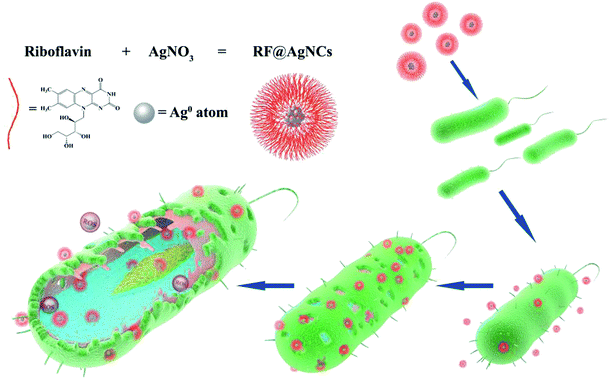
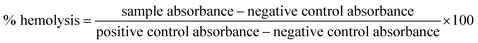Open Access Article
Open Access ArticleRiboflavin-protected ultrasmall silver nanoclusters with enhanced antibacterial activity and the mechanisms†
Xizhe Li,
Tao Fu ,
Bingyu Li,
Peng Yan and
Yayan Wu
,
Bingyu Li,
Peng Yan and
Yayan Wu *
*
Key Laboratory of Biomedical Information Engineering of Ministry of Education, School of Life Science and Technology, Xi'an Jiaotong University, 710049, Xi'an, PR China. E-mail: yayanwu@mail.xjtu.edu.cn
First published on 30th April 2019
Abstract
Developing silver nanomaterials with efficient antimicrobial properties is of importance for combating bacteria. Here, we report ultrasmall riboflavin-protected silver nanoclusters (RF@AgNCs) that can effectively kill or suppress the growth of Gram-positive S. aureus, Gram-negative E. coli, and fungi C. albicans. Riboflavin (RF) with intrinsic biocompatibility was used as a surface ligand to synthesize silver nanoclusters. TEM revealed that the synthesized RF@AgNCs were ultrasmall (2.4 ± 1.2 nm), spherical and well-dispersed. Antibacterial activity tests showed that RF@AgNCs possessed superior antibacterial efficacy in comparison with RF, AgNPs and mixed RF and AgNPs (RF + AgNPs). The mechanisms of antibacterial activity of RF@AgNCs were studied by fluorescence microscopy-based Live/Dead cell staining assays and ROS measurement. And the results illustrated that the integrity of the bacteria membrane was disrupted and intracellular high level ROS generation was induced by RF@AgNCs. The cytotoxic activities were also assessed and RF@AgNCs were found to be non-toxic to human red blood cells and mammalian cells. With the highly efficient antibacterial activity and acceptable biocompatibility, RF@AgNCs hold great promise in biomedical applications as well as in water sterilization and the textile industry.
1. Introduction
Silver (Ag) and its compounds have been accepted as antimicrobial agents in medicine and public health for centuries. In ancient times, people used silver containers to store water or food to keep it fresh. Then silver foil was used to cover wounds for the prevention of ulceration. Silver nitrate (AgNO3) was introduced to prevent newborn gonorrhoeal eye infections in 1884. In recent years, a new generation of antimicrobials, silver nanoparticles (AgNPs),1–4 has drawn significant attention owing to the intrinsic broad-spectrum antibacterial properties, non-drug resistance, good biocompatibility,5 and low dosage. They show more advantages than bulk Ag. It is reported that a lower concentration of AgNPs can inhibit or kill a wide range of bacteria in a few minutes.6 Recent studies show that AgNPs are able to improve wound healing.7–9 With the emergence of antibiotic-resistant bacteria, it is highly desirable to develop new efficacious broad-spectrum antibacterial agents.10,11 All these make AgNPs or their complexes one of the most promising antibacterial agents.12 Now they have been extensively used in medical applications including wound dressings and treatment, food packaging, the textile industry, water treatment, and environmental management.Some studies have reported that the antibacterial activities of AgNPs depend on their size,13,14 shape,15 surface charge,16 and surface coating.17 AgNPs with different sizes exerted different antibacterial properties.1,18 AgNPs below 10 nm exhibited surface-dependent ion release and toxicity. They can effectively penetrate the bacterial wall and affect the integrity of membrane and the viability of bacteria, exhibiting higher potency against bacteria cells.19,20 Xie J. P. et al. have reported that silver nanoclusters (AgNCs) with 2–3 nm possess higher activities than AgNPs.19 In addition, surface coating (surface ligand) plays a prominent role in toxicity of nanoparticles (NPs). AgNPs are typically synthesized through surface coating which reduces AgNO3 as well as stabilizes AgNPs in suspension. Hinton D. E. et al. have demonstrated that dissolution of AgNPs was largely dependent on their particle size, but their aggregation behavior and toxicity were more dependent on coating materials.21,22 Doktycz M. J. et al. have indicated that bacteria growth inhibition depended on the surface coating after the comparison of protein-protected AgNPs, colloidal AgNPs, and oleate-protected AgNPs.23 Zhou Q. F. synthesized AgNPs with four different coatings and illustrated that surface coating controlled silver ion release and their antibacterial activity.24 Moreover, surface coating determines potential cytotoxicity of AgNPs to human cells or the effect on environmental health.25 Therefore, it is fundamentally important to optimize surface coating to develop small or ultrasmall AgNPs with high antibacterial properties but a proper human biocompatibility which is essential for daily life application.
