Open Access Article
Open Access ArticleCu(II)/Ni(II)–organic frameworks constructed from the homometallic clusters by 5-(2-carboxyphenoxy)isophthalic acid and N-ligand: synthesis, structures and visible light-driven photocatalytic properties†
Qi-Wei Xu,
Qiu-Shuang Wang,
Shan-Shan Li and
Xia Li *
*
Department of Chemistry, Capital Normal University, Beijing, 100048, China. E-mail: xiali@cnu.edu.cn; Fax: +86 1068902320; Tel: +86 1068902320
First published on 24th May 2019
Abstract
Four new complexes, namely, Cu2(O-cpia)(btb)0.5·(OH) (1), Cu3(O-cpia)2(bpy)2 (2), [Ni2(O-cpia)(phen)·(OH)·H2O]·2H2O (3) and [Ni3(O-cpia)2(bpy)3·2H2O]·2H2O (4) (O-cpia = 5-(2-carboxyphenoxy)isophthalic acid, btb = 1,4-bis(1,2,4-triazol-1-yl)butane, bpy = 4,4′-bipyridine) were successfully isolated under hydrothermal conditions. The four complexes exhibit different architectures constructed from different homometallic clusters varying from mononuclear, binuclear to tetranuclear metal(II) polyhedra as Second Building Blocks (SBUs). 1 features a 3D framework constructed from the tetranuclear clusters [Cu4(μ3-OH)2] as SBUs, linked with Cu(1)O4N and Cu(2)O5 polyhedra by O-cpia/btb mixed linkers. 2 also exhibits a 3D structure based on trinuclear clusters [Cu3(COO)4] SBUs, bridged with Cu(1)O3N2 and Cu(2)O4 polyhedra via O-cpia/bpy mixed ligands. 3 shows a 2D network consisting of tetranuclear clusters [Ni4(μ3-OH)2] SBUs, which are bridged with Ni(1)O4N2 and Ni(2)O6 through O-cpia ligands. It is worth noting that 4, with a 3D structure, is generated from the binuclear clusters [Ni2(COO)4] (Ni(1)O4N) and mononuclear metal Ni(2) cores (Ni(2)O4N2) as SBUs, and bridged by O-cpia/bpy mixed ligands. Meanwhile, the degradation of dyes (RhB) by the complexes under visible light irradiation was studied. 1–4 are semiconducting in nature, with Eg of 1.30 eV (1), 1.78 eV (2), 2.85 eV (3) and 2.14 eV (4). Cu(II) complexes 1 and 2 are highly efficient photocatalysts for the degradation of RhB under visible light irradiation.
Introduction
Metal–organic frameworks (MOFs) are prepared by the self-assembly of metal ions or clusters with organic ligands. A variety of structural types have been reported, such as one-, two- and three- dimensional structures, which are based on M–O–M, homometallic clusters as Second Building Blocks (SBUs).1–4 MOFs, with interesting architectures, also have good physicochemical properties, which can be potentially applied to many fields such as catalysis, luminescence, gas adsorption, magnetic materials, etc.3,5–10 In the past few decades, MOFs have attracted wide attention. Transition metal cations have various coordination geometries which can be used in various crystal architectures.6,11,12 For example, Zn(II), Cd(II), Cu(II), Ni(II), and Co(II), with different electron configurations and coordination numbers, may form compounds in various architectures, which can be applied to catalysis and luminescence.13–18 Among them, Cu(II) and Ni(II) complexes have shown to be effective chemical photocatalysts under visible light irradiation.19–22 The development of MOFs has provided more opportunities for new photocatalysts, because of their diverse structures containing variable valence electrons and empty orbits of the metal center, high thermal and chemical stability.23,24 Recently, MOFs have been demonstrated with a good photocatalytic activity for the degradation of organic dyes such as methyl orange, methylene blue and rhodamine B (RhB) under visible light irradiation.15,19,25–29 Due to the high importance of environment protection, how to effectively remove organic pollutants has become a hot social issue, leading to high demands for high-efficiency photocatalysts.We are interested in synthesizing new MOFs with diverse structures, and in studying their photocatalytic properties. Metal polyhedra in hybrid frameworks with polycarboxylate ligands and N-ligands have led to numerous examples of MOFs materials with interesting structures and remarkable physical properties. This has promotes the synthesis of new MOFs using a tricarboxylate ligand with N-donor co-ligands to construct new MOFs with Cu(II)/Ni(II) metal nodes and the study of their catalytic performance. 5-(2-Carboxyphenoxy)isophthalic acid (O–H3cpia), semi-rigid ligand is composed of tricarboxylate groups, which has diverse binding capability as bridging ligand by multiple coordination modes, leading to interesting MOFs. Some studies have been reported regarding the synthesis and structural analysis of MOFs with O–H3cpia.30–33 N-donor molecules are important building blocks in crystal engineering because of their ability of coordinating to metal centres and forming networks.34 A number of N-donor ligands have been used for the synthesis of MOFs with carboxylic acids to tune the dimensionality of the MOFs.35,36 Phen has high affinities for metal ions and has been used to prepare complexes with excellent photophysical and chemical properties.37,38 Neutral N-ligands, such as 4,4′-bipyridine (bpy)39,40 and 1,4-bis(1,2,4-triazol-1-yl)butane (btb),41,42 can pillar metal-carboxylate coordination polymer motifs into higher dimensions structures. In addition, the btb flexible ligand can adopt TTT, GTG, and GTT conformations in N4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N4′-mode and act as a bridge between two central metal atoms to form new and unpredictable structural motifs.
N4′-mode and act as a bridge between two central metal atoms to form new and unpredictable structural motifs.
