results, the concentrations of cytokines (IL-6, IL-1β, IL-10, and TNF-α, as shown in Fig. 4A–D) and chemokines (MCP-1, Rantes, and MIP-1, as shown in Fig. 4E–G) in the culture medium were obviously elevated after the LPS treatment (p < 0.01). Pre-treatment with XQEO significantly reduced the overproduction of these cytokines (Fig. 4A–D) and chemokines (Fig. 4E–G) in the RAW264.7 macrophages (p < 0.05 and p < 0.01).Effects of the XQEO on the nuclear translocation of NF-κB, AP-1, and IRF3 in the RAW264.7 macrophages
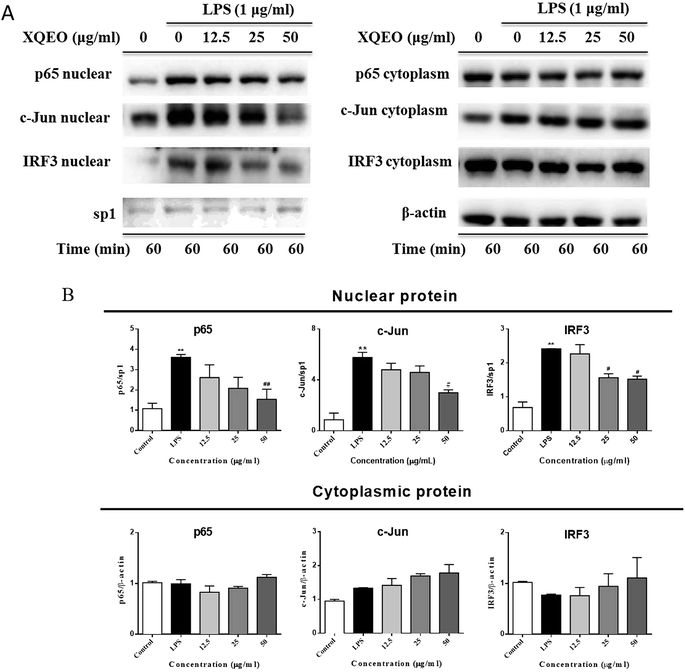
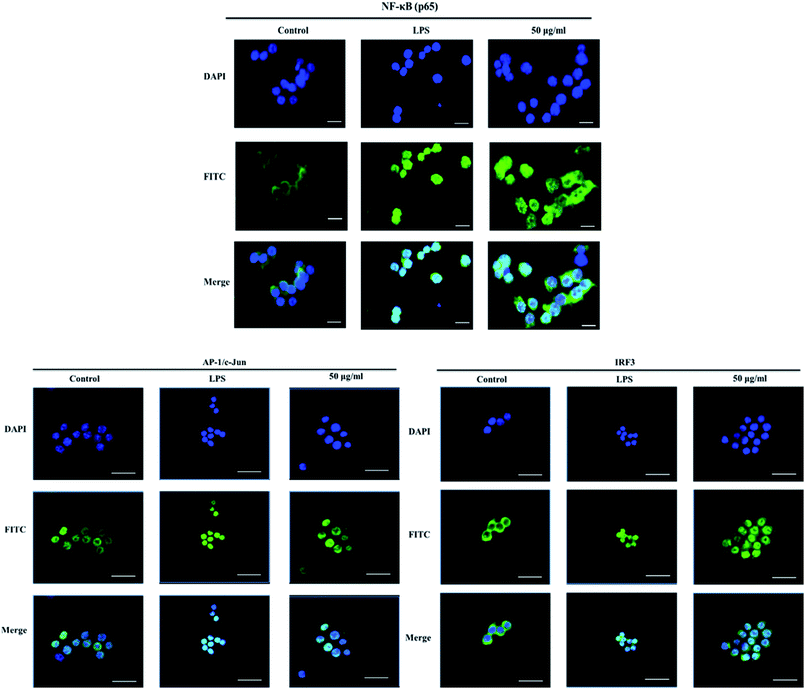
The nuclear translocation of transcription factors, such as NF-κB, AP-1, and IRF3, is considered a prerequisite for the transcription of genes associated with inflammatory processes.23 Therefore, in the present study, we also explored the effect of XQEO on the nuclear translocation of these transcription factors in the RAW264.7 cells. As shown in Fig. 5A and B, the nuclear protein levels of NF-κB/p65, AP-1/c-Jun, and IRF3 were markedly increased after LPS-stimulation (p < 0.01). However, stimulation of the cells with LPS in the presence of XQEO decreased the nuclear protein levels of these three transcription factors (p < 0.05 or p < 0.01 Fig. 5A and B) in a concentration-dependent manner. Moreover, the cytoplasmic protein levels of NF-κB/p65, AP-1/c-Jun, and IRF3 were not significantly changed after the XQEO treatment (p > 0.05 Fig. 5A and B). In addition, LPS enhanced the nuclear accumulation of NF-κB/p65, AP-1/c-Jun, and IRF3, as evidenced by immunofluorescence staining. However, the nuclear localization of these transcription factors was effectively reduced by the XQEO pre-treatment (Fig. 6). These results suggest that XQEO acts as a negative regulator of LPS-stimulated NF-κB, AP-1, and IRF3 activation in the RAW264.7 macrophages.Effect of XQEO on the molecular components of NF-κB, AP-1, and IRF3 signalling in the RAW264.7 macrophages
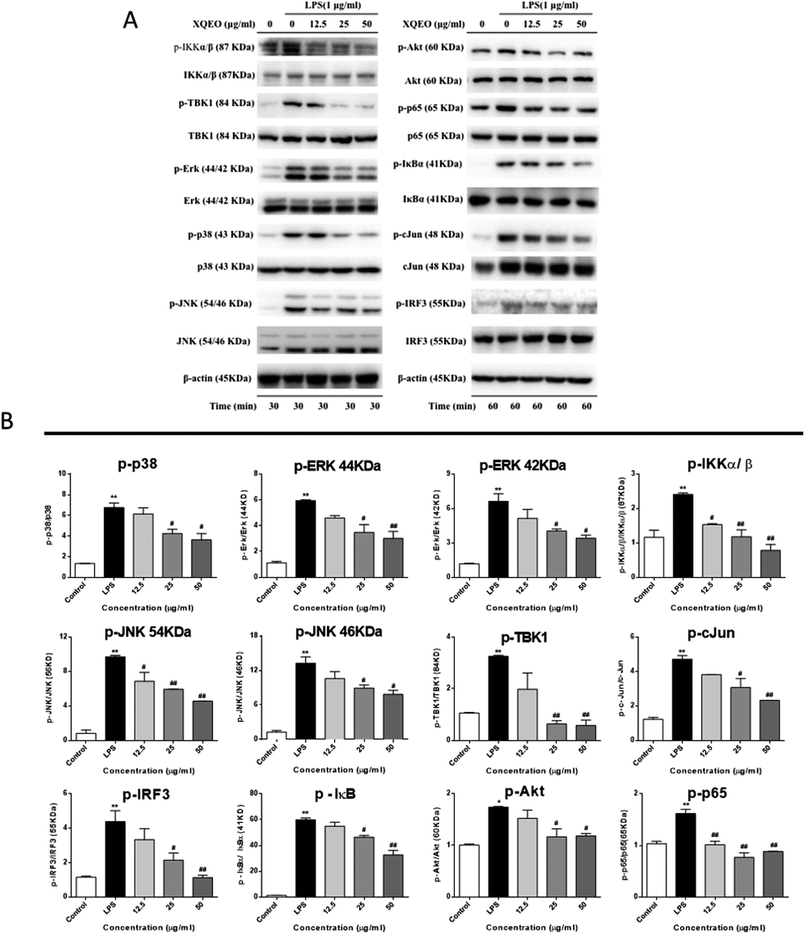