Riboflavin (RF, vitamin B2), an important component of cofactors FAD (flavin adenine dinucleotide) and FMN (flavin mononucleotide), is crucial for biological oxidation metabolism. RF can act as growth inhibitor and fungicide because riboflavin molecule can intercalate into DNA or RNA, making pathogens unable to replicate. Some studies have reported that RF effectively enhanced resistance against virus and fungi in plant.26,27 Others have indicated that RF can promote the disappearance of inflammation and can be used to treat burns or stomach ulcers and prevent upper oral ulcers. RF/light combination or their complex with other compounds are often used as antimicrobial or antiviral agents.28,29 Moreover, there is a conjugated double bond of isoalloxazine ring in RF molecular structure which can be involved in redox reactions. RF can be used for the reduction of AgNO3 because the reduction potential is lower than Ag.30,31 Therefore, using riboflavin as surface coating to synthesize AgNPs may integrate the characteristics of RF and antimicrobial effects of silver nanoparticles which may show more efficient antimicrobial than RF or AgNPs alone. Furthermore, RF has intrinsic biocompatibility and RF as surface ligand may have no toxicity to human or environment.
In this study, riboflavin-protected silver nanoclusters (RF@AgNCs) were prepared by RF reducing AgNO3. RF has certain antibacterial properties and serving as surface coating can reduce the cytotoxicity of silver nanoclusters. The antimicrobial activity against Gram-positive bacteria (S. aureus), Gram-negative bacteria (E. coli), and fungus (C. albicans) were evaluated. Fluorescence images showed that RF@AgNCs could disrupt bacteria cell membranes. The antibacterial mechanism was discussed by intracellular reactive oxygen species (ROS) level assay. The schematic diagram of RF@AgNCs against bacteria is shown in Fig. 1. In vitro biocompatibility of AgNCs was evaluated by hemolysis assay and MTT test. The results show that the RF@AgNCs have superior antimicrobial and antifungal effects and low toxicity to normal human cells.
2. Materials and methods
2.1. Reagents and materials
Riboflavin and AgNO3 were purchased from Aladdin Bio-Chem Technology Co., LTD (Shanghai, China). Phosphate-buffered saline (PBS, pH 7.2) was bought from Hyclone, Thermo Scientific (Massachusetts, USA). Hoechst 33342 and propidium iodide (PI) were purchased from Beyotime Biotechnology (Shanghai, China). 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) and 2′,7′-dichlorofluorescin diacetate (DCFH-DA) were bought from Sigma-Aldrich (St. Louis, USA). The Gram-positive Staphylococcus aureus (S. aureus) (CGMCC 1.0089), Gram-negative Escherichia coli (E. coli) (CMCC (B) 44102), and fungi Candida albicans (C. albicans) (CGMCC 2.2086) were bought from the Microbiology Institute of Shaanxi Province (Xi'an, China). Human Umbilical Vein Endothelial (HUVE) cells (KCB 2012087YJ) and Human Kidney (HK-2) cells (KCB 200815YJ) were purchased from the Conservation Genetics, the Chinese Academy of Sciences Kunming Cell Bank (Kunming, China). The human blood sample was supplied by Xi'an No. 3 Hospital. All experiments performed were in compliance with relevant laws, as well as the guidelines of Biomedical Research Ethics Committee of the School of Life Science & Technology, Xi'an Jiaotong University. And the experiments were approved by all the above mentioned institutes. The volunteer had provided his informed consent for the experiments. Ultrapure water (18.2 MΩ) was used throughout the experiment. All glassware and magnetic stirring bars were soaked in aqua regia, rinsed with ultrapure water, and dried in an oven before use.2.2. Preparation of and characterization of RF@AgNCs
Riboflavin-protected silver nanoclusters (RF@AgNCs) were synthesized according to the previously reported method30 with some modification. In brief, 1 mL 1 mM homogeneous riboflavin solution was added into 8 mL water under stirring at room temperature. Then, 1 mL AgNO3 solution (0.1 mM) was introduced dropwise to the above solution. The mixture was stirred for 3 h at 4 °C. The color of the mixture changed from bright yellow to reddish orange, indicating the formation of AgNCs. The morphologies of RF@AgNCs were observed with transmission electron microscopy (TEM, JEM-2100F, JEOL). The measurement of UV-vis absorption (Tecan Infinite M200 Pro, Switzerland) and zeta potential (Mastersizer Nano ZS90, Malvern) were performed to characterize the surface modification of RF@AgNCs.Silver nanoparticles (AgNPs) were synthesized based on the previous method.32 Briefly, 10 mM Tollen reagent was firstly prepared by AgNO3 and ammonium hydroxide at a certain proportion. Then, 0.5 g β-cyclodextrin and 2 mL 50 mM glucose were added into 1 mL 10 mM Tollen reagent and ultrapure water was added to the total volume of 50 mL. When the mixture was heated to be boiled, it was continuously stirred and refluxed for 15 min until the solution turned to light yellow. The synthesized AgNPs was analyzed by the Microplate Reader and the Malvern Mastersizer.
2.3. Bacteria growth and antimicrobial activity test
S. aureus, E. coli, and C. albicans were recovered from frozen glycerol stocks on Luria–Bertani (LB) agar plates or potato dextrose (PD) agar plates at 37 °C. Single colony of bacteria were then incubated for 24 h to exponential phase. Then, the bacteria suspension was diluted by PBS to achieve the concentration of 1 × 107 colony forming unit (CFU) per mL (OD600 = 0.05) prior to antibacterial test.Antibacterial activities of RF@AgNCs against each kind of bacteria were determined by the colony counting method. PBS, RF (0.28 μg mL−1), AgNPs (0.125 μg mL−1, on the basis of Ag atoms), mixed RF and AgNPs (RF + AgNPs, 0.125 μg mL−1 Ag atoms and 0.28 μg mL−1 RF) were used as control groups. Each bