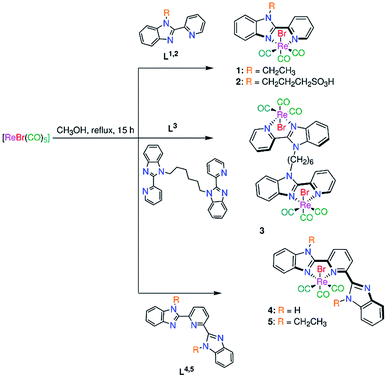
Open Access Article
Open Access ArticlePyridylbenzimidazole based Re(I)(CO)3 complexes: antimicrobial activity, spectroscopic and density functional theory calculations†
Ahmed M. Mansour *
*
Department of Chemistry, Faculty of Science, Cairo University, Gamma Street, Giza, Cairo 12613, Egypt. E-mail: mansour@sci.cu.edu.eg; inorganic_am@yahoo.com
First published on 14th May 2019
Abstract
fac-[ReBr(CO)3(L1,2)] L1 = 1-ethyl-2-(pyridin-2-yl)benzimidazole (1) and L2 = 1-[(pyridin-2-yl) benzimidazole]-propyl-sulfonic acid (2), fac-[Re2Br2(CO)6L3] (3) L3 = 1,1′-(hexane-1,6-diyl)bis[2-(pyridin-2-yl)1H-benzimidazole] and fac-[ReBr(CO)3(L4,5-κ2N1N2)] (L4 = 2,6-bis(benzimidazol-2′-yl)pyridine (4) and L5 = 2,6-bis(1-ethyl-benzimidazol-2′-yl)pyridine (5) were synthesized and fully characterized using different spectrocopic and analytical tools. The spectrocopic data showed coordination of L1–3 to fac-ReBr(CO)3 via the benzimidazole and pyridine N-atoms. For 4 and 5, the absence of a two-fold axis of symmetry for L4,5 in the 1H NMR spectra reflect the κ2N1,N2 mode of coordination. The electronic properties of 1–5 were investigated by time-dependent density functional theory calculations in the singlet and triplet states. The ligands and their Re(I) complexes were assessed for their potential antimicrobial activity. Compound 5 was screened against non-malignant cell line (noncancerous human embryonic kidney cell line (HEK293)) as well as evaluated for its blood compatibility.
Introduction
Rhenium(I) tricarbonyl complexes (fac-[Re(N–N)X(CO)3]) bearing N,N-bidentate ligands e.g. 2,2′-bipyridine,1 κ2N1,N2-terpyridine2 and 1,10-phenanthroline derivatives3 exhibit interesting photophysical properties and diverse applications such as luminescent biological molecular probes, organic light emitting diodes (OLEDs)4 (e.g. chemosensors and biotechnology probes) electro-5 and photo-catalysis (e.g. reduction of CO2).5 The luminescence properties of fac-[Re(N–N)L(CO)3] are ascribed to the metal-ligand-charge transfer (MLCT) bands that originate from the lowest triplet excited states as a result of the rapid vibrational relaxation and the intersystem crossing (ISC) from the upper vibrational levels. The photophysical properties of fac-Re(CO)3 diamine complexes are sensitive to the nature of bidentate ligand and/or the ancillary “spectator” ligand (halogen, pyridine, tetrazole derivatives, etc.).1–3,6 fac-[Re(N–N)L(CO)3] are characterized by large Stokes' shifts (difference between the absorbed and emitted wavelengths), long-lived excited state and high quantum emission efficiency; which decrease the self-quenching effects frequently detected by using organic dyes as fluorescent labelling of biomolecules. In the analyses of tissue samples, interferences of short lifetime biomolecule auto-fluorescence from the desired signal could be diminished by using compounds with large Stoke' shift.7Some tricarbonyl Re(I) complexes were evaluated as efficient photo-sensitizers capable of generation 1O2 molecules for photo-dynamic therapy of cancer.8 Some heterotrimetallic Re(I)(CO)3 complexes attached to a peptide nucleic acid (PNA) backbone have been recognised as novel antibacterial agents against only Gram-positive bacteria such as Bacillus subtilis and Staphylococcus aureus.9 Cationic tricarbonyl Re(I) complexes of 4-(2-pyridyl)-1,2,3-triazole derivatives exhibited better activity against Staphylococcus aureus than the neutral one.10 Furthermore, some photo induced Re(I) carbonyl complexes were used to deliver CO into tissues and cells.11–14 Phenanthroline tricarbonyl Re(I) complexes capable of release CO upon the exposure to UV light have been published by Ford13 and Mascharak.14 Exchange of the halide with a π-acid labilizes the axial CO ligand.7 Interestingly, a nontoxic water-soluble complex, [Re(bpy)(CO)3(thp)](CF3SO3) (thp = tris(hydroxymethyl)phosphine) released one CO upon illumination at 405 nm.12
To find new motivating photophysical properties of tricarbonyl Re(I) complexes relative to that observed by bipyridine and 1,10-phenanthroline complexes, new mononuclear fac-[ReBr(CO)3(L1,2)] (1, 2) (Scheme 1) (L1 = 1-ethyl-2-(pyridin-2-yl)benzimidazole (1) and L2 = 1-[(pyridin-2-yl) benzimidazole]-propyl-sulfonic acid (2)) and binuclear fac-[Re2Br2(CO)6(L3)] (3) (L3 = 1,1′-(hexane-1,6-diyl)bis[2-(pyridin-2-yl)1H-benzimidazole] (Scheme 1)) complexes have been synthesized and fully characterized by elemental analysis, IR, ESI-MS and NMR (1H, 13C, {1H, 1H} COS90 and {1H, 13C} HSQC). Referenced to bipyridine ligand system, a more conjugated ligand (pyridylbenzimidazole) has been chosen to motive the electronic properties of this class of Re(I) tricarbonyl compounds. To get an insight into the influence of presence of free benzimidazole arm close to the coordination sphere on the electronic structure of 1, fac-[ReBr(CO)3(L4,5-κ2N1,N2)] (L4 = 2,6-bis(benzimidazol-2′-yl)pyridine15 (4) and L5 = 2,6-bis(1-ethyl-benzimidazol-2′-yl) pyridine16 (5)) (Scheme 1)) have been synthesized in a similar way to 1–3. With the aim of understanding the electronic transitions of 1–5, time-dependent density functional theory calculations (TDDFT) were undertaken in both singlet and triplet states. Referenced to free ligands L1–5, compounds 1–5 were screened for their potential antimicrobial activity against some representative bacteria and fungi. Compound 5 was screened against non-malignant cell line (noncancerous human embryonic kidney cell line (HEK293)) as well as evaluated for its blood compatibility.