Previous studies have reported that NF-κB, AP-1, and IRF3 as well as their upstream proteins are involved in the LPS-induced inflammation in macrophages.20 Therefore, the possible involvement of these signalling in the XQEO-mediated inhibition of the LPS-induced inflammatory responses was investigated in the present study. Fig. 7 shows that the phosphorylation of IKKα/β and IκBα was markedly up-regulated by the LPS treatment (p < 0.01). XQEO decreased the LPS-induced elevation of phosphorylated IKKα/β (Ser176/180) and IκBα in the RAW264.7 cells (p < 0.01) in a concentration-dependent manner. Moreover, we observed that XQEO obviously attenuated the LPS-induced phosphorylation of NF-κB/p65 (ser536) in a concentration-dependent manner (p < 0.01). These results show that XQEO has a potential inhibitory effect on the NF-κB signalling. Since Akt also plays a pivotal role in the LPS-induced inflammation response in macrophages, the inhibitory effect of XQEO on the Akt signalling has also been determined. We found that the phosphorylation of Akt (ser473) was significantly elevated after LPS-stimulation (p < 0.05), and the elevated phosphorylation of Akt induced by LPS was suppressed by the XQEO pre-treatment (p < 0.05). Moreover, Fig. 7 and 8 indicate that the LPS treatment substantially promotes the phosphorylation of three MAPKs, i.e. JNK (Thr183/Tyr185), p38 (Thr180/Tyr182) and ERK (Thr202/Tyr204), as well as AP-1/cJun (ser73) (p < 0.05 or p < 0.01). However, these effects were completely abrogated by pre-treatment with XQEO; this indicated that XQEO also suppressed AP-1 signalling. Moreover, the phosphorylation levels of TBK1 and IRF3 were significantly up-regulated after the LPS treatment (p < 0.01). XQEO remarkably decreased the phosphorylation of TBK1 and IRF3 in a concentration-dependent manner (p < 0.05 or p < 0.01); this indicates that the IRF3 signalling is also inhibited by the XQEO treatment.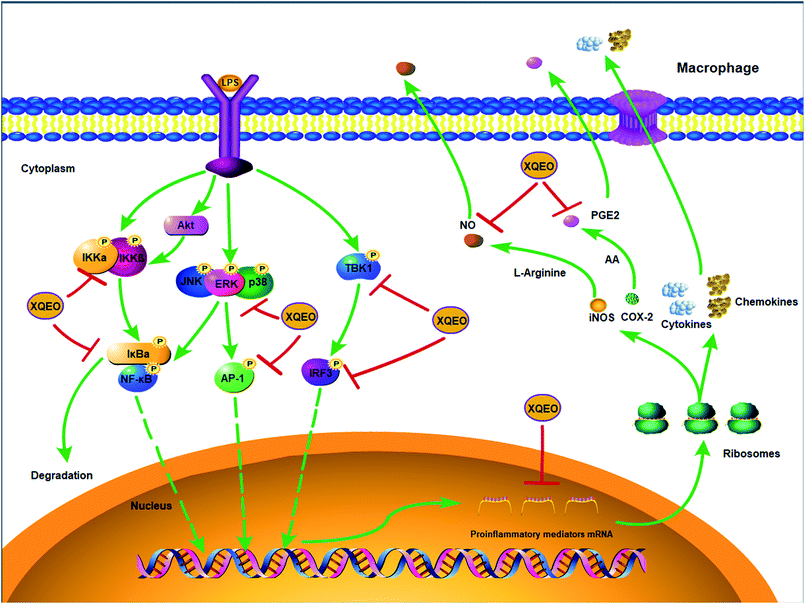 | ||
| Fig. 8 The molecular components of NF-κB, AP-1, and IRF3 signalling targeted by XQEO in macrophages. | ||
Discussion
Essential oils are complex mixtures of volatile, lipophilic and odiferous substances produced via the secondary metabolism of plants.24 Many essential oils have been used as therapeutic agents since ancient times, and some of them have been scientifically proven to possess medicinal properties, especially anti-inflammatory activity.25 In the present study, it has been identified by GC-MS that the five main compounds of XQEO are safrole, methyl eugenol, camphene, α-terpineol, and endo-Borneol, which