Driven by the interest to synthesize new MOFs with diverse structures, and to study their photocatalytic properties, the O–H3cpia and phen/bpy/btb ligands were introduced in the reaction system of Cu(II) and Ni(II) metal salt in H2O solvent, and new complexes, i.e., Cu2(O-cpia)(btb)0.5·(OH) (1), Cu3(O-cpia)2(bpy)2 (2), [Ni2(O-cpia)(phen)·(OH)·H2O]·2H2O (3) and [Ni3(O-cpia)2(bpy)3·2H2O]·2H2O (4) were synthesized. We also studied the structures and ability of the complexes as photocatalysts for the degradation of the organic dyes RhB. Interestingly, these new compounds show different interesting architectures based on the different homometallic clusters varying from mononuclear, binuclear to tetranuclear clusters as Second Building Blocks (SBUs) by different organic linkers. The catalytic properties of the complexes to the degradation of rhodamine B (RhB) organic pollutant under visible light irradiation were studied as well. Complexes 1 and 2 are proved to be highly efficient photocatalysts for the degradation of the RhB under visible light irradiation.
Experimental
Materials and methods
All experimental materials are commercially available and could be used as received without further purification. Elemental analyses (C, H, and N) were run on an Elementar Vario EL elemental analyzer. Powder X-ray diffraction (PXRD) was carried out on a PANaytical X'Pert PRO MPD diffractometer for Cu Kα radiation (λ = 1.5406 Å). Infrared (IR) spectra were recorded on a Bruker Tensor 37 spectrophotometer using the KBr pellets in the range 4000–400 cm−1. The thermogravimetric analyses (TGA) were performed on a HCT-2 thermal analyzer under air atmosphere from the temperature of 25 to 800 °C at a heating rate of 10 °C min−1. Solid UV-visible spectra were obtained on a Japan Shimadzu UV-2600 spectrophotometer. UV-vis spectra were recorded on a 2550 UV-vis spectrophotometer (Shimadzu, Japan) at room temperature.Synthetic procedures
Crystallography
The X-ray single crystal data collection for complexes 1–4 was performed on a Bruker Smart Apex II CCD diffractometer equipped with graphite monochromated Mo Kα radiation (λ = 0.71073 Å) at 293(2) K. Absorption corrections were applied using the program SADABS. The structures were solved by direct methods and refined by a full matrix least squares method on F2 using SHELXS 97 and SHELXL 97 programs.43,44 The hydrogen atoms were generated geometrically and treated by a mixture of independent and constrained refinement. The crystallographic data are summarized in Table 1, while selected bond lengths and angles are given in Table S1.† More details of the crystal structures could be known from the Cambridge Crystallographic Data Centre.| Complex | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Empirical formula | C19H14N3O8Cu2 | C50H30N4O14Cu3 | C27H22N2O11Ni2 | C60H46N6O18Ni3 |
| Formula weight | 539.41 | 1101.4 | 667.89 | 1315.16 |
| Crystal system | Triclinic | Triclinic | Triclinic | Triclinic |
| Space group | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
| a (Å) | 9.5272(4) | 10.0683(8) | 11.1024(8) | 8.034(2) |
| b (Å) | 10.6671(5) | 10.8967(9) | 11.5542(9) | 9.981(2) |
| c (Å) | 11.6450(5) | 11.0913(9) | 12.4520(9) | 18.072(4) |
| α (°) | 100.8070(10) | 89.393(3) | 72.963(2) | 87.730(8) |
| β (°) | 98.5420(10) | 85.366(2) | 72.622(2) | 83.662(8) |
| γ (°) | 115.0540(10) | 64.932(2) | 67.195(2) | 76.257(8) |
| Volume (Å3) | 1017.95(8) | 1098.24(15) | 1376.45(18) | 1398.9(5) |
| Z | 2 | 1 | 2 | 1 |
| Calculated density/Mg m−3 | 1.760 | 1.665 | 1.611 | 1.561 |
| Absorption coefficient/mm−1 | 2.143 | 1.518 | 1.433 | 1.082 |
| F(000) | 542 | 557 | 684 | 676 |
| Crystal size/mm3 | 0.176 × 0.116 × 0.114 | 0.268 × 0.103 × 0.054 | 0.347 × 0.313 × 0.122 | 0.317 × 0.048 × 0.045 |
| Theta range for data collection/(°) | 3.03 to 25.01 | 3.02 to 25.01 | 2.95 to 25.01 | 2.95 to 25.01 |
| Limiting indices | −11 ≤ h ≤ 11 | −11 ≤ h ≤ 11 | −13 ≤ h ≤ 13 | −9 ≤ h ≤ 9 |
| −12 ≤ k ≤ 12 | −12 ≤ k ≤ 12 | −13 ≤ k ≤ 13 | −11 ≤ k ≤ 11 | |
| −13 ≤ l ≤ 13 | −13 ≤ l ≤ 13 | −14 ≤ l ≤ 14 | −21 ≤ l ≤ 21 | |
| Reflections collected/unique | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 268/3582 [R(int) = 0.0741] 268/3582 [R(int) = 0.0741] |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 635/3866 [R(int) = 0.0765] 635/3866 [R(int) = 0.0765] |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 992/4836 [R(int) = 0.0713] 992/4836 [R(int) = 0.0713] |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 796/4929 [R(int) = 0.1398] 796/4929 [R(int) = 0.1398] |
