Open Access Article
Open Access ArticleA molecular dynamics study on the resilience of Sec61 channel from open to closed state†
Sujuan Suna,
Shuangshuang Wangb,
Zhangfa Tong c,
Xingdong Yao*a and
Jian Gao
c,
Xingdong Yao*a and
Jian Gao *b
*b
aKey Laboratory of Forest Chemistry & Engineering, School of Chemistry and Chemical Engineering, Guangxi University for Nationalities, Nanning 530006, China. E-mail: yaoxingdong@gxun.edu.cn
bGuangxi Colleges and Universities Key Laboratory of Beibu Gulf Oil and Natural Gas Resource Effective Utilization, College of Petroleum and Chemical Engineering, Beibu Gulf University, Qinzhou 535011, China. E-mail: jgao12@163.com
cSchool of Chemistry and Chemical Engineering, Guangxi University, Nanning 530004, China
First published on 14th May 2019
Abstract
When the nascent chain is released from the ribosome, its packing into the apolar environment of the lipid bilayer in the endoplasmic reticulum is facilitated by the Sec61 translocon. In this process, coupling of the conformational change of the channel is essential to transport the nascent chain and meanwhile maintain the membrane permeability barrier. Two molecular dynamics simulations were performed in the current work to investigate the resilience of the lateral gate and the linkage mechanism of the lateral gate, pore ring and plug. The results affirmed that the lateral gate is able to recover its partially-closed state rapidly after the nascent chain segment enters the bilayer. This triggers subsequent motions of the pore ring and plug, which prevent the small molecules passing through the pore. The pore diameter in the partially-closed state is about 6–7 Å. The plug would move upward ∼2 Å if the lateral gate could not close. Two waters permeate through the channel when the lateral gate was open. Water molecules could go across the bilayer via the gap of the open lateral gate due to the occluding of the pore ring and plug.
1. Introduction
Secreted and integral membrane proteins make up to one-third of the biological proteome.1 The secretion proteins are translocated across lipid membranes to the external side through a series of protein-conducting channels2 while the integral membrane proteins also utilize such channels as a means of transfer from the cytoplasm into the lipid bilayer.3 The process is extremely complex and may involves many partners, such as SecA and SecD/F in bacteria and Sec62/Sec63, and Bip in eukaryotes, if it is a posttranslational translocation.4 In cotranslational translocation, the ribosome binds to the Sec61/SecY channel and provides a driving force for the insertion of the nascent peptide into the channel. This pathway exists in all cells and is used for the translocation of secretion proteins as well as for the integration of most membrane proteins.5 When Sec61/SecY combines with a ribosome, the channel undergoes a series of conformational changes so that the polypeptide can go across or into the bilayer while preventing the passage of small molecules.4,5The Sec61/SecY conducting channel is formed by three subunits, α, β, and γ and is conserved in evolution from bacteria to eukaryotic cells.6,7 The α-subunit is comprised of 10 transmembrane (TM) helices that form an hourglass-shaped pore. This pore undertakes the function of protein translocation.7–9 The β-subunit does not interact significantly with other subunits. The γ-subunit has a shape of clamp and stabilizes the α-subunit. The β and γ-subunit are not essential to the translocation. In the hourglass-shaped pore, six bulky hydrophobic residues project their side chains radially inward forming a bottleneck region that is referred to as the “pore ring”. It seals the pore when the channel is in an idle state so that water and small molecules cannot cross the bilayer. When the polypeptide is inserted into the channel, the pore ring closes contact around the polypeptide to prevent small molecules from entering. A short helix called “plug” locates below the pore ring. It moves out of the way as the polypeptide is inserted into the channel, and returns back as the polypeptide exits the channel. Among the 10 TM helices of the α-subunit, TM2 and TM7 form a lateral gate that opens allowing the polypeptide to enter the bilayer. Lateral gate opening might be induced by the interaction between hydrophobic segments of the polypeptide and the lateral gate.10 The secretion protein will go through the channel to the other side of the membrane because the hydrophobicity is insufficient to open the lateral gate.