exert strong inhibitory effects on the inflammatory responses by modulating the signalling molecules. Safrole has been reported to exhibit anti-inflammatory properties in the LPS-stimulated spleen cells.26 Methyleugenol has been demonstrated to reduce the IgE-mediated allergic inflammation in mast cells.27 Camphene has been reported to inhibit the LPS-induced TNF-α production in the RAW264.7 macrophages.28 α-Terpineol suppresses the production of inflammatory mediators in LPS-stimulated human macrophages by regulating NF-κB and MAPKs.28 Moreover, borneol exhibits remarkable anti-inflammatory effects by reducing the level of inflammatory factors in the LPS-induced RAW264.7 macrophages.29 These reports suggest that the complexes of these compounds in XQEO contribute to the anti-inflammatory properties of XQEO.Several studies have reported that macrophages perform an important function in the inflammatory reaction by restoring homeostasis through the release of inflammatory mediators, such as NO and PGE2, and pro-inflammatory cytokines and chemokines.23,30 In the present study, we found that the XQEO remarkably suppressed the production of NO and PGE2 in the LPS-stimulated RAW264.7 macrophages. It has been reported that iNOS and COX-2 are the inducible enzymes for the NO and PGE2 production, which are mainly responsible for the increased levels of NO and PGE2, respectively.31,32 Our results showed that the XQEO pre-treatment effectively reduced the protein levels and mRNA expression of iNOS and COX-2, suggesting that the inhibitory effect of the XQEO on the secretion of NO and PGE2 might be due to the suppression of iNOS and COX-2, respectively. In addition, mPGES-1 is a terminal synthase of PGE2, which catalyzes COX-1 and COX-2-derived PGH2 conversion to PGE2.33 Therefore, mPGES-1 is regarded as a novel target for anti-inflammatory agents to suppress the PGE2 production.34 Our study indicates that at high concentrations (25 and 50 μg mL−1), XQEO significantly inhibits the gene expression of mPGES-1 in the LPS-stimulated RAW264.7 cells. Hence, we conclude that the XQEO treatment inhibits PGE2 production via a double blocking action on the expressions of COX-2 and mPGES-1 in the LPS-stimulated RAW264.7 cells.