Results and discussion
Synthesis of ligands and complexes
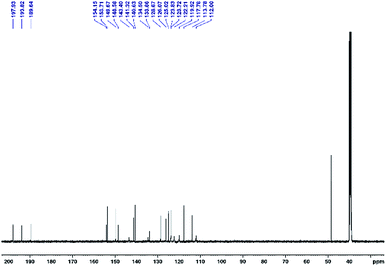
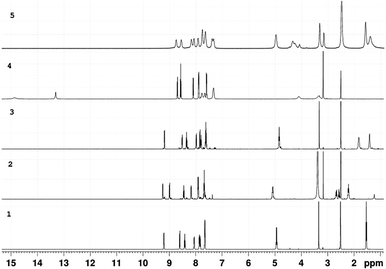
Bromo tricarbonyl Re(I) complexes were prepared in the dark from the reaction of N,N-bidentate benzimidazole ligands and [ReBr(CO)5] (Scheme 1). The complexes were fully characterized by elemental analysis, IR (Fig. S1†), NMR (1H, 13C, {1H, 1H} COS90, and {1H, 13C} HSQC) (Fig. S2–S6†) and ESI-MS. The NMR chemical shifts of 1–5 are given in Table S1.† The IR spectra (Fig. S1†) of 1–5 (except 3) show two strong stretches in the ranges of 2022–2008 and 1904–1886 cm−1 assigned to the symmetrical and anti-symmetrical modes of the CO molecules, respectively. These vibrational modes appear at 2016, 1902 and 1883 cm−1 in the spectrum of 3. Three characteristic 13CO signals of fac-Re(CO)3 moiety are observed at δ = (198.3, 197.7, 189.2) (1), (197.0, 196.5, 191.3) (2), (198.3, 197.7, 189.2) (3), (197.9, 193.8, 189.6) (4) (Fig. 1), and (197.8, 192.8, 188.8) ppm (5) in the 13C NMR spectra (Fig. S2–S6†). The downfield shift of pyridine-H6 (δ = 8.76 ppm (L1), 8.93 ppm (L2)16 and 8.62 ppm (L3)17) and benzimidazole-H4 (δ = 7.90 (L1), 7.89 (L2)16 and 7.70 ppm (L3)17) upon the complex formation (δ = 9.19–9.25 ppm and 7.92–8.05 ppm (1–3)) reveals the bidentate nature of the L1–3 ligands.The 1H NMR spectra of 4 and 5 (in DMSO-d6) (Fig. 2 and S6†) consist of eight aromatic signals due to absence of a two-fold axis of symmetry in the κ2N1,N2 mode of coordination. For 4, the NH signals are observed at two chemical shifts, δ = 14.83 and 13.31 ppm (Fig. 2). Similar, the signals of the ethyl group in 5 are shown at δ = (4.97, 4.26) and (1.58, 1.40) ppm for –CH3 and –CH2 in that order. The ESI-MS spectra show a characteristic peak of the suggested structures at m/z = 494.0509 {[M − Br]}+ (1), 665.9320 {[M − H]}− (2), 1091.0342 {[M − Br]}+ (3), 661.9832 {[M + H]}+ (4), and 638.1192 {[M − Br]}+ (5).
Absorption spectroscopy and time-dependent DFT calculations
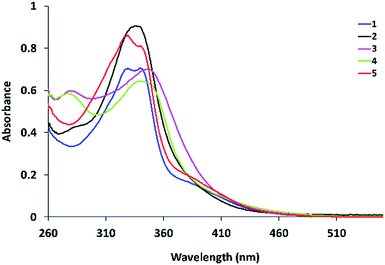
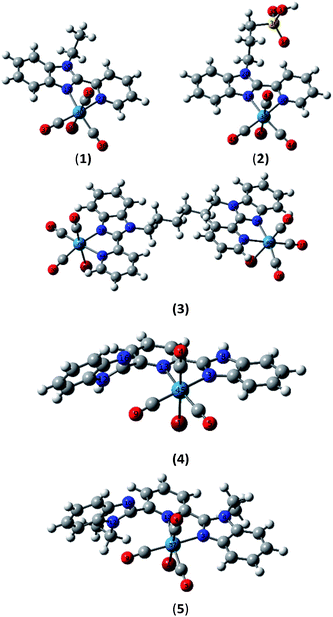
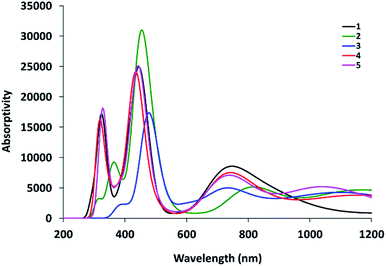
The electronic absorption spectra of Re(I) complexes in DMSO are shown in Fig. 3. Compounds 1–3 are characterized by a broad band around 334 nm. In addition, a shoulder at 390 nm is observed in the absorption spectrum of 1. The latter broad band is 10 nm red-shifted upon the extension of ligand conjugation π-system of 1–3 by free benzimidazole arm in compounds 4 and 5.Ground-state geometry optimization and TDDFT calculations were carried out on models representing the suggested molecular structures of 1–5. First, fac-[ReBr(L1–5)(CO)3] were fully optimised in the ground-state using Becke 3-parameter (exchange) Lee–Yang–Parr (B3LYP)18 and the effective core potential (ECP) of the Hady and Wadt, LANL2DZ basis set.19 The ground-state local minimum structures, with the labelling scheme used, are shown in Fig. 4.
The atomic coordinates of the optimized structures and energy values are given in Tables S2–S6.† Selected calculated bond lengths are given in Table S7.† The Re–NBz (Bz: benzimidazole) bond distances are uniform in the optimized structures except 5, which has shorter bond distance (2.14634 Å) than 1–4. The Re–NPy (Py = pyridine) bonds in 4 (2.24130 Å) and 5 (2.25661 Å) are longer than the corresponding one in 1–3 (2.18461–2.18901 Å). This may be accounted for the steric hindrance presented by the free benzimidazole arm, which is rotated from the equatorial plane by 58° (4) and 75° (5). The conjugation system of 2,6-bis (benzimidazol-2′-yl)pyridine in 4 is decreased upon the alkylation with ethyl groups.