Since the structural change is essential for the translocon-assisted protein insertion, the detailed study of the structural change is fundamental in elucidation of the molecular nature of the complex process. Many experiments have shown various structure snapshots at the idle and active state of Sec61/SecY that reveal the detail structure of the lateral gate, pore ring and plug.11–13 However, the continued conformational transition process is still unclear, especially the coupling among these structural changes. The crystal structures of the channel at the idle state indicate that the diameter of the pore ring is barely large enough to allow the passage of an extended polypeptide chain, but it can widen when the polypeptide enters the pore.14 The side chains of the pore ring residues also move outwards in the channel accordingly. Do these motions coordinate the subsequent lateral gate opening for integral protein? How does the lateral gate close after the helix leaves and is inserted into the lipid bilayer? In other words, the plasticity and resilience of the channel are not understood very well.
Moreover, the hydrophobicity of the polypeptide likely controls the open state of the lateral gate. However, a molecular dynamics simulation study did not observe the presumed correlation between hydrobobicity and opening of the lateral gate.10 The binding of partners especially a ribosome with Sec61/SecY could open slightly the lateral gate, subsequently fully opening allows the insertion of hydrophobic polypeptide into the bilayer.1,12 Finally, the lateral gate should close because of the hydrophilic environment of the channel. The lateral gate possesses remarkable plasticity,15 but the closing rate may be influenced by the retraction of the pore ring and displacement of the plug. In this paper, we performed coarse-grained (CG) molecule dynamics simulations of the Sec61α/bilayer system to investigate the plasticity and resilience of the channel.
2. Method
The simulation system contains a translocating Sec61 channel (PDB id: 4cg6) and membrane in aqueous solution. The Sec61 complex is comprise of three subunits. The α-subunit forms the channel. The β-subunit does not interact significantly with α and γ-subunit. The γ-subunit clamps the two halves of the α-subunit together, stabilizes the structure of channel.15 We think the main function of both subunits is maintain the whole structure stable. They do not play key role on the series of inside conformational change of channel during translocation. From our results, we did not find broken of the channel either. Given the β and γ-subunit are not necessary for the function of the channel, only the fully open state α-subunit was selected to model, and the signal peptide was removed from the system. The MARTINI coarse-grained (CG) models were built for the Sec61α and model membrane, 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC).16–18 To maintain the helical structure of Sec61α, harmonic restraints were imposed19 on the dihedral angles along the backbone particles with a force constant of 100 kcal per mol per radian for those helices showed as the crystal structure. The strength of the force constant produced rigid helical structures, and thus preserved a stable tertiary structure in the 2 μs simulations. A patch of 392 lipid molecules (196 per leaflet) was equilibrated in a Martini water box for 20 ns at 323 K. Then, the Sec61α was inserted into the DPPC bilayer. An additional 20 chlorine and 18 potassium ions respectively were added in the aqueous solution to neutralize the whole system and mimic cellular electrolyte concentration (0.15 M salt). The overlapped CG DPPC and water particles were deleted. The number (158) of the remaining DPPC molecules in the top leaflet is roughly close to that (146) of the bottom leaflet. The system was minimized using the steepest descent method and then equilibrated via constant particle, pressure, and temperature (NPT) ensemble molecular dynamics simulations in CHARMM20 for 2 ns at 1 atm and 310 K. A harmonic restraint was imposed on the backbone particles of the Sec61α to maintain the open state of the lateral gate. The resulting system consists of a rectangular box of dimensions 105 × 105 × 116 Å3, which yields about a 4 nm thick slab of lipid surrounded by bulk water and ions, and contains approximately 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 CG particles.
800 CG particles.
Afterwards, the simulations were ran using NAMD21 for higher parallel computational efficiency. The system was equilibrated for an additional 2 ns to suppress the subtle differences between CHARMM and NAMD. Similarly, a harmonic constraint was imposed on the same particles to maintain the open state of the lateral gate. Subsequently, two long time (2 μs) NPT production simulations at 1 atm and 310 K were carried out at a time step of 20 fs. In simulation A (SA), we removed the constraints of Sec61 backbone particles to investigate the resilience of the channel structure. For simulation B (SB), we maintained constraints on TM2 and TM7 only allowing the remainder of the protein to move freely. The equilibrated system is shown in Fig. 1. Analyses of the trajectories were performed using CHARMM. All snapshots were made using VMD.22
3. Results and discussion
3.1 Resilience of the lateral gate
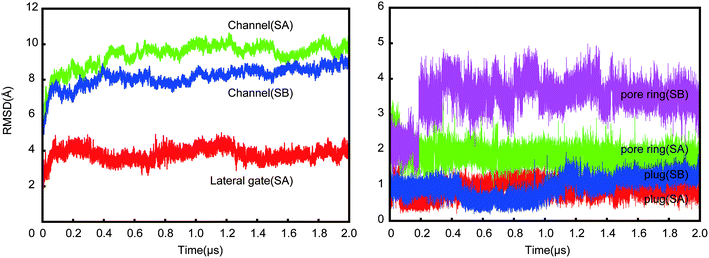
The lateral gate can close in SA, but cannot in SB due to the constraints applied. The RMSDs of the lateral gate and the channel (Fig. 2) show that the channel structure was stable after ∼0.4 μs in SB. In contrast, it stabilized slower (∼1 μs) in SA. The conformation change of the channel in SB was limited by the constrained gate. Although the conformation of the channel in SA stabilized more slowly, the lateral gate changed faster than other parts of the channel. It became a partially closed structure at ∼0.2 μs despite a little fluctuation later. This is in agreement with the prediction of hundreds of nanoseconds for the lateral gate recovery. Recovery of the lateral gate prior to the other parts of the channel implies that the conformation recovery of the other parts might be chain events triggered by the lateral gate. Fig. 3 shows the initial and final snapshots of the SA system. The TM helices were initially almost perpendicular to the bilayer membrane. However, they leaned toward the channel at the cytoplasmic side by the end of simulation; the entrance of the pore became narrow, and the pore at the external side widened resulting perhaps in allowing the entering of small molecules. The unusual pyramidal shaped structure does not resemble the structures reported by experiments.8 It remained stable during additional 0.1 μs simulation (see Fig. S1E†). In addition, three independent 0.1 μs simulations beginning with the SA structure at 1.9 μs were performed. The final structures are all similar pyramidal shape (see Fig. S1B–D†). The closing process of the lateral gate was mainly accomplished by the rotations of TM7 and TM2, which made the lateral gate close at the cytoplasmic side, and open at the external side. The conformational changes of the lateral gate resulted in the pyramidal shape of channel.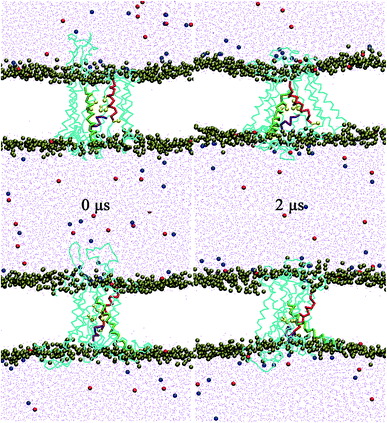 | ||
| Fig. 3 Snapshots of the lateral gate at the initial and the final states in SA. Top are side views from the lateral gate, and bottom are rotated by 90° around the channel axis. Six CG pore ring residues are shown as yellow ball-sticks. The other coloring scheme are same as in Fig. 1. The channel is in the open state at 0 μs and in a partially closed state at 2 μs. | ||
We calculated the distance between TM2 and TM7, which was measured by the distance between the center of mass of backbone of V85-I90 and I292-L297. As shown in Fig. 4, the lateral gate underwent a rapid closure initially, and reached a partially closed state at ∼0.2 μs, then a stable state at ∼0.8 μs after a small fluctuation. The lateral gate maintained a partially closed state with ∼0.5 Å fluctuation. The lateral gate closed about 4 Å (from ∼14 Å at 0 μs to ∼10 Å at 2 μs). A fully closed state was not achieved within 2 μs, but the partially closed state is a stable intermediate state in our simulation. The series of changes resembled those observed in the simulations of Zhang et al.19 In their simulation systems, the translocon contains a hydrophilic substrate with the lateral gate undergoing a change from a fully to a partially open state in 0.5 μs CG simulation. For our simulation (SA), the lateral gate was empty. Because the interior of the channel is a hydrophilic environment,7,23 the free energies of the translocon with and without hydrophilic peptide are roughly equivalent in partially close state of lateral gate.19 They think the slow timescale for relaxation is related to the conformation of the plug. However, no significant conformation change of plug was observed in our simulation. It seems the plug did not affect the open extent of lateral gate. A few of microseconds simulation is not enough to observe the fully close process. Furthermore, the angle between TM2 and TM7 had been fluctuating between ∼60° and ∼80° (Fig. S2†) in our simulation. It is apparently independent on the open degree of the lateral gate. Our simulation shows that the structure of the lateral gate has a good resilience after the peptide inserts into the bilayer, which enables the subsequent movement of the pore ring and plug. The closing of the lateral gate without a ribosome bound also supports a role of the channel-partner binding in inducing the partially open state.