The imbalance in the production of inflammatory cytokines, such as IL-6, TNF-α, IL-1β and IL-10, contributes to immune dysfunction and also mediates inflammation of the tissues and organ damage.35 Chemokines are a family of chemoattractant molecules that play pivotal roles in the pathogenesis of various inflammatory diseases such as asthma,36 rheumatoid arthritis,37 and pneumonia,38 and chemokine inhibition may have beneficial effects in these diseases. In the present study, we found that the gene expression and production of cytokines (IL-6, TNF-α, IL-1β and IL-10) and chemokines (MCP-1, Rantes, and MIP-1α) were obviously elevated after LPS stimulation. However, the XQEO treatment down-regulated the gene expressions and secretions of cytokines and chemokines in a concentration-dependent manner; this indicates that XQEO has strong anti-inflammatory activity and the potential to act as a good anti-inflammatory agent.
On stimulation with LPS, the downstream proteins of TLR4, such as IKKs (IKKα, IKKβ and IKKγ) are activated; this in turn phosphorylates IκB that results in the degradation of IκB and the translocation of NF-κB into the nucleus followed by its binding to the cognate DNA binding sites for the promotion of the transcription of cytokines such as iNOS, COX-2, IL-1β, IL6, and TNF-α.39,40 Hence, blocking the NF-κB signalling is considered as an important target for anti-inflammatory agents. The present study has shown that XQEO remarkably inhibits the phosphorylation of IKKα/β and IκBα, suggesting that XQEO can block the IKKα/β/IκBα axis. We also found that phosphorylation and the expression of the nuclear protein NF-κB were remarkably increased in the LPS-stimulated RAW264.7 macrophages, and XQEO reduced the phosphorylation and the expression of the nuclear protein NF-κB in a concentration-dependent manner. Note that Akt has also been reported to target TLR signalling following LPS-stimulation and primes the NF-κB activation.41 Our results indicate that XQEO significantly down-regulates the LPS-induced elevation of phosphorylated Akt; this may, at least partially, contribute to the inhibitory effects of XQEO on NF-κB signalling. Overall, these results indicate that XQEO could potentially inhibit the activation of the NF-κB signalling, thereby inhibiting the expression of pro-inflammatory mediators.
It has been reported that MAPKs are mainly composed of three characteristic subfamilies, i.e. ERK1/2, JNK and p38 MAPK, and regulate the synthesis of inflammatory mediators (MCP-1, MIP1α, iNOS, COX-2, etc.) at the level of transcription and translation through the activation of the NF-κB and AP-1 transcription factors.20,23,30 Our results revealed that the phosphorylation of MAPKs (p38, ERK, and JNK) was elevated in the LPS-stimulated RAW264.7 macrophages, and XQEO markedly inhibited the phosphorylation of these three kinases in a concentration-dependent manner. Moreover, the nuclear translocation and phosphorylation of AP-1 (c-Jun) were inhibited by the XQEO treatment; this showed that the suppressive effect on the AP-1 signalling also contributed to the anti-inflammatory activity of XQEO.
In addition to NF-κB and AP-1, IRF3 is another major transcription factor that is responsible for the induction of type-I interferon production in LPS-activated RAW264.7 macrophages.42 After LPS binds to TLR4, it activates the phosphorylation of TBK1 and further phosphorylates IRF3. The phosphorylated IRF3 is released and translocated into the nucleus to regulate the transcription of target genes including IFN-β and Rantes.43 Our data showed that the XQEO treatment negatively influenced the phosphorylated protein levels of IRF3 as well as the phosphorylation of the upstream protein TBK1 in the LPS-activated RAW264.7 cells. These results indicate that the down-regulation of IRF3 signalling partially contributes to the inhibitory effects of XQEO on the production of inflammatory mediators.
In conclusion, the suppressive effects of XQEO on the production of inflammatory mediators were investigated in this study using an LPS-stimulated macrophages model. It was shown that XQEO notably inhibited the production of inflammatory mediators including NO, PGE2, TNF-α, IL-6. The underlying mechanism of this action may be related to the inhibitory effect of XQEO on NF-κB, AP-1, and IRF3 signalling in the LPS-activated RAW264.7 macrophages, as summarized in Fig. 8. These findings provide a molecular insight into the effect of XQEO on inflammation and suggest that XQEO can be developed as a novel therapeutic agent for inflammatory diseases; further research should focus on validating XQEO as an anti-inflammatory agent in animal models.
Author contributions
Conceptualization: Yi Zhang, formal analysis: Gan Luo, Jing Kong, Brian Chi-Yan Cheng, Hui Zhao and Shuo-Feng Zhang, funding acquisition: Yi Zhang, investigation: Gan Luo, Li-Shan Yan, Yu Ding, Yan-Ling Liu and Yi Zhang, methodology: Jing Kong, project administration: Yi Zhang, software: Xiu Qiong Fu, writing – original draft, Gan Luo and Yi Zhang, writing – review & editing, Brian Chi-Yan Cheng and Si-Yuan Pan.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
This study was supported by the National Natural Science Foundation of China (81803793) and the Fundamental Research Funds for the Central Universities (2017-JYB-JS-057, 2018-JYBZZ-XJSJJ008).References
- W. Liao, X. He, Z. Yi, W. Xiang and Y. Ding, Biomed. Pharmacother., 2018, 107, 1151–1159 CrossRef CAS PubMed.
- L. Meng, L. Li, S. Lu, K. Li, Z. Su, Y. Wang, X. Fan, X. Li and G. Zhao, Mol. Immunol., 2018, 94, 7–17 CrossRef CAS PubMed.
- M. Zhou, W. Xu, J. Wang, J. Yan, Y. Shi, C. Zhang, W. Ge, J. Wu, P. Du and Y. Chen, EBioMedicine, 2018, 35, 345–360 CrossRef PubMed.