To understand the electronic absorption spectra observed in 1–5, TDDFT calculations were done, in the singlet-state, at CAM-B3LYP20/LANL2DZ level of theory using PCM model to introduce the effect of the solvent. Based on the optimized geometries, the first 30 singlet excited states have been calculated. The theoretical spectra (Fig. S7†) of 1 and 2 are characterized by a broad band at 343 nm with oscillator strength of 0.1026 and 0.1097, respectively. The broad band is theoretically assigned to HOMO−1 → LUMO with ground-state composed of d(Re)/π(pyridine)/π(Br) character and excited state contained upon d(Re)/π(CO)/π(Br). The band at 343 nm deviates by 9 nm from the experimental one. The lowest energy transition at 360 nm in 1 and 2 is assigned to HOMO → LUMO (Fig. 5) and allocated for HOMO → LUMO+1 in 3. Although the position and assignment of the lowest energy band does not change on going from 1 to 3, the oscillator strength increases twice.
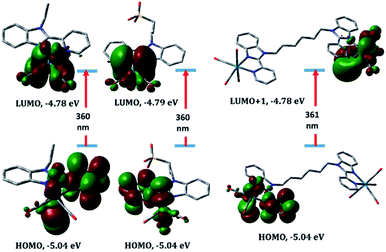 | ||
| Fig. 5 Selected molecular orbitals involved in the lowest energy transition of complexes 1–3 (from left to right) calculated at PCM/CAM-B3LYP/LANL2DZ level of theory, including LANL2DZ ECP. | ||
Compared to 1–3, the slightly increase in the ligand conjugation π-system leads to a red-shift of the main theoretical electronic transition to 368 (4) and 369 nm (5). As shown in Fig. 6, the latter transition, assigned to HOMO → LUMO, is MLCT/d–d in nature. HOMO–LUMO gap in 4 is 0.22 eV higher than 5.
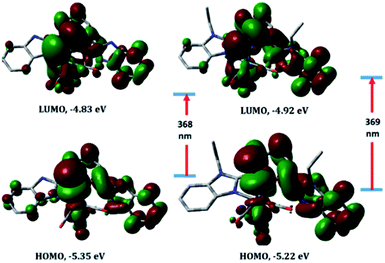 | ||
| Fig. 6 Selected molecular orbitals involved in the lowest energy transition of complexes 4 (left) and 5 (right) calculated at PCM/CAM-B3LYP/LANL2DZ level of theory, including LANL2DZ ECP. | ||
TDDFT calculations of 1–5 in their lowest energy lying excited triplet state (T1) have been performed at PCM/CAM-B3LYP/LANL2DZ level of theory. The calculated spectrum of 1 (Fig. 7) shows a broad band at 764 nm due to transitions from HSOMO-3/-4(β) orbitals to LSOMO(β). The HSOMO and LSOMO orbitals involved in the theoretical calculations are available in the ESI (Fig. S8†). The S0 → T1 transition at 764 nm is MLCT in nature (Fig. S8†). Compared to 1, the broad band at 836 nm in 5 is assigned to HSOMO-6(α) → LSOMO(α). LMCT and 3IL dominate the character of the band at 836 nm in 5.
The photoinduced carbon monoxide releasing properties of 1–5 was investigated by solution 1H NMR and myoglobin assay.21 During exposure of the DMSO solution of Mn(I) and Ru(II) carbonyl analogues,16,22 the characteristic signals of the κN1N2 bidentate ligand mode disappear and a new set of signals assigned to the meridional tridentate manner grows with time. In other words, the coordination mode of L4,5 is changed during the illumination, which facilitates the CO release process. For 4 and 5, the bidentate mode is persevered up to the end of the illumination time (Fig. S9†). No CO release is detected from the myoglobin solutions of 1–5 in the dark and upon the illumination with a light source in the range of 410–365 nm. In tricarbonyl rhenium(I) complexes, exposure at the longest wavelength band induces excitation that rapidly undergoes inter-system crossing into a triplet-state, which is non-productive of CO release. Therefore, the tricarbonyl Re(I) complexes 1–5 were not further examined for CO phototherapeutic applications. On the other hand, significant change (Fig. 8) is monitored upon the photolysis of DMSO solution of 4, incorporating free benzimidazole arm, at 365 nm. A blue shift of the broad band (344 nm) to 336 nm with an increase in the intensity as well as appearance of two isosbestic points at 350 and 293 nm are the main features of the illumination process. This may be accounted for the free rotation of the pendant benzimidazolyl arm from the plane of the pyridine ring.23
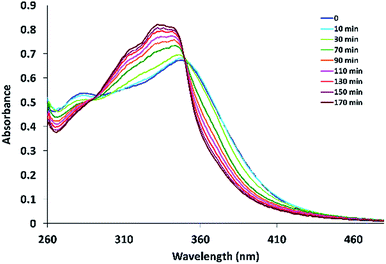 | ||
| Fig. 8 UV/Vis spectral changes of 4 (in DMSO) upon the photolysis at 365 nm for 0–170 min, respectively, after pre-incubation in the dark for 16 h. | ||
Antimicrobial and cytotoxic activity
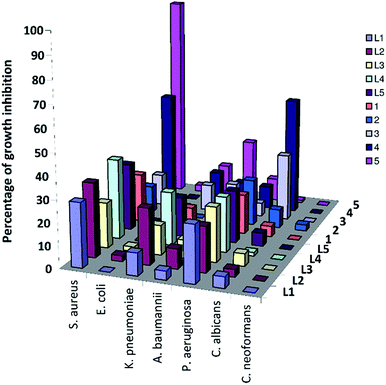
The antimicrobial activity of the benzimidazole ligands and their complexes 1–5 was investigated using cultures of Gram-positive bacterium (Staphylococcus aureus ATCC 43300), Gram-negative bacteria (Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603, Acinetobacter baumannii ATCC 19606, Pseudomonas aeruginosa ATCC) as well as two fungal species (Candida albicans ATCC 90028 and Cryptococcus neoformans var. grubii H99; ATCC 208821) according to the standard broth microdilution assays24 (ESI†). Initial screening was carried out at 32 μg mL−1. The compounds exhibited partially or no inhibitory activity at the tested concentration (Table S2†). Coordination of L4,5 to ReBr(CO)3 moiety led to an improvement in the antimicrobial activity (Fig. 9). Compounds 4 and 5 inhibited 70% and 93% of the growth of Staphylococcus aureus ATCC 43300 with MIC value > 32 μg mL−1 (determined according to CLSI guidelines). The toxicity of 4 and 5 against the Gram-positive bacterium compares well with the previously published data.9,10 Referenced to 1–3, presence of benzimidazolyl free arm improved the toxicity against the tested microbes. Alkylation of benzimidazole NH group increases the toxicity by about 25%. The cytotoxicity of 5 was assessed against the noncancerous human embryonic kidney cells (HEK293) (the evaluation was done by CO-ADD (The Community for Antimicrobial Drug Discovery)). Moderate cell viability (67%) for 5 was reported with CC50 > 32 μg mL−1. Interaction of 5 with the blood components RBCs was evaluated for blood compatibility. The HC10 and HC50 (the concentration at 10% and 50% haemolysis, respectively) were calculated by curve fitting the inhibition values versus log(concentration). Incubation of complex 5 with RBCs caused negligible haemolysis with HC10 and HC50 values > 32 μg mL−1. Therefore, compound 5 exhibited moderate antibacterial activity against Staphylococcus aureus ATCC 43300, perfect compatibility with the blood components, and slightly cytotoxicity to non-malignant cell HEK293.Conclusions
Mono- and binuclear tricarbonyl rhenium(I) complexes based on 2-(2′-pyridyl)benzimidazole derivatives have been synthesized, characterized and tested for their potential antimicrobial against cultures of Gram-positive bacterium (Staphylococcus aureus ATCC 43300), Gram-negative bacteria (Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603, Acinetobacter baumannii ATCC 19606, Pseudomonas aeruginosa ATCC) as well as two fungal species (Candida albicans ATCC 90028 and Cryptococcus neoformans var. grubii H99; ATCC 208821). Singlet-state TDDFT calculations allocated the lowest energy electronic transition of the Re(I) complexes to MLCT. Triplet-state TDDFT calculations indicated that the complexes have emission spectra in the red spectral region allocated to MLCT and/or 3IL. Although the MLCT absorption transition matches the wavelength of the illumination wavelength, no CO release is detected as assessed by the myoglobin assay. The influence of presence of free benzimidazole arm close to Re(I) coordination sphere on the electronic structure and CO release process has been studied via the synthesis of fac-[ReBr(CO)3(L-κ2N1N2)] (L = 2,6-bis (benzimidazole-2′-yl)pyridine derivatives). In contrast to the previously published photoactivable Mn(I) and Ru(II) analogues, the bidentate mode of 2,6-bis(benzimidazole-2′-yl)pyridine is persevered up to the end of the illumination time with no probability of CO release. In tricarbonyl rhenium(I) complexes, exposure at the longest wavelength band induces excitation that rapidly undergoes intersystem crossing into a triplet-state, which is non-productive of CO release. The markedly spectral change observed in the absorption spectrum of fac-[ReBr(CO)3(L-κ2N1N2)] upon the illumination at 365 nm may be attributed to free rotation of the pendant benzimidazolyl arm from the plane of the pyridine ring. The synthesized Re(I) compounds exhibited partially or no inhibitory activity at the tested concentration. Incubation of the complexes with RBCs caused negligible haemolysis with HC10 and HC50 values > 32 μg mL−1.Experimental section
Materials and instruments
The experimental manipulations were carried out in the dark using oven-dried Schlenk glassware. Degassed solvents and argon atmosphere were used. The chemicals were purchased from the commercial suppliers and used without primary purification. The ligands, 1-ethyl-2-(pyridin-2-yl)benzimidazole (L1),25 1-[(pyridin-2-yl) benzimidazole]-propyl-sulfonic acid (L2),17 2,6-bis(benzimidazole-2′-yl)pyridine (L3),15 2,6-bis(1-ethyl-benzimidazol-2′-yl) pyridine (L4),16 and 1,1′-(hexane-1,6-diyl)bis[2-(pyridin-2-yl)1H-benzimidazole] (L5)26 were synthesized following the published procedures. 1H and 13C NMR spectra were recorded with Bruker-Avance 500 (1H, 500.13 MHz; 13C{1H}, 125.77 MHz) and Bruker-Avance 400 (1H, 400.40 MHz; 13C{1H}, 100.70 MHz) spectrometers. Assignments were done with the aid of {1H, 1H} COS90 and {1H, 13C} HSQC. Electrospray mass spectra were run with a Thermo Fisher Exactive Plus instrument with an Orbitrap mass analyzer at a resolution of R = 70.000 and a solvent flow rate of 5 μL min. Elemental microanalysis was performed with a Vario Micro Cube analyzer of Elementar Analysensysteme or an EA 3000 elemental analyzer from HEKtech. Illumination process was performed by a UV/Vis hand lamp (365 nm, UVIlite LF-206LS, 6 W, UVItec Ltd, Cambridge, UK). The cuvette was also positioned perpendicular to the LED source light at a distance of 3 cm, where the illumination was interrupted in regular intervals to take UV/Vis spectra on an Agilent 8453 diode array spectrophotometer until no more spectral changes were observed.Synthesis of complexes
1: Colour: yellow powder. Yield: 84% (174 mg, 0.30 mmol). IR (ATR): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) = 3049, 2008 (vs., C
= 3049, 2008 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1886 (vs., C
O), 1886 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1603 (m, CN/CC), 1440, 1350, 1135, 766. 1H NMR ([D6]DMSO, 500.13 MHz): δ = 9.21 (d, 3JH,H = 5.9 Hz, 1H, py-H6), 8.61 (d, 3JH,H = 8.4 Hz, 1H, py-H3), 8.41 (t, 3JH,H = 7.8 Hz, 1H, py-H4), 8.06 (m, 1H, bim-H4), 7.87 (m, 1H, bim-H7), 7.83 (m, 1H, py-H5), 7.65 (m, 2H, bim-H5/H6), 4.93 (q, 3JH,H = 7.3 Hz, 2H, NCH2), 1.53 (t, 3JH,H = 7.2 Hz, 3H, CH3) ppm. 13C-NMR ([D6]DMSO, 125.75 MHz): δ = 198.3 (C
O), 1603 (m, CN/CC), 1440, 1350, 1135, 766. 1H NMR ([D6]DMSO, 500.13 MHz): δ = 9.21 (d, 3JH,H = 5.9 Hz, 1H, py-H6), 8.61 (d, 3JH,H = 8.4 Hz, 1H, py-H3), 8.41 (t, 3JH,H = 7.8 Hz, 1H, py-H4), 8.06 (m, 1H, bim-H4), 7.87 (m, 1H, bim-H7), 7.83 (m, 1H, py-H5), 7.65 (m, 2H, bim-H5/H6), 4.93 (q, 3JH,H = 7.3 Hz, 2H, NCH2), 1.53 (t, 3JH,H = 7.2 Hz, 3H, CH3) ppm. 13C-NMR ([D6]DMSO, 125.75 MHz): δ = 198.3 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 197.7 (C
O), 197.7 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 189.2 (C
O), 189.2 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 155.3 (py-C6), 152.3 (py-C2), 146.7 (bim-C2), 141.4 (py-C4), 140.1 (bim-C3a), 135.7 (bim-C7a), 128.4 (py-C5), 126.6 (bim-C5), 126.1 (bim-C6), 125.7 (py-C3), 118.5 (bim-C7), 112.9 (bim-C4), 41.2 (NCH2), 15.2 (CH3) ppm. ESI-MS (positive mode, methanol): 597.9570 {M + Na}+, 494.0509 {M − Br}+. C17H13BrN3O3Re: C 35.61, H 2.29, N 7.33, found C 35.66, H 2.84, N 7.10.
O), 155.3 (py-C6), 152.3 (py-C2), 146.7 (bim-C2), 141.4 (py-C4), 140.1 (bim-C3a), 135.7 (bim-C7a), 128.4 (py-C5), 126.6 (bim-C5), 126.1 (bim-C6), 125.7 (py-C3), 118.5 (bim-C7), 112.9 (bim-C4), 41.2 (NCH2), 15.2 (CH3) ppm. ESI-MS (positive mode, methanol): 597.9570 {M + Na}+, 494.0509 {M − Br}+. C17H13BrN3O3Re: C 35.61, H 2.29, N 7.33, found C 35.66, H 2.84, N 7.10.
2: Colour: yellow powder. Yield: 77% (191 mg, 0.26 mmol). IR (ATR): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) = 3066 (w, CH), 3019 (w, CH), 2976 (w, CH), 2022 (vs., C
= 3066 (w, CH), 3019 (w, CH), 2976 (w, CH), 2022 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1905 (vs., C
O), 1905 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1604 (m, CN/CC), 1514, 1488, 1440, 1209, 1135, 1025, 751. 1H NMR ([D6]DMSO, 500.13 MHz): δ = 9.26 (dd, 3JH,H = 5.5 Hz, 4JH,H = 1.1 Hz, 1H, py-H6), 9.00 (d, 3JH,H = 8.2 Hz, 1H, py-H3), 8.46 (td, 3JH,H = 1.5 Hz, 4JH,H = 7.9 Hz, 1H, py-H4), 8.18 (m, 1H, py-H5), 7.91 (m, 2H, bim-H4/H7), 7.69 (m, 2H, bim-H5/H6), 5.10 (m, 2H, NCH2), 2.63 (m, 2H, CH2SO3H), 2.23 (quintet, 3JH,H = 7.6 Hz, 2H, CH2CH2CH2) ppm. 13C-NMR ([D6]DMSO, 125.75 MHz): δ = 197.0 (C
O), 1604 (m, CN/CC), 1514, 1488, 1440, 1209, 1135, 1025, 751. 1H NMR ([D6]DMSO, 500.13 MHz): δ = 9.26 (dd, 3JH,H = 5.5 Hz, 4JH,H = 1.1 Hz, 1H, py-H6), 9.00 (d, 3JH,H = 8.2 Hz, 1H, py-H3), 8.46 (td, 3JH,H = 1.5 Hz, 4JH,H = 7.9 Hz, 1H, py-H4), 8.18 (m, 1H, py-H5), 7.91 (m, 2H, bim-H4/H7), 7.69 (m, 2H, bim-H5/H6), 5.10 (m, 2H, NCH2), 2.63 (m, 2H, CH2SO3H), 2.23 (quintet, 3JH,H = 7.6 Hz, 2H, CH2CH2CH2) ppm. 13C-NMR ([D6]DMSO, 125.75 MHz): δ = 197.0 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 196.6 (C
O), 196.6 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 191.3 (C
O), 191.3 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 155.1 (py-C6), 153.1 (py-C2), 146.2 (bim-C2), 142.1 (py-C4), 139.2 (bim-C3a), 135.7 (bim-C7a), 128.7 (bim-C4), 126.5 (bim-C5), 126.4 (bim-C6), 126.2 (py-C3), 117.4 (bim-C7), 113.0 (py-C5), 48.6 (CH2SO3H), 47.6 (NCH2), 25.6 (CH2CH2CH2) ppm. ESI-MS (negative mode, methanol): m/z = 665.9320 {M − H}−. C18H15BrN3O6SRe: C 32.39, H 2.27, N 6.30, S 4.80, found C 32.53, H 2.69, N 6.77, S 4.82.
O), 155.1 (py-C6), 153.1 (py-C2), 146.2 (bim-C2), 142.1 (py-C4), 139.2 (bim-C3a), 135.7 (bim-C7a), 128.7 (bim-C4), 126.5 (bim-C5), 126.4 (bim-C6), 126.2 (py-C3), 117.4 (bim-C7), 113.0 (py-C5), 48.6 (CH2SO3H), 47.6 (NCH2), 25.6 (CH2CH2CH2) ppm. ESI-MS (negative mode, methanol): m/z = 665.9320 {M − H}−. C18H15BrN3O6SRe: C 32.39, H 2.27, N 6.30, S 4.80, found C 32.53, H 2.69, N 6.77, S 4.82.
3: Colour: yellow powder. Yield: 68% (289 mg, 0.15 mmol). IR (ATR): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) = 2940 (w, CH), 2018 (vs., C
= 2940 (w, CH), 2018 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1904 (vs., C
O), 1904 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1883 (vs., C
O), 1883 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1604 (m, CN/CC), 1439, 1338, 752. 1H NMR ([D6]DMSO, 500.13 MHz): δ = 9.19 (d, 3JH,H = 6.1 Hz, 1H, py-H6), 8.52 (d, 3JH,H = 8.1 Hz, 1H, py-H3), 8.35 (t, 3JH,H = 7.9 Hz, 1H, py-H4), 7.99 (d, 3JH,H = 7.0 Hz, 1H, bim-H4), 7.85 (d, 3JH,H = 7.3 Hz, 1H, bim-H7), 7.81 (t, 3JH,H = 6.6 Hz, 1H, py-H5), 7.62 (m, 2H, bim-H5/H6), 4.85 (t, 3JH,H = 7.4 Hz, 2H, NCH2), 1.83 (m, 2H, NCH2CH2), 1.43 (m, 2H, NCH2CH2CH2) ppm. 13C-NMR ([D6]DMSO, 125.75 MHz): δ = 198.3 (C
O), 1604 (m, CN/CC), 1439, 1338, 752. 1H NMR ([D6]DMSO, 500.13 MHz): δ = 9.19 (d, 3JH,H = 6.1 Hz, 1H, py-H6), 8.52 (d, 3JH,H = 8.1 Hz, 1H, py-H3), 8.35 (t, 3JH,H = 7.9 Hz, 1H, py-H4), 7.99 (d, 3JH,H = 7.0 Hz, 1H, bim-H4), 7.85 (d, 3JH,H = 7.3 Hz, 1H, bim-H7), 7.81 (t, 3JH,H = 6.6 Hz, 1H, py-H5), 7.62 (m, 2H, bim-H5/H6), 4.85 (t, 3JH,H = 7.4 Hz, 2H, NCH2), 1.83 (m, 2H, NCH2CH2), 1.43 (m, 2H, NCH2CH2CH2) ppm. 13C-NMR ([D6]DMSO, 125.75 MHz): δ = 198.3 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 197.7 (C
O), 197.7 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 189.2 (C
O), 189.2 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 155.3 (py-C6), 152.4 (py-C2), 146.8 (bim-C2), 141.2 (py-C4), 139.9 (bim-C3a), 136.1 (bim-C7a), 128.3 (py-C5), 126.6 (bim-C5), 126.1 (bim-C6), 125.6 (py-C3), 118.6 (bim-C7), 113.1 (bim-C4), 45.7 (NCH2), 29.5 (NCH2CH2), 25.9 (NCH2CH2CH2) ppm. ESI-MS (positive mode, acetone): m/z = 1091.0342, 1093.0353 and 1095.0364 {M − Br}+. C36H28Br2Re2N6O6·CH3OH: C 36.87, H 2.41, N 7.17, found C 36.88, H 2.68, N 6.97.
O), 155.3 (py-C6), 152.4 (py-C2), 146.8 (bim-C2), 141.2 (py-C4), 139.9 (bim-C3a), 136.1 (bim-C7a), 128.3 (py-C5), 126.6 (bim-C5), 126.1 (bim-C6), 125.6 (py-C3), 118.6 (bim-C7), 113.1 (bim-C4), 45.7 (NCH2), 29.5 (NCH2CH2), 25.9 (NCH2CH2CH2) ppm. ESI-MS (positive mode, acetone): m/z = 1091.0342, 1093.0353 and 1095.0364 {M − Br}+. C36H28Br2Re2N6O6·CH3OH: C 36.87, H 2.41, N 7.17, found C 36.88, H 2.68, N 6.97.
4: Colour: yellow powder. Yield: 77% (184 mg, 0.26 mmol). IR (ATR): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) = 3057 (w, CH), 2019 (vs., C
= 3057 (w, CH), 2019 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1895 (vs., C
O), 1895 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1426, 1315, 767. 1H NMR ([D6]DMSO, 500.13 MHz): δ = 14.83 (br, 1H, NH), 13.31 (br, 1H, NH), 8.70 (d, 3JH,H = 7.0 Hz, 1H, py-H3), 8.58 (t, 3JH,H = 8.6 Hz, 1H, py-H4), 8.10 (d, 3JH,H = 7.7 Hz, 1H, py-H5), 7.89 (m, 2H, bim-H4/H7), 7.77 (m, 1H, bim-H4′), 7.67 (m, 1H, bim-H7′), 7.60 (m, 2H, bim-H5/H6), 7.33 (m, 2H, bim-H5′/H6′) ppm. 13C-NMR ([D6]DMSO, 125.75 MHz): δ = 197.9 (C
O), 1426, 1315, 767. 1H NMR ([D6]DMSO, 500.13 MHz): δ = 14.83 (br, 1H, NH), 13.31 (br, 1H, NH), 8.70 (d, 3JH,H = 7.0 Hz, 1H, py-H3), 8.58 (t, 3JH,H = 8.6 Hz, 1H, py-H4), 8.10 (d, 3JH,H = 7.7 Hz, 1H, py-H5), 7.89 (m, 2H, bim-H4/H7), 7.77 (m, 1H, bim-H4′), 7.67 (m, 1H, bim-H7′), 7.60 (m, 2H, bim-H5/H6), 7.33 (m, 2H, bim-H5′/H6′) ppm. 13C-NMR ([D6]DMSO, 125.75 MHz): δ = 197.9 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 193.8 (C
O), 193.8 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 189.6 (C
O), 189.6 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 154.2 (py-C2), 153.7 (py-C6), 149.7 (bim-C2), 148.6 (bim-C2′), 143.3 (bim-C3a), 141.3 (bim-C7a), 140.6 (py-C4), 134.5 (bim-C3a′), 133.9 (bim-C7a′), 128.7 (py-C5), 126.1 (bim-C6), 125.0 (bim-C5), 123.8 (py-C3), 123.7 (bim-C6′), 122.2 (bim-C5′), 119.9 (bim-C7′), 117.8 (bim-C7), 113.8 (bim-C4), 112.0 (bim-C4′) ppm. ESI-MS (positive mode, methanol): 661.9832 {M + H}+. C22H13BrN5O3Re·H2O: C 38.89, H 2.23, N 10.31, found C 38.98, H 2.68, N 10.28.
O), 154.2 (py-C2), 153.7 (py-C6), 149.7 (bim-C2), 148.6 (bim-C2′), 143.3 (bim-C3a), 141.3 (bim-C7a), 140.6 (py-C4), 134.5 (bim-C3a′), 133.9 (bim-C7a′), 128.7 (py-C5), 126.1 (bim-C6), 125.0 (bim-C5), 123.8 (py-C3), 123.7 (bim-C6′), 122.2 (bim-C5′), 119.9 (bim-C7′), 117.8 (bim-C7), 113.8 (bim-C4), 112.0 (bim-C4′) ppm. ESI-MS (positive mode, methanol): 661.9832 {M + H}+. C22H13BrN5O3Re·H2O: C 38.89, H 2.23, N 10.31, found C 38.98, H 2.68, N 10.28.