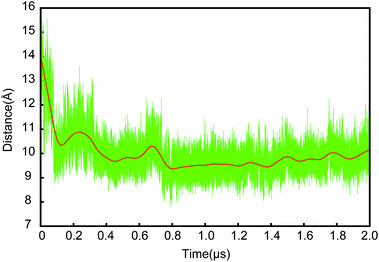 | ||
| Fig. 4 The time evolution of distance between TM2 and TM7. The distance is measured by the center of mass of backbone of V85-I90 and I292-L297. | ||
3.2 Coupling effect of the lateral gate on the pore ring and plug
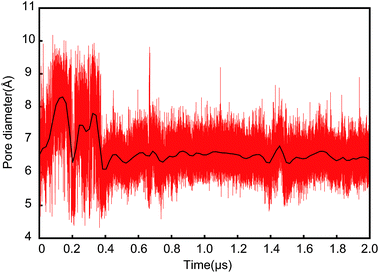
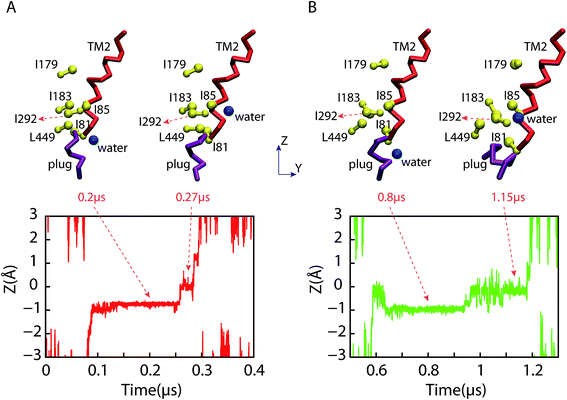
The relative positions of the six pore ring residues in SA fluctuated in ∼0.2 μs (see green profile in Fig. 2 right panel) due to the evolution of the lateral gate. Obviously, a partially closure of the lateral gate causes retraction of the pore. Both pore ring residues and plug seal the channel in idle state to prevent the permeation of small molecules.24 Our simulation provides the trajectory of pore ring residues during the conformation change from open to partially close state. As shown in Fig. 3 (snapshot at 2 μs), when the lateral gate close partially, the top part of TM2 approached to the TM7, the bottom part went away from TM7. The I81 is at the bottom part of TM2. So, it actually located under and out-ring of the other five pore ring residues. As shown in Fig. 5A, six residue side chains moved around the pore ring along with the closure of the lateral gate in simulation. But, the I81 side chain (magenta) was located in the outermost region of the pore, and moved maximum with ∼25 Å on X direction and ∼15 Å on Y direction. Besides, it was ∼5 Å under the pore ring (Fig. 5C). Actually, the pore ring structure is formed mainly by the other five residue side chains at the center of the pore. Among the five residues, the I183, I292 and L449 located in the center of the pore and played important role in maintaining the membrane permeability barrier of the partially closed state of the channel. The diameter of the pore was shown in Fig. 6. Because the initial conformation in SA is that the signal peptide has left the pore and reached the area of the open lateral gate. The diameter with ∼6.5 Å of the pore in the open state (0 μs) was almost the same as that in the partially closed state (2.0 μs). This implies that the pore ring residue side chains went back to the center of the pore as soon as the signal peptide left the pore for the lateral gate. However, the close of the lateral gate led to fluctuations of ∼1.5 Å of the pore diameter before ∼0.4 μs. The largest diameter is over 8 Å. The time of the pore diameter recovery is ∼100 ns, which is longer than 4 ns probed by virtual soft ball.15 Diffusion of ions and water across open membrane pores can occur on timescales not much longer than nanoseconds.25 Therefore, small molecules could go though the pore ring in the duration of fluctuation. The diameter with ∼6.5 Å is smaller than the fluorescence-quenching experiments of 9–15 Å.26,27For the case of the constrained lateral gate in SB, the I179 underwent a leaping shift outwards in the pore (see Fig. 5B), which corresponded to the sudden increase of RMSD of the pore ring at ∼0.2 μs (magenta profile in Fig. 2 right panel). Furthermore, as shown in Fig. 5D, it moved to ∼10 Å upward in the pore ring that was formed mainly by I183, I292 and L449. This dilation of the pore ring is in response to the opened lateral gate.28 Because the lateral gate was constrained to the open state, the six pore ring residues did not form a perfect ring despite their retraction. I292 of TM7 was located in the center of the pore between I183 and L449, and played an important role in preventing passage of small molecules. It requires closure of the lateral gate quickly so that the pore ring recovers to its ring structure. I292 can occlude the center of the pore before the lateral gate closes. The position of L449 also increase ∼5 Å forming a plane with I183 and I292 that is better for occluding the pore.