- Q. Shi, J. Cao, L. Fang, H. Zhao, Z. Liu, J. Ran, X. Zheng, X. Li, Y. Zhou, D. Ge, H. Zhang, L. Wang, Y. Ran and J. Fu, Int. Immunopharmacol., 2014, 20, 298–306 CrossRef CAS PubMed.
- S. Wei, D. Yang, J. Yang, X. Zhang, J. Zhang, J. Fu, G. Zhou, H. Liu, Z. Lian and H. Han, Eur. J. Cell Biol., 2019, 98, 36–50 CrossRef CAS PubMed.
- X. Chen, X. Zheng, M. Zhang, H. Yin, K. Jiang, H. Wu, A. Dai and S. Yang, Inflammation Res., 2018, 67, 903–911 CrossRef CAS PubMed.
- F. Chen, X. Zhu, Z. Sun and Y. Ma, Front. Pharmacol., 2018, 9, 1187 CrossRef PubMed.
- Z. Belshaw, L. Asher and R. S. Dean, Prev. Vet. Med., 2016, 131, 121–126 CrossRef PubMed.
- J. R. Perkins, M. Sanak, G. Canto, M. Blanca and J. A. Cornejo-Garcia, Trends Pharmacol. Sci., 2015, 36, 172–180 CrossRef CAS PubMed.
- S. Takayama and K. Iwasaki, Geriatr. Gerontol. Int., 2017, 17, 679–688 CrossRef PubMed.
- R. Deng, F. Li, H. Wu, W. Y. Wang, L. Dai, Z. R. Zhang and J. Fu, Front. Pharmacol., 2018, 9, 105 CrossRef PubMed.
- X. M. Li and L. Brown, J. Allergy Clin. Immunol., 2009, 123, 297–306 CrossRef PubMed.
- L. Liu and Z. W. Liu, Essentials of Chinese Medicine, Springer, Dordrecht, Heidelberg, London, New York, 2009, 2, pp. 417–418 Search PubMed.
- L. Zhou, Q. Zhang, W. Qi, S. Yan, J. Qu, T. Makino and D. Yuan, J. Sep. Sci., 2017, 40, 3582–3592 CrossRef CAS PubMed.
- H. W. Kim, C. Y. Lim, B. Y. Kim and S. I. Cho, Pharmacogn. Mag., 2014, 10, S506–S511 CrossRef PubMed.
- S. T. Kao, S. D. Wang, J. Y. Wang, C. K. Yu and H. Y. Lei, Allergy, 2000, 55, 1127–1133 CrossRef CAS PubMed.
- G. A. Nogueira de Melo, R. Grespan, J. P. Fonseca, T. O. Farinha, E. L. da Silva, A. L. Romero, C. A. Bersani-Amado and R. K. Cuman, J. Nat. Med., 2011, 65, 241–246 CrossRef CAS PubMed.
- J. L. Funk, J. B. Frye, J. N. Oyarzo, J. Chen, H. Zhang and B. N. Timmermann, PharmaNutrition, 2016, 4, 123–131 CrossRef CAS PubMed.
- Z. Rao, F. Xu, T. Wen, F. Wang, W. Sang and N. Zeng, Biomed. Pharmacother., 2018, 101, 304–310 CrossRef CAS PubMed.
- Y. Zhang, B. Chi-Yan Cheng, R. Xie, B. Xu, X. Y. Gao and G. Luo, RSC Adv., 2019, 9, 8912–8925 RSC.
- Y. M. Ma, X. Z. Zhang, Z. Z. Su, N. Li, L. Cao, G. Ding, Z. Z. Wang and W. Xiao, J. Ethnopharmacol., 2015, 173, 91–99 CrossRef PubMed.
- X. Chen, K. Mu and D. D. Kitts, Food Chem., 2019, 271, 248–258 CrossRef CAS PubMed.
- G. Luo, B. C. Cheng, H. Zhao, X. Q. Fu, R. Xie, S. F. Zhang, S. Y. Pan and Y. Zhang, Molecules, 2018, 23, 3319 CrossRef PubMed.