5: Colour: orange powder. Yield: 76% (197 mg, 0.24 mmol). IR (ATR): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) = 3056 (w, CH), 2981 (w, CH), 2019 (vs., C
= 3056 (w, CH), 2981 (w, CH), 2019 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1907 (vs., C
O), 1907 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1605 (w, C
O), 1605 (w, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1407, 1338, 1154, 1097, 819, 749. 1H NMR ([D6]DMSO, 400 MHz): δ = 8.76 (m, 1H, py-H3), 8.57 (m, 1H, py-H4), 8.18 (m, 1H, py-H5), 8.07 (m, 1H, bim-H4), 7.92 (m, 1H, bim-H7), 7.77 (m, 2H, bim-H4′/H7′), 7.65 (m, 2H, bim-H5/6), 7.34 (m, 2H, bim-H5′/H6′), 4.97 (m, 2H, CH2), 4.26 (m, 2H,
N), 1407, 1338, 1154, 1097, 819, 749. 1H NMR ([D6]DMSO, 400 MHz): δ = 8.76 (m, 1H, py-H3), 8.57 (m, 1H, py-H4), 8.18 (m, 1H, py-H5), 8.07 (m, 1H, bim-H4), 7.92 (m, 1H, bim-H7), 7.77 (m, 2H, bim-H4′/H7′), 7.65 (m, 2H, bim-H5/6), 7.34 (m, 2H, bim-H5′/H6′), 4.97 (m, 2H, CH2), 4.26 (m, 2H, 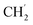 ), 1.58 (m, 3H, CH3), 1.40 (m, 3H,
), 1.58 (m, 3H, CH3), 1.40 (m, 3H, 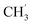 ) ppm. 13C-NMR ([D6]DMSO, 100.68 MHz): δ = 197.8 (C
) ppm. 13C-NMR ([D6]DMSO, 100.68 MHz): δ = 197.8 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 192.8 (C
O), 192.8 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 188.8 (C
O), 188.8 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 153.4 (py-C2), 152.4 (py-C6), 150.4 (bim-C2), 148.1 (bim-C2′), 142.8 (bim-C3a), 141.0 (py-C4), 140.0 (bim-C7a), 135.3 (bim-C3a′), 134.4 (bim-C7a′), 129.1 (py-C5), 126.3 (py-C3), 125.8 (bim-C5), 125.7 (bim-C6), 123.6 (bim-C5′), 122.6 (bim-C6′), 120.1 (bim-C7′), 118.2 (bim-C7), 112.5 (bim-C4), 111.4 (bim-C4′), 48.6 (CH2), 41.0 (CH2), 14.9 (CH3), 14.7 (CH3) ppm. ESI-MS (positive mode, methanol): m/z = 638.1192 {M − Br}+. C26H21BrReN5O3·H2O: C 42.45, H 3.15, N 9.52; found C 42.22, H 3.31, N 9.54.
O), 153.4 (py-C2), 152.4 (py-C6), 150.4 (bim-C2), 148.1 (bim-C2′), 142.8 (bim-C3a), 141.0 (py-C4), 140.0 (bim-C7a), 135.3 (bim-C3a′), 134.4 (bim-C7a′), 129.1 (py-C5), 126.3 (py-C3), 125.8 (bim-C5), 125.7 (bim-C6), 123.6 (bim-C5′), 122.6 (bim-C6′), 120.1 (bim-C7′), 118.2 (bim-C7), 112.5 (bim-C4), 111.4 (bim-C4′), 48.6 (CH2), 41.0 (CH2), 14.9 (CH3), 14.7 (CH3) ppm. ESI-MS (positive mode, methanol): m/z = 638.1192 {M − Br}+. C26H21BrReN5O3·H2O: C 42.45, H 3.15, N 9.52; found C 42.22, H 3.31, N 9.54.
Density functional theory calculations
Density functional theory (DFT) calculations were carried out using Gaussian 03.27 Geometry optimization was performed by Becke 3-parameter (exchange) Lee–Yang–Parr functional and the effective core potential (ECP) of the Hady and Wadt, LANL2DZ basis set. Harmonic frequency analysis was done. Time-dependent DFT calculations were carried out in both the singlet and triplet state at CAM-B3LYP18/LANL2DZ level of theory using the default continuum model (PCM) to introduce the solvent effect.Biological activity testing
The details of the biological (cytotoxicity and antimicrobial activity) assays done here are given in ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. Mansour thanks the Alexander von Humboldt Foundation for Georg Förster postdoctoral fellowship. Antimicrobial screening was performed by CO-ADD (The Community for Antimicrobial Drug Discovery), funded by the Wellcome Trust (UK) and The University of Queensland (Australia). Thanks also Dr Ola Shehab, Cairo University, Faculty of Science, Egypt for the mass spectrometry measurements.References
- J. L. Smithback, J. B. Helms, E. Schutte, S. M. Woessner and B. Patrick Sullivan, Inorg. Chem., 2006, 45, 2163 CrossRef CAS PubMed; O. S. Wenger, L. M. Henling, M. W. Day, J. R. Winkler and H. B. Gray, Inorg. Chem., 2004, 43, 2043 CrossRef PubMed.
- P. Bulsink, A. Al-Ghamdi, P. Joshi, L. Korobkov, T. Woo and D. Richeson, Dalton Trans., 2016, 45, 8885 RSC; T. Klemens, A. Switlicka-Olszewska, B. Machura, M. Grucela, E. Schab-Balcerzak, K. Smolarek, S. Mackowski, A. Szlapa, S. Kula, S. Krompiec, P. Lodowski and A. Chrobok, Dalton Trans., 2016, 45, 1746 RSC; B. Milette, C. Lachance-Baris and G. S. Hanan, Dalton Trans., 2015, 44, 41 RSC; B. A. Frenzel, J. E. Schumaker, D. R. Black and S. E. Hightower, Dalton Trans., 2013, 42, 12240 RSC; D. Wang, Q. Xu, S. Zhang, H. Li, C. Wang, T. Li, Y. Jing, W. Hyang, Y. Zheng and G. Accorsi, Dalton Trans., 2013, 42, 2716 RSC; E. R. Civitello, G. Bierach, J. F. O'Connell and T. D. Westmoreland, Inorg. Chem., 1993, 32, 237 CrossRef CAS; A. Juris, S. Campagna, I. Bidd, J. M. Lehn and R. Ziessel, Inorg. Chem., 1988, 27, 4007 CrossRef.
- M. P. Coogan and J. A. Platts, Chem. Commun., 2016, 52, 12498 RSC; A. Casula, V. Nairi, V. Fernández-Moreira, A. Laguna, V. Lippolis, A. Garau and M. Concepción-Gimeno, Dalton Trans., 2015, 44, 18506 RSC; X. Yi, J. Zhao, W. Wu, D. Huang, S. Ji and J. Sun, Dalton Trans., 2012, 41, 8931 RSC; J. M. Villegas, S. R. Stoyanov, W. Huang and D. Paul Rillema, Dalton Trans., 2005, 1042 RSC.