| Data/restraints/parameters | 3582/20/297 | 3866/0/322 | 4836/27/390 | 4929/7/398 |
| Goodness-of-fit on F2 | 1.032 | 1.022 | 1.047 | 0.996 |
| Final R indices [I > 2σ(I)] | R1 = 0.0355, wR2 = 0.0901 | R1 = 0.0403 wR2 = 0.0919 | R1 = 0.0355 wR2 = 0.0955 | R1 = 0.0560 wR2 = 0.0921 |
| R Indices (all data) | R1 = 0.0473 wR2 = 0.0966 | R1 = 0.0578 wR2 = 0.1000 | R1 = 0.0388 wR2 = 0.1016 | R1 = 0.1268 wR2 = 0.1124 |
| Largest diff. peak and hole/e Å−3 | 1.191 and −0.528 | 0.465 and −0.444 | 0.670 and −0.453 | 0.448 and −0.498 |
Results and discussion
Structural description of complexes
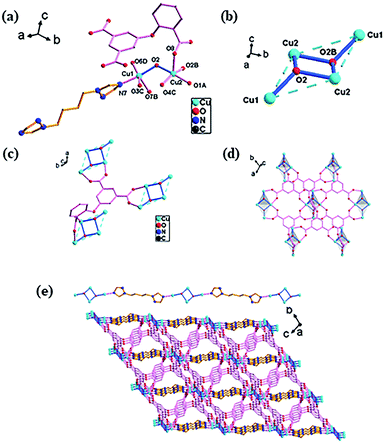
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group and has a 3D framework structure. It can be seen from Fig. 1a that the asymmetric unit consists of two crystallographically independent Cu(II) ions, one O-cpia ligand, half a btb ligand and a hydroxide ion. The copper center is in the five-coordinated geometry of CuO4N for Cu1 and CuO5 for Cu2, forming a square-pyramidal environment. Cu1 atom is coordinated with three oxygen atoms (O7B, O3C, O6D) from three O-cpia ligands, one oxygen atom (O2) from the hydroxide ion and one nitrogen atom (N7) from a btb ligand. The Cu1–O bond lengths are in the range of 1.949(2) to 2.128(3) Å while the bond angles of O–Cu–O/N are in the range of 84.37(11)° to 172.00(11)°. Cu2 is completed by three oxygen atoms (O1A, O4C, O8) from O-cpia ligands, and two oxygen atoms from hydroxide ions(O2, O2B) with the Cu–O distances ranging from 1.944(2) to 2.135(2) Å and the bond angles of O–Cu2–O ranging from 84.38(10)° to 175.13(11)°. The OH group as 3-connected node (μ3-OH) connects the Cu atoms to generate a tetranuclear cluster [Cu4(μ3-OH)2], which is in the shape of a chair (Fig. 1b). In the tetranuclear unit, the four Cu(II) atoms are coplanar while the distance of Cu1–Cu2 is 3.4461(7) Å, Cu2–Cu2 is 3.0405(5) Å, and Cu1–Cu1 is 6.038 Å. In 1, the O-cpia acting as a μ6-bridging ligand adopts μ2:η1η1/μ2:η1η1/μ2:η1η1 coordination mode and links six Cu atoms. As shown in Fig. 1c, each O-cpia bridges three tetranuclear [Cu4(μ3-OH)2] units in turn and each [Cu4(μ3-OH)2] unit is connected to six surrounding units and expands infinitely into the Cu–O-cpia network (Fig. 1d). The btb ligand adopts the TTT conformation with bidentate coordination mode to link Cu1 atoms to form a Cu1-btb chain. Thus, the [Cu4(μ3-OH)2] units as SBUs, which are bridged by O-cpia and btb mixture ligands, finally resulting in a 3D framework (Fig. 1e).
space group and has a 3D framework structure. It can be seen from Fig. 1a that the asymmetric unit consists of two crystallographically independent Cu(II) ions, one O-cpia ligand, half a btb ligand and a hydroxide ion. The copper center is in the five-coordinated geometry of CuO4N for Cu1 and CuO5 for Cu2, forming a square-pyramidal environment. Cu1 atom is coordinated with three oxygen atoms (O7B, O3C, O6D) from three O-cpia ligands, one oxygen atom (O2) from the hydroxide ion and one nitrogen atom (N7) from a btb ligand. The Cu1–O bond lengths are in the range of 1.949(2) to 2.128(3) Å while the bond angles of O–Cu–O/N are in the range of 84.37(11)° to 172.00(11)°. Cu2 is completed by three oxygen atoms (O1A, O4C, O8) from O-cpia ligands, and two oxygen atoms from hydroxide ions(O2, O2B) with the Cu–O distances ranging from 1.944(2) to 2.135(2) Å and the bond angles of O–Cu2–O ranging from 84.38(10)° to 175.13(11)°. The OH group as 3-connected node (μ3-OH) connects the Cu atoms to generate a tetranuclear cluster [Cu4(μ3-OH)2], which is in the shape of a chair (Fig. 1b). In the tetranuclear unit, the four Cu(II) atoms are coplanar while the distance of Cu1–Cu2 is 3.4461(7) Å, Cu2–Cu2 is 3.0405(5) Å, and Cu1–Cu1 is 6.038 Å. In 1, the O-cpia acting as a μ6-bridging ligand adopts μ2:η1η1/μ2:η1η1/μ2:η1η1 coordination mode and links six Cu atoms. As shown in Fig. 1c, each O-cpia bridges three tetranuclear [Cu4(μ3-OH)2] units in turn and each [Cu4(μ3-OH)2] unit is connected to six surrounding units and expands infinitely into the Cu–O-cpia network (Fig. 1d). The btb ligand adopts the TTT conformation with bidentate coordination mode to link Cu1 atoms to form a Cu1-btb chain. Thus, the [Cu4(μ3-OH)2] units as SBUs, which are bridged by O-cpia and btb mixture ligands, finally resulting in a 3D framework (Fig. 1e).
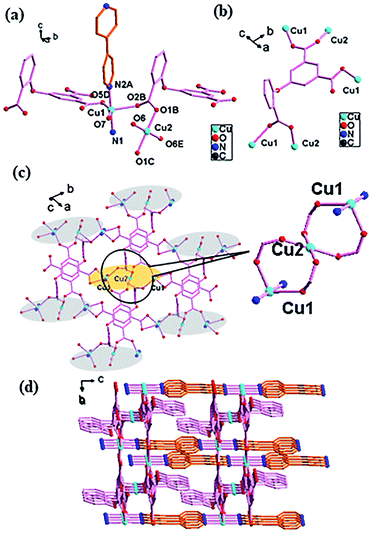
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group with one asymmetric unit containing two crystallographically independent Cu(II) atoms (CuO3N2 for Cu1, CuO4 for Cu2), one O-cpia ligand and one bpy ligand. Cu1 is five-coordinated with three oxygen atoms (O2B, O5D, O7) from O-cpia ligands, and two nitrogen atoms (N1 and N2) of bpy ligand, forming a square-pyramidal geometry. Cu2 is four-coordinated with four O atoms (O1B, O1C, O6, O6E) from four O-cpia ligands in a planar quadrilateral configuration, while the angles of O–Cu2–O are 180.00(19)°, 90.16(13)°, 89.84(13)°, 89.84(13)°, 90.16(13)° and 180.00(16)° (Fig. 2a). The Cu–O/N bond lengths are comparable to those in 1, that the bond lengths of Cu–O are in the range of 1.906(2)–2.252(2) Å, and the bond lengths of Cu–N are 1.993(2) and 2.036(3) Å. The difference is in 2, the O-cpia adopts μ2:η1η1/μ1:η1η0/μ2:η1η1 coordination mode as a μ5-bridging ligand linking five Cu(II) atoms, which can be seen in Fig. 2b. The adjacent three Cu atoms (Cu1, Cu2 and Cu1) are connected by COO groups, forming a trinuclear clusters [Cu3(COO)4] with the Cu1–Cu2 distance of 3.6636(5) Å. The [Cu3(COO)4] clusters as SUBs are connected by O-cpia ligands to Cu–O-cpia network parallel to the ab plane (Fig. 2c). The bpy ligand adopts bidentate coordination mode and links Cu1 atom, forming a Cu1-bpy chain with the Cu1–Cu1 distance of 11.0913(10) Å along the a axis. As shown in Fig. 2d, the Cu–O-cpia network interweave with Cu1-bpy chains, while the trinuclear clusters [Cu3(COO)4] act as SBUs, forming a 3D framework.
space group with one asymmetric unit containing two crystallographically independent Cu(II) atoms (CuO3N2 for Cu1, CuO4 for Cu2), one O-cpia ligand and one bpy ligand. Cu1 is five-coordinated with three oxygen atoms (O2B, O5D, O7) from O-cpia ligands, and two nitrogen atoms (N1 and N2) of bpy ligand, forming a square-pyramidal geometry. Cu2 is four-coordinated with four O atoms (O1B, O1C, O6, O6E) from four O-cpia ligands in a planar quadrilateral configuration, while the angles of O–Cu2–O are 180.00(19)°, 90.16(13)°, 89.84(13)°, 89.84(13)°, 90.16(13)° and 180.00(16)° (Fig. 2a). The Cu–O/N bond lengths are comparable to those in 1, that the bond lengths of Cu–O are in the range of 1.906(2)–2.252(2) Å, and the bond lengths of Cu–N are 1.993(2) and 2.036(3) Å. The difference is in 2, the O-cpia adopts μ2:η1η1/μ1:η1η0/μ2:η1η1 coordination mode as a μ5-bridging ligand linking five Cu(II) atoms, which can be seen in Fig. 2b. The adjacent three Cu atoms (Cu1, Cu2 and Cu1) are connected by COO groups, forming a trinuclear clusters [Cu3(COO)4] with the Cu1–Cu2 distance of 3.6636(5) Å. The [Cu3(COO)4] clusters as SUBs are connected by O-cpia ligands to Cu–O-cpia network parallel to the ab plane (Fig. 2c). The bpy ligand adopts bidentate coordination mode and links Cu1 atom, forming a Cu1-bpy chain with the Cu1–Cu1 distance of 11.0913(10) Å along the a axis. As shown in Fig. 2d, the Cu–O-cpia network interweave with Cu1-bpy chains, while the trinuclear clusters [Cu3(COO)4] act as SBUs, forming a 3D framework.
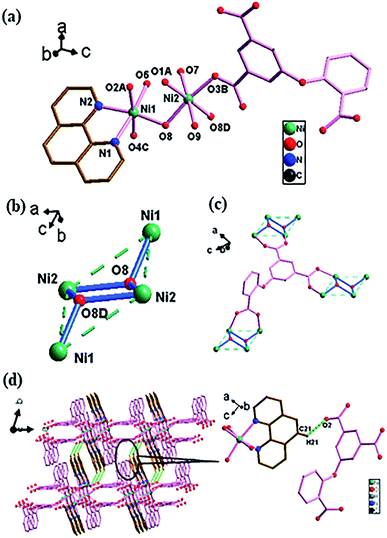
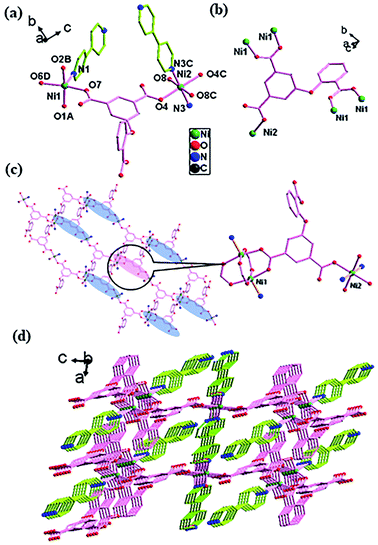
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group (Fig. 4). There are two crystallographically independent Ni(II) atoms (NiO4N for Ni1 and NiO4N2 for Ni2) in an asymmetric unit, with one O-cpia ligand, one and a half of bpy ligands, one coordinated water molecule and free water molecules. The Ni1 is surrounded by four oxygen atoms (O1A, O2B, O6D and O7) from four O-cpia ligands and one nitrogen atom (N1) of one bpy ligand, forming a square-pyramidal geometry. The Ni2 is attached with two oxygen atoms (O4, O4C) from two O-cpia ligands, two oxygen atoms (O8, O8C) from coordinated water molecules, and two nitrogen atoms (N3, N3C) of 4,4′-bpy ligands, forming a octahedral geometries (Fig. 4a). The Ni–O/N bond lengths are similar to that in 3, with the bond lengths of Ni–O in the range of 1.997(3)–2.084(3) Å and the lengths of Ni–N in the range of 2.012(3)–2.070(4) Å. The bond angles of O–Ni–O/N are between 88.04(14)° and 167.36(12)°. As shown in Fig. 4b, the O-cpia acts as μ5-bridging ligand in μ2:η1η1/μ2:η1η1/μ1:η1η0 coordination mode, linking five Ni atoms to form Ni–O-cpia network (Fig. 4c). The two adjacent Ni1 atoms are connected by COO groups, forming a binuclear cluster [Ni2(COO)4] with the Ni1–Ni1 distance of 2.6602(12) Å. Different from complex 2, in 4, bpy ligands adopt two kinds of coordination modes. One bpy as terminal ligand coordinates with Ni1 atom by its one N atom, the other bpy as bidentate-bridging ligand connects two Ni2 atoms by its two N atoms to form Ni2-bpy chain with the Ni2–Ni2 distance of 11.2279 Å along a axis. The two kinds of bpy ligands are almost perpendicular. Thus, a 3D architecture, as shown in Fig. 4d, is built by the bpy ligands between Ni–O-cpia networks, in which the [Ni2(COO)4] binuclear clusters and mononuclear metal Ni2 cores act as SUBs.
space group (Fig. 4). There are two crystallographically independent Ni(II) atoms (NiO4N for Ni1 and NiO4N2 for Ni2) in an asymmetric unit, with one O-cpia ligand, one and a half of bpy ligands, one coordinated water molecule and free water molecules. The Ni1 is surrounded by four oxygen atoms (O1A, O2B, O6D and O7) from four O-cpia ligands and one nitrogen atom (N1) of one bpy ligand, forming a square-pyramidal geometry. The Ni2 is attached with two oxygen atoms (O4, O4C) from two O-cpia ligands, two oxygen atoms (O8, O8C) from coordinated water molecules, and two nitrogen atoms (N3, N3C) of 4,4′-bpy ligands, forming a octahedral geometries (Fig. 4a). The Ni–O/N bond lengths are similar to that in 3, with the bond lengths of Ni–O in the range of 1.997(3)–2.084(3) Å and the lengths of Ni–N in the range of 2.012(3)–2.070(4) Å. The bond angles of O–Ni–O/N are between 88.04(14)° and 167.36(12)°. As shown in Fig. 4b, the O-cpia acts as μ5-bridging ligand in μ2:η1η1/μ2:η1η1/μ1:η1η0 coordination mode, linking five Ni atoms to form Ni–O-cpia network (Fig. 4c). The two adjacent Ni1 atoms are connected by COO groups, forming a binuclear cluster [Ni2(COO)4] with the Ni1–Ni1 distance of 2.6602(12) Å. Different from complex 2, in 4, bpy ligands adopt two kinds of coordination modes. One bpy as terminal ligand coordinates with Ni1 atom by its one N atom, the other bpy as bidentate-bridging ligand connects two Ni2 atoms by its two N atoms to form Ni2-bpy chain with the Ni2–Ni2 distance of 11.2279 Å along a axis. The two kinds of bpy ligands are almost perpendicular. Thus, a 3D architecture, as shown in Fig. 4d, is built by the bpy ligands between Ni–O-cpia networks, in which the [Ni2(COO)4] binuclear clusters and mononuclear metal Ni2 cores act as SUBs.
Power X-ray diffraction (PXRD) and thermal analysis
In order to confirm the phase purity, these complexes were checked by PXRD analysis. It can be seen that experimental PXRD patterns agree with the simulated PXRD result very well. The result indicates that pure crystals have been successfully synthesized (Fig. S1†). To examine the thermal stabilities of the complexes, thermogravimetric analysis (TGA) were employed at a heating rate of 10 °C min−1 in air atmosphere. The results are shown in Fig. S2.† It can be seen that complexes 1 and 2 are stable up to 282 and 320 °C, respectively. With the increase of temperature, the organic ligands in the complexes decomposed. The residue can be attributed to CuO (for 1, calcd: 70.2%, found: 69.6%; for 2, calcd: 78.3%, found: 78.1%). For complex 3, a weight loss of 8.5% is observed from 51 to 172 °C, while for 3, a weight loss of 4.4% is observed between 58 and 201 °C. The weight loss can be attributed to the loss of the coordinated water molecules and lattice water molecules (calcd 8.1% for 3 and 5.4% for 4). The skeleton began to collapse at 299 °C for 3 and 324 °C for 4, and the residue may be NiO (calcd: 77.0%, found: 82.0% for 3 and calcd: 83.0%, found: 84.1% for 4).Photocatalytic activities
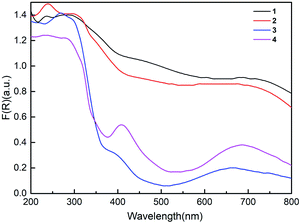
To explore the photocatalytic potential of the compounds, the solid UV diffuse reflectance spectra of the complexes were recorded at room temperature, as shown in Fig. 5. The spectra of these compounds show different absorption activities in the visible region from 200 to 800 nm. According to the equation (αhν = K(hν–Eg)1/n, where α is the absorption coefficient, hν is the discrete photo energy, K is a constant, Eg is the band gap energy, n equals 1/2 for indirect transition), the extrapolated values of hν at α = 0 give absorption edge energies (Eg) of complexes.45–47 The absorption edge energies are 1.30 eV for 1 and 1.78 eV for 2, which are lower than those for 3 (2.85 eV) and 4 (2.14 eV) in Fig. S3.† The band gap sizes of 1–4 indicate that they may have potential capacity for photocatalytic reactions.In recent years, coordination polymers, as a kind of photocatalyst, have attracted much attention due to their potential applications in purifying waste water under visible light irradiation by decomposing organic dyes. To study the photocatalytic activity of the complexes as catalysts, rhodamine B (RhB) was selected as a representative dye, and the test was carried under visible light illumination. For this experiment, 30 mg compound and 500 mL RhB (1.0 × 10−5 mol L−1) were mixed and stirred for half an hour to achieve the equilibrium condition in the dark environment. Then, 4 mL H2O2 was added under visible light irradiation at a temperature of 298.15 K. Equivalent samples from the reaction system were taken for the measurement every 10 min, and the degradation progress was monitored by observing the intensity of the characteristic absorption band of RhB at 553 nm. Blank experiments in the presence of H2O2 and the absence of the photocatalyst were also performed.