- A. C. Rivas da Silva, P. M. Lopes, M. M. Barros de Azevedo, D. C. Costa, C. S. Alviano and D. S. Alviano, Molecules, 2012, 17, 6305–6316 CrossRef PubMed.
- J. Koudou, A. A. Abena, P. Ngaissona and J. M. Bessiere, Fitoterapia, 2005, 76, 700–703 CrossRef CAS PubMed.
- B.-S. Kim, T.-C. Jeong, S.-Y. Choe and K.-H. Yang, Toxicol. Res., 1992, 8, 191–203 Search PubMed.
- F. Tang, F. Chen, X. Ling, Y. Huang, X. Zheng, Q. Tang and X. Tan, Mediators Inflammation, 2015, 2015, 463530 Search PubMed.
- M. Mitoshi, I. Kuriyama, H. Nakayama, H. Miyazato, K. Sugimoto, Y. Kobayashi, T. Jippo, K. Kuramochi, H. Yoshida and Y. Mizushina, Int. J. Mol. Med., 2014, 33, 1643–1651 CrossRef CAS PubMed.
- L. Zou, Y. Zhang, W. Li, J. Zhang, D. Wang, J. Fu and P. Wang, Molecules, 2017, 22, 1446 CrossRef PubMed.
- Y. Zhang, H. Guo, B. C. Cheng, T. Su, X. Q. Fu, T. Li, P. L. Zhu, K. W. Tse, S. Y. Pan and Z. L. Yu, Drug Des., Dev. Ther., 2018, 12, 2731–2748 CrossRef PubMed.
- D. S. Chi, M. Qui, G. Krishnaswamy, C. Li and W. Stone, Nitric Oxide, 2003, 8, 127–132 CrossRef CAS PubMed.
- J. Y. Park, M. H. Pillinger and S. B. Abramson, Clin. Immunol., 2006, 119, 229–240 CrossRef CAS PubMed.
- B. Samuelsson, R. Morgenstern and P.-J. Jakobsson, Pharmacol. Rev., 2007, 59, 207–224 CrossRef CAS PubMed.
- H. H. Chang and E. J. Meuillet, Future Med. Chem., 2011, 3, 1909–1934 CrossRef CAS PubMed.
- B. R. Lauwerys and F. A. Houssiau, Adv. Exp. Med. Biol., 2003, 520, 237–251 CrossRef CAS PubMed.
- S. T. Holgate, K. S. Bodey, A. Janezic, A. J. Frew, A. P. Kaplan and L. M. Teran, Am. J. Respir. Crit. Care Med., 1997, 156, 1377–1383 CrossRef CAS PubMed.
- T. Iwamoto, H. Okamoto, Y. Toyama and S. Momohara, FEBS J., 2008, 275, 4448–4455 CrossRef CAS PubMed.
- M. C. Dessing, K. F. van der Sluijs, S. Florquin and T. van der Poll, Clin. Immunol., 2007, 125, 328–336 CrossRef CAS PubMed.
- M. Karin and Y. Ben-Neriah, Annu. Rev. Immunol., 2000, 18, 621–663 CrossRef CAS PubMed.
- B. C. Cheng, X. Q. Ma, H. Y. Kwan, K. W. Tse, H. H. Cao, T. Su, X. Shu, Z. Z. Wu and Z. L. Yu, J. Ethnopharmacol., 2014, 153, 922–927 CrossRef PubMed.
- O. Nidai Ozes, L. D. Mayo, J. A. Gustin, S. R. Pfeffer, L. M. Pfeffer and D. B. Donner, Nature, 1999, 401, 82 CrossRef PubMed.
- B. C. Cheng, H. Yu, H. Guo, T. Su, X. Q. Fu, T. Li, H. H. Cao, A. K. Tse, Z. Z. Wu, H. Y. Kwan and Z. L. Yu, Sci. Rep., 2016, 6, 20042 CrossRef CAS PubMed.
- Y. Kim, H. J. Shim, S. Y. Kim, S. Heo and H. S. Youn, Int. Immunopharmacol., 2018, 64, 1–9 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2019 |