- X. Gong, P. K. Ng and W. K. Chan, Adv. Mater., 1998, 10, 1337 CrossRef CAS; T. Yu, D. P. K. Tsang, V. K. M. Au, W. H. Lam, M. Y. Chan and V. W. W. Yam, Chem.–Eur. J., 2013, 19, 13418 CrossRef PubMed.
- C. Sun, S. Prosperini, P. Quagliotto, G. Viscardi, S. S. Yoon, R. Gobetto and C. Nervi, Dalton Trans., 2016, 45, 14678 RSC; T. Jin, D. He, W. Li, C. J. Stanton, S. A. Pantovich, G. F. Majetich, H. F. Schaefer, J. Agarwal, D. Wang and G. Li, Chem. Commun., 2016, 52, 14258 RSC; M. V. Vollmer, C. W. Machan, M. L. Clark, W. E. Antholine, J. Agarwal, H. F. Schafer, C. P. Kubiak and J. R. Walensky, Organometallics, 2015, 34, 3 CrossRef CAS PubMed.
- M. V. Werrett, G. S. Huff, S. Muzzioli, V. Fiorini, S. Zacchini, B. W. Skelton, A. Maggiore, J. M. Malicka, M. Cocchi, K. C. Gordon, S. Stagni and M. Massi, Dalton Trans., 2015, 44, 8379 RSC.
- N. Billinton and A. W. Knight, Anal. Biochem., 2001, 291, 17 CrossRef PubMed.
- K. K. Lo, Acc. Chem. Res., 2015, 48, 2985 CrossRef CAS PubMed; G. Gasser and A. Leonidova, ACS Chem. Biol., 2014, 9, 2180 CrossRef PubMed.
- L. C. Lee, K. Leung and K. K. Lo, Dalton Trans., 2017, 46, 16357 RSC; M. Wenzel, M. Patra, C. H. R. Senges, I. Ott, J. J. Stepanek, A. Pinto, P. Prochnow, C. Vuong, S. Langklotz, N. Metzler-Nolte and J. E. Bandow, ACS Chem. Biol., 2013, 8, 1442 CrossRef CAS PubMed.
- S. V. Kumar, W. K. C. Lo, H. J. L. Brooks, L. R. Hanton and J. D. Crowley, Aust. J. Chem., 2016, 69, 489 CrossRef CAS.
- K. Koike, N. Okoshi, H. Hori, K. Takauchi, O. Ishitani, H. Tsubaki, I. P. Clark, M. W. George, F. P. A. Johnson and J. J. Turner, J. Am. Chem. Soc., 2002, 124, 11448 CrossRef CAS PubMed.
- A. E. Pierri, A. Pallaoro, G. Wu and P. C. Ford, J. Am. Chem. Soc., 2012, 134, 18197 CrossRef CAS PubMed.
- S. C. Marker, S. N. MacMillan, W. R. Zipfel, Z. Li, P. C. Ford and J. J. Wilson, Inorg. Chem., 2018, 57, 1311 CrossRef CAS PubMed.
- I. Chakraborty, J. Jimenez, W. M. C. Sameera, M. Kato and P. K. Mascharak, Inorg. Chem., 2017, 56, 2863 CrossRef CAS PubMed; J. Jimenez, I. Chakraborty, A. Dominguez, J. Martinez-Gonzalez, W. M. Chamil Sameera and P. K. Mascharak, Inorg. Chem., 2018, 57, 1766 CrossRef PubMed.
- A. W. Addison and P. J. Burke, J. Heterocycl. Chem., 1981, 18, 803 CrossRef CAS.
- A. M. Mansour and O. R. Shehab, Eur. J. Inorg. Chem., 2017, 4299 CrossRef CAS.
- A. M. Mansour and O. R. Shehab, Dalton Trans., 2018, 47, 3459 RSC.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648 CrossRef CAS.
- P. J. Hay and W. R. Wadt, J. Chem. Phys., 1985, 82, 270 CrossRef CAS.
- T. Yanai, D. P. Tew and N. C. Handy, Chem. Phys. Lett., 2004, 393, 51 CrossRef CAS.
- D. Nguyen and C. Boyer, ACS Biomater. Sci. Eng., 2015, 1(10), 895 CrossRef CAS; S. Mclen, B. E. Mann and R. K. Poole, Anal. Biochem., 2012, 427, 36 CrossRef PubMed.
- A. M. Mansour and A. Friedrich, Inorg. Chem. Front., 2017, 4, 1517 RSC.
- A. Gelling, K. G. Orrell, A. G. Osborne, V. Šik, M. B. Hursthouse, D. E. Hibbs and K. M. Abdul Malik, Polyhedron, 1998, 17(13–14), 2141 CrossRef CAS.
- M. A. Blaskovich, J. Zuegg, A. G. Elliott and M. A. Cooper, ACS Infect. Dis., 2015, 1(7), 285 CrossRef CAS PubMed.
- L. Huang, K.-Z. Wang, C.-H. Huang, F.-Y. Li and Y.-Y. Huang, J. Mater. Chem., 2001, 11, 790 RSC.
- A. M. Mansour and A. Friedrich, New J. Chem., 2018, 42, 18418 RSC.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, V. G. Zakrzewski, J. A. Montgomery, R. E. Stratmann, J. C. Burant, S. Dapprich, J. M. Millam, A. D. Daniels, K. N. Kudin, M. C. Strain, O. Farkas, J. Tomasi, V. Barone, M. Cossi, R. Cammi, B. Mennucci, C. Pomelli, C. Adamo, S. Clifford, J. Ochterski, G. A. Petersson, P. Y. Ayala, Q. Cui, K. Morokuma, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. Cioslowski, J. V. Ortiz, A. G. Baboul, B. B. Stefanov, G. Liu, A. Liashenko, P. Piskorz, I. Komaromi, R. Gomperts, R. L. Martin, D. J. Fox, T. Keith, M. A. Al-Laham, C. Y. Peng, A. Nanayakkara, C. Gonzalez, M. Challacombe, P. M. W. Gill, B. G. Johnson, W. Chen, M. W. Wong, J. L. Andres, M. Head-Gordon, E. S. Replogle, and J. A. Pople, Gaussian 03 (Revision A.9), Gaussian, Inc., Pittsburgh, 2003 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra01566c |
| This journal is © The Royal Society of Chemistry 2019 |