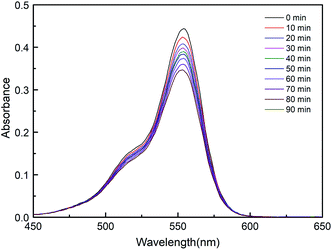
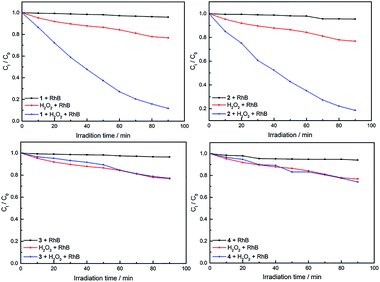
As can be seen in Fig. S4,† with the presence of compound only, the concentration of RhB shows very little reduction after 1.5 h, which is 4.2% (for 1), 4.6% (for 2), 3.6% (for 3) and 6.0% (for 4), respectively. Compared with the presence of compound and H2O2, different results for degradation was observed in the case of the complexes, as shown in Fig. 6. When using an equimolar amount of 1 as the catalyst for degradation of dichromate under the same conditions, 28%, 52%, 80% and 88% reduction of RhB were observed after 20, 40, 70 and 90 min, respectively. Similarly, when using 2 as the catalyst, 25%, 48%, 73% and 81% of RhB were degradated after 20, 40, 70 and 90 min, respectively. However, 3 and 4 have low photocatalytic efficiency. In the same time of 1.5 h, about 5.0% RhB (for 20 min), 9.0% (for 40 min), 19% (for 70 min) and 23% (for 90 min) are decomposed under visible light irradiation when 3 used as the catalyst. For 4, about 6.0% RhB (for 20 min), 11% (for 40 min), 20% (for 70 min) and 26% (for 90 min) are decomposed. The results indicate that 1 and 2 possess better photocatalytic efficiency than 3 and 4, as shown in Fig. 7.
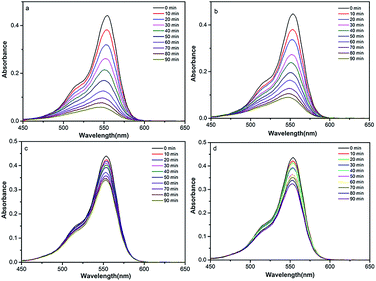 | ||
| Fig. 6 Absorption spectra of RhB degraded with complex and H2O2 (1 (a), 2 (b), 3 (c) and 4 (d)) under visible light illumination. | ||
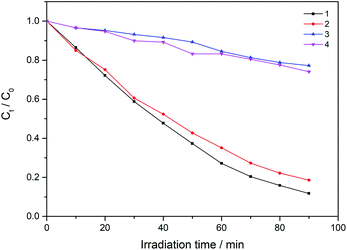 | ||
| Fig. 7 Change of absorption spectra of the solution of RhB with H2O2 and different complexes under visible light irradiation. Ct and C0 stand for the RhB concentrations after and before irradiation. | ||
In addition, the blank experiments of the degradation of organic pollutants were finished in the same reaction condition without using the addition of compound. As shown in Fig. 8, in the presence of only H2O2, the degradation of RhB are merely 23% after 90 min under visible light irradiation.
The photocatalytic behaviors of 1–4 for the degradation of RhB under visible light irradiation are summarized in Fig. 9. As can be seen, the catalytic efficiency for the degradation of RhB is 1 > 2 > 4 > 3. Interestingly, the order is opposite to that of their Eg (3 > 4 > 2 > 1). The similar relationship of Eg and catalytic rate has been reported previously.48 These results indicate that the Cu(II) complexes have a faster degradation rate for the RhB than the Ni(II) complex, which can be attributed to the nature of the metal cation used, that Cu(II) is a more active metal cation than Ni(II).49 Meanwhile, Cu(II) and Ni(II) have different electronic configurations and geometries, which may also influence the photocatalytic activity.50,51 In addition, based on the good catalytic performance of complexes 1–2, the catalytic performance of 1–2 for methylene blue (MB) and methyl orange (MO) was measured. The experimental results are shown in Fig. S5–S8.† When complexes 1 and 2 used as the catalyst, respectively, the 94% MB/61% MO and 89% MB/42% MO (90 min) were decomposed under visible light irradiation.
The reaction mechanism for dyes photodegradation has been discussed based on semiconductor theory.26,48,49,52 When the complex is irradiated by a xenon lamp, the electrons in the valence band are easily excited into the conduction band, which increases the amount of holes (h+) in the valence band. Then electrons may combine with O2 on the surface of the complex to form oxygen radical, which will further produce hydroxyl free (·OH). Meanwhile, hydrogen peroxide itself could also decompose to generate oxygen (O2) and hydroxide (OH−), the O2 and OH− present in the solution are adsorbed on the surface of the complex. At the same time, (OH−) adsorpted on the surface of the complex interacts with holes (h+), generating (·OH). As the most effective group for catalytically degrading organic dyes, (·OH) can degrade organic dyes into inorganic ions. Therefore, the synergistic catalysis of complexes 1, 2 and hydrogen peroxide improves the efficiency of photocatalytic degradation of RhB.