As shown in Fig. 7, the motion of the plug along the direction perpendicular to the bilayer are different in SA and SB. In SB, the opened lateral gate destabilized the plug,15 and led to its motion upward ∼3 Å at ∼1 μs, approaching the pore ring. I183, I292, L449 and the plug occluded the pore together. The tighter interactions between the plug and the three residues compensated the imperfect of the pore ring, and maintained the membrane permeability barrier.11 In SA, the plug stayed near the original position all the time despite small fluctuations while the lateral gate closed rapidly (Fig. S3†). It is significantly stable throughout SA, which agrees with the observations in simulations of Tian et al.15
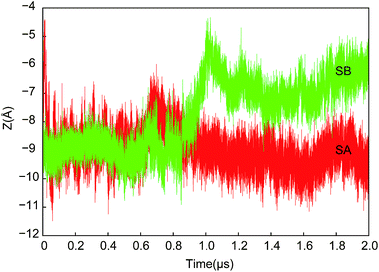 | ||
| Fig. 7 Z coordinates of center of mass of the plug as a function of time in both SA and SB. The plug in SB moved upward to approach the pore ring and occluded the pore. | ||
3.3 Water permeation through the channel and the membrane
The pore ring and plug play a key role on preventing small molecules from passing through the bilayer during the closure of the lateral gate.24 In both SA and SB, there was no water permeation through the pore formed by the pore ring residues. However, we observed two water particles crossing the bilayer via the lateral gate gap between TM2 and TM7 in SB. Similar permeation was not observed in SA. That means it is possible to pass through the bilayer for the small molecules if the lateral gate closes slowly. The slow closing of the lateral gate might provide a pathway for small molecules to pass through the membrane. Fig. 8 shows the permeation process. A water particle went into the pore through the open gate from the external side, but did not pass via the center of the pore due to the occlusion of the plug and pore ring residues. In the whole permeation process, the water particle was between TM2 and TM7. It went to the cytoplasmic side along TM2 after arriving at the center of the channel. Both permeations occurred from the external to the cytoplasmic side. We found the plug and open gate formed a more commodious entrance at the external side than cytoplasmic side. | ||
| Fig. 8 A series of representative snapshots from a CG water permeation trajectory passing through the channel when the lateral gate is in an open state. The top are side views, and the bottom corresponds to top views. The permeation occurred in 0.08–0.3 μs. Coloring scheme is same as in Fig. 3. Snapshots show that the CG water entered the channel between TM2 and TM7 at the external side, skirted I81 and I85, and arrived at the cytoplasmic side along TM2 helix. | ||
We checked the two permeation processes. Fig. 9 shows the positions of the water particles for the two permeations. The water particle reached the position rapidly that is ∼1 Å below the center of the channel, remaining there for a long time before it exited the pore. This position is just above the plug and under the pore ring residue side chains (refer to Fig. 5 and 7). The pore ring actually prevented the water from passing through the pore with the result that it went along the TM2 helix to reach the external side. The first permeation happened at ∼0.08 μs and finished at ∼0.3 μs. Once the water particle skirted I81 and I85, it rapidly reached the surface between bilayer and water along TM2. Similarly, a water particle needed to skirt I81 and I85 in the second permeation.
4. Conclusions
After the signal peptide left the lateral gate and started to insert into the bilayer, the TM2 and TM7 approached each other quickly resulting in the close of the lateral gate. Both TM2 and TM7 leaned into the channel and formed an angle with 60–80°. The pore retracted at the cytoplasmic side, and expanded at the external side, which resulted in the outward movement of I81 residue. But the other pore ring residues and plug always remained in the pore. The pore diameter measured with the pore ring residues reduced along with the close of the lateral gate. The simulation showed that the lateral gate recovered to a partially closed state from the fully open state in ∼0.2 μs. Subsequently, it resulted in a retraction of the pore. The pore ring was formed, and I183, I292 and L449 located at the center. They played important role on preventing small molecules from passing through the pore. If the lateral gate could not close (SB), a ringed structure could not be formed either. The I292 located at the center of the pore, is able to partly occlude the pore. Moreover, the plug would go upward to approach the pore ring. In summary, the lateral gate can recover structure rapidly, causing a conformational change of the pore ring. The pore could be occluded by the pore ring and plug regardless of whether the lateral gate was closed or not, which is in agreement with experimental data.29Although the pore is occluded, it is still possible for transporting small molecules across the apolar membrane via the gap of open lateral gate. Two permeation events were observed in our simulation (SB). Owing to the expanding of the pore at the external side, the water molecules are prone to enter the channel from this side. The two water particles entered the channel between the TM2 and TM7, skirting I81 and I85, and finally arriving at the other side along TM2. No similar permeation process is observed in the simulation with the closed lateral gate (SA).
In this work, we performed two 2 μs CG molecular dynamics simulations. Based in the analysis of the channel conformation, we aimed to provide insight into the resilience of the lateral gate and its linkage mechanism with the pore ring and plug. The permeation barrier of the membrane was maintained by the pore ring and plug during Sec61-assisted peptide translocation. However, it was still possible for water molecules to pass through the membrane via the gap of the open lateral gate if the lateral gate closed slowly.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
Computations were performed at the high-performance computing platform of Guangxi University. This work was supported by Guangxi Natural Science Foundation (No. 2016GXNSFCA380011) and Beibu Gulf University Scientific Research Foundation for the Introduction of Talent (No. 2018KYQD04).References
- A. Tsirigotaki, J. De Geyter, N. Šoštarić, A. Economou and S. Karamanou, Nat. Rev. Microbiol., 2017, 15, 21–36 CrossRef CAS PubMed.
- K. E. S. Matlack, W. Mothes and T. A. Rapoport, Cell, 1998, 92, 381–390 CrossRef CAS PubMed.
- S. H. White and G. von Heijne, Curr. Opin. Struct. Biol., 2004, 14, 397–404 CrossRef CAS PubMed.
- T. A. Rapoport, Nature, 2007, 450, 663–669 CrossRef CAS PubMed.
- E. Park and T. A. Rapoport, Annu. Rev. Biophys., 2012, 41, 21–40 CrossRef CAS PubMed.
- A. R. Osborne, T. A. Rapoport and B. van den Berg, Annu. Rev. Cell Dev. Biol., 2005, 21, 529–550 CrossRef CAS PubMed.
- B. van den Berg, W. M. Jr Clemons, I. Collinson, Y. Modis, E. Hartmann, S. C. Harrison and T. A. Rapoport, Nature, 2004, 427, 36–44 CrossRef CAS PubMed.