Four complexes derived from O-cpia, Cu(II)/Ni(II) and various co-N-ligands show several interesting structural features. In the frameworks, the tetranuclear clusters [Cu4(μ3-OH)2] and [Ni4(μ3-OH)2] (in 1 and 3), trinuclear cluster [Cu3(COO)4] (in 2), and the binuclear unit [Ni2(COO)4]/mononuclear metal (Ni2) unit (in 4) are found, which act as SBUs to be assembled into different framework structures by O-cpia and co-N-ligands. Moreover, each asymmetric unit contains two crystallographically independent metal(II) center atoms with different metal polyhedra. Ni(II) forms a the octahedral coordination geometry while Cu(II) forms square-pyramidal or planar quadrilateral geometry due to the larger radius of Ni(II) than that of Cu(II). O-cpia ligand exhibits different bridging coordination modes and acts as μ6-bridging (in 1 and 3) and μ5-bridging (in 2 and 4) ligand, respectively, while N-ligands, phen, bpy and btb are used to tune the dimensionality of the complexes. The bpy and btb ligands have bridging function to form high dimensional architectures (3D, 1, 2 and 4). When the terminal ligand phen is introduced into the system, the resulting complex 3 shows a 2D network structure. The auxiliary N-ligands play an important role in governing the formation of the final frameworks. Meanwhile, the degradation of RhB by the complexes under visible light irradiation were studied. 1–4 are semiconducting in nature, with Eg of 1.30 eV (1), 1.78 eV (2), 2.85 eV (3) and 2.14 eV (4). 1–2 are universal and highly efficient photocatalysts for the degradation of RhB under visible light irradiation. They are stable and can be easily separated from the reaction system for reuse. Photocatalytic activity of Cu(II) and Ni(II) complexes against dyes under visible light irradiation were studied and the Cu(II) complexes (1–2) exhibit a higher photocatalysts activity than the Ni(II) complexes (3–4).
Conclusions
In summary, four metal–organic frameworks, namely, Cu2(O-cpia)(btb)0.5·(OH) (1), Cu3(O-cpia)2(bpy)2 (2), [Ni2(O-cpia)(phen)·(OH)·H2O]·2H2O (3) and [Ni3(O-cpia)2(bpy)3·2H2O]·2H2O (4) were synthesized through hydrothermal reactions. Compounds 1, 2, and 4 possess different complicated 3D frameworks whereas 3 exhibits a 2D structure. They display different homometallic clusters varying from mononuclear, binuclear to tetranuclear metal(II) units as SBUs within the frameworks. O-cpia ligand exhibits bridging coordination modes for coordination polymer structure, while the structural characteristics of auxiliary N-ligands also play an important role in governing the structures of such complexes. 1 and 2 are universal and more efficient photocatalysts than 3 and 4 for the degradation of RhB under visible light irradiation. This work provides new information regarding polynuclear metal(II) chemistry and the MOFs prepared in this paper can be applied to the degradation of organic dye.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful to the Key Project of Science and Technology plan of Beijing Education Commission (KZ201910028038) and the National Natural Science Foundation of China (21471104).Notes and references
- Z. J. Lin, J. Lv, M. Hong and R. Cao, Chem. Soc. Rev., 2014, 43, 5867–5895 RSC.
- G. Maurin, C. Serre, A. Cooper and G. Ferey, Chem. Soc. Rev., 2017, 46, 3104–3107 RSC.
- J. Poater, M. Gimferrer and A. Poater, Inorg. Chem., 2018, 57, 6981–6990 CrossRef CAS PubMed.
- H. C. Zhou and S. Kitagawa, Chem. Soc. Rev., 2014, 43, 5415–5418 RSC.
- N. Stock and S. Biswas, Chem. Rev., 2012, 112, 933–969 CrossRef CAS PubMed.
- S. Seth, G. Savitha and J. N. Moorthy, Cryst. Growth Des., 2018, 18, 2129–2137 CrossRef CAS.
- G. Mínguez Espallargas and E. Coronado, Chem. Soc. Rev., 2018, 47, 533–557 RSC.
- Y. Wang, M. He, X. Gao, Y. Zhang, H. Zhong, P. Long, X. Wang and Y. He, Inorg. Chem. Front., 2018, 5, 2811–2817 RSC.
- J. Yu, L. H. Xie, J. R. Li, Y. Ma, J. M. Seminario and P. B. Balbuena, Chem. Rev., 2017, 117, 9674–9754 CrossRef CAS PubMed.
- E. A. Dolgopolova, A. M. Rice, C. R. Martin and N. B. Shustova, Chem. Soc. Rev., 2018, 47, 4710–4728 RSC.
- M. C. Bernini, A. E. Platero-Prats, N. Snejko, E. Gutiérrez-Puebla, A. Labrador, R. Sáez-Puche, J. Romero de Paz and M. A. Monge, CrystEngComm, 2012, 14, 5493 RSC.
- X.-L. Qu, X.-L. Zheng and X. Li, RSC Adv., 2016, 6, 69007–69015 RSC.
- T. Devic and C. Serre, Chem. Soc. Rev., 2014, 43, 6097–6115 RSC.
- Y. Q. Sun, J. C. Zhong, L. Ding and Y. P. Chen, Dalton Trans., 2015, 44, 11852–11859 RSC.
- Y. Qiao, Y. Ma, W. Jiang, X. Wang, W. Guan, G. Che, W. Li and F. Qin, CrystEngComm, 2018, 20, 7782–7794 RSC.
- Z. J. Wang, L. J. Han, X. J. Gao and H. G. Zheng, Inorg. Chem., 2018, 57, 5232–5239 CrossRef CAS PubMed.
- Z. Li, J. Guo, F. Xiang, Q. Lin, Y. Ye, J. Zhang, S. Chen, Z. Zhang and S. Xiang, CrystEngComm, 2018, 20, 7567–7573 RSC.
- F. Wan, L. X. Qiu, L. L. Zhou, Y. Q. Sun and Y. You, Dalton Trans., 2015, 44, 18320–18323 RSC.
- N. Ahmad, H. A. Younus, A. H. Chughtai, K. Van Hecke, Z. A. K. Khattak, Z. Gaoke, M. Danish and F. Verpoort, Catal. Sci. Technol., 2018, 8, 4010–4017 RSC.
- R. Kaur, A. Kaur, A. Umar, W. A. Anderson and S. K. Kansal, Mater. Res. Bull., 2019, 109, 124–133 CrossRef CAS.
- S. Kongchoo, K. Chainok, A. Kantacha and S. Wongnawa, Inorg. Chem. Commun., 2017, 83, 97–102 CrossRef CAS.
- S. Sanram, J. Boonmak and S. Youngme, Polyhedron, 2016, 119, 151–159 CrossRef CAS.
- Y. Li, J. Jiang, Y. Fang, Z. Cao, D. Chen, N. Li, Q. Xu and J. Lu, ACS Sustainable Chem. Eng., 2018, 6, 16186–16197 CrossRef CAS.
- L. Lu, J. Wang, S.-L. Cai, B. Xie, B.-H. Li, J.-H. Man, Y.-X. He, A. Singh and A. Kumar, J. Coord. Chem., 2017, 70, 3409–3421 CrossRef CAS.
- N. Ahmad, H. A. Younus, A. H. Chughtai, K. Van Hecke, Z. A. K. Khattak, Z. Gaoke, M. Danish and F. Verpoort, Catal. Sci. Technol., 2018, 8, 4010–4017 RSC.
- H. Chen, P. X. Liu, N. Xu, X. Meng, H. N. Wang and Z. Y. Zhou, Dalton Trans., 2016, 45, 13477–13482 RSC.
- Y. Zhou, L. Qin, M.-K. Wu and L. Han, Cryst. Growth Des., 2018, 18, 5738–5744 CrossRef CAS.
- F. Wei, D. Chen, Z. Liang and S. Zhao, Nanomaterials, 2018, 8, 248 CrossRef CAS PubMed.
- X. R. Wang, Z. Huang, J. Du, X. Z. Wang, N. Gu, X. Tian, Y. Li, Y. Y. Liu, J. Z. Huo and B. Ding, Inorg. Chem., 2018, 57, 12885–12899 CrossRef CAS PubMed.
- R. Mishra, M. Ahmad, M. R. Tripathi and R. J. Butcher, Polyhedron, 2013, 50, 169–178 CrossRef CAS.
- P. Lama, J. Mrozinski and P. K. Bharadwaj, Cryst. Growth Des., 2012, 12, 3158–3168 CrossRef CAS.
- X.-m. Meng, X.-y. Zhang, X.-p. Wang, R.-x. Wu, X. Zhang, F. Jin and Y.-h. Fan, Polyhedron, 2017, 137, 81–88 CrossRef CAS.
- R. Ma, Z. Chen, S. Wang, C. Zhou, Y. Li, J. Lu, D. Li and J. Dou, Inorg. Chem. Commun., 2017, 81, 59–66 CrossRef CAS.
- Ž. Zupanek, M. Tramšek, A. Kokalj and G. Tavčar, Inorg. Chem., 2018, 57, 13866–13879 CrossRef PubMed.
- W. Q. Zhang, R. D. Wang, Z. B. Wu, Y. F. Kang, Y. P. Fan, X. Q. Liang, P. Liu and Y. Y. Wang, Inorg. Chem., 2018, 57, 1455–1463 CrossRef CAS PubMed.
- J. L. Zhang, J. Yang, X. Wang, H. Y. Zhang, X. L. Chi, Q. Yang, Y. Chen and D. R. Xiao, Inorg. Chim. Acta, 2016, 447, 66–76 CrossRef CAS.
- J.-M. Li, R. Li and X. Li, CrystEngComm, 2018, 20, 4962–4972 RSC.
- X. Pang, L. Li, Y. Wei, X. Yu and Y. Li, Dalton Trans., 2018, 47, 11530–11538 RSC.
- Y. Zhao, L. Wang, N.-N. Fan, M.-L. Han, G.-P. Yang and L.-F. Ma, Cryst. Growth Des., 2018, 18, 7114–7121 CrossRef CAS.
- M. B. Coban, J. Mol. Struct., 2019, 1177, 331–337 CrossRef CAS.
- X.-L. Qu, X.-L. Zheng and X. Li, RSC Adv., 2016, 6, 69007–69015 RSC.
- M.-N. Zhang, T.-T. Fan, Q.-S. Wang, H.-L. Han and X. Li, J. Solid State Chem., 2018, 258, 744–752 CrossRef CAS.
- G. M. Sheldrick, SHELXS-97, Program for Crystal Structure Refinement, University of Göttingen, 1997 Search PubMed.
- G. M. Sheldrick, SHELXL-97, Program for Crystal Structure Solution, University of Göttingen, 1997 Search PubMed.
- W. Hu, L. Li, G. Li, C. Tang and L. Sun, Cryst. Growth Des., 2009, 9, 3676–3682 CrossRef CAS.
- Y.-S. Chang, M. Choi, M. Baek, P.-Y. Hsieh, K. Yong and Y.-J. Hsu, Appl. Catal., B, 2018, 225, 379–385 CrossRef CAS.
- Y.-H. Chiu, K.-D. Chang and Y.-J. Hsu, J. Mater. Chem. A, 2018, 6, 4286–4296 RSC.
- J. Liu, Y. H. Tang, F. Wang and J. Zhang, CrystEngComm, 2018, 20, 1232–1236 RSC.
- P. M. Krishna, N. B. Reddy, N. Kottam, B. C. Yallur and H. R. Katreddi, Sci. World J., 2013, 828313 Search PubMed.
- H. R. Mardani, A. Esmaeili and A. Malekzadeh, Res. Chem. Intermed., 2018, 44, 6183–6195 CrossRef CAS.
- X. T. Zhang, H. T. Chen, B. Li, G. Z. Liu and X. Z. Liu, Dalton Trans., 2018, 47, 1202–1213 RSC.
- T. R. Zheng, L. L. Qian, M. Li, Z. X. Wang, K. Li, Y. Q. Zhang, B. L. Li and B. Wu, Dalton Trans., 2018, 47, 9103–9113 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1811807, 1832419, 1832425, 1832426. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ra01496a |
| This journal is © The Royal Society of Chemistry 2019 |