- T. Tsukazaki, H. Mori, S. Fukai, R. Ishitani, T. Mori, N. Dohmae, A. Perederina, Y. Sugita, D. G. Vassylyev, K. Ito and O. Nureki, Nature, 2008, 455, 988–991 CrossRef CAS PubMed.
- J. Zimmer, Y. Nam and T. A. Rapoport, Nature, 2008, 455, 936–943 CrossRef CAS PubMed.
- J. C. Gumbart, I. Teo, B. Roux and K. Schulten, J. Am. Chem. Soc., 2013, 135, 2291–2297 CrossRef CAS PubMed.
- M. Gogala, T. Becker, B. Beatrix, J.-P. Armache, C. Barrio-Garcia, O. Berninghausen and R. Beckmann, Nature, 2014, 506, 107–110 CrossRef CAS PubMed.
- R. M. Voorhees, I. S. Fernández, S. H. W. Scheres and R. S. Hegde, Cell, 2014, 157, 1632–1643 CrossRef CAS PubMed.
- R. M. Voorhees and R. S. Hegde, Science, 2016, 351, 88–91 CrossRef CAS PubMed.
- S. Haider, B. A. Hall and M. S. Sansom, Biochemistry, 2006, 45, 13018–13024 CrossRef CAS PubMed.
- P. Tian and I. Andricioaei, Biophys. J., 2006, 90, 2718–2730 CrossRef CAS PubMed.
- S. J. Marrink, A. H. de Vries and A. E. Mark, J. Phys. Chem. B, 2004, 108, 750–760 CrossRef CAS.
- S. J. Marrink, H. J. Risselada, S. Yefimov, D. P. Tieleman and A. H. de Vries, J. Phys. Chem. B, 2007, 111, 7812–7824 CrossRef CAS PubMed.
- L. Monticelli, S. K. Kandasamy, X. Periole, R. G. Larson, D. P. Tieleman and S. J. Marrink, J. Chem. Theory Comput., 2008, 4, 819–834 CrossRef CAS PubMed.
- B. Zhang and T. F. Miller, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 5399–5404 CrossRef CAS PubMed.
- B. R. Brooks, C. L. Brooks, A. D. Mackerell, L. Nilsson, R. J. Petrella, B. Roux, Y. Won, G. Archontis, C. Bartels, S. Boresch, A. Caflisch, L. Caves, Q. Cui, A. R. Dinner, M. Feig, S. Fischer, J. Gao, M. Hodoscek, W. Im, K. Kuczera, T. Lazaridis, J. Ma, V. Ovchinnikov, E. Paci, R. W. Pastor, C. B. Post, J. Z. Pu, M. Schaefer, B. Tidor, R. M. Venable, H. L. Woodcock, X. Wu, W. Yang, D. M. York and M. Karplus, J. Comput. Chem., 2009, 30, 1545–1614 CrossRef CAS PubMed.
- J. C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kal and K. Schulten, J. Comput. Chem., 2005, 26, 1781–1802 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graph., 1996, 14, 33–38 CrossRef CAS.
- K. S. Crowley, S. Liao, V. E. Worrell, G. D. Reinhart and A. E. Johnson, Cell, 1994, 78, 461–471 CrossRef CAS PubMed.
- E. Park and T. A. Rapoport, Nature, 2011, 473, 239–242 CrossRef CAS PubMed.
- F. Zhu, E. Tajkhorshid and K. Schulten, Biophys. J., 2004, 86, 50–57 CrossRef CAS PubMed.
- B. D. Hamman, J. C. Chen, E. E. Johnson and A. E. Johnson, Cell, 1997, 89, 535–544 CrossRef CAS.
- B. D. Hamman, L. M. Hendershot and A. E. Johnson, Cell, 1998, 92, 747–758 CrossRef CAS PubMed.
- D. Hizlan, A. Robson, S. Whitehouse, V. A. Gold, J. Vonck, D. Mills, W. Kühlbrandt and I. Collinson, Cell Rep., 2012, 1, 21–28 CrossRef CAS PubMed.
- P. F. Egea and R. M. Stroud, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 17182–17187 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra01684h |
| This journal is © The Royal Society of Chemistry 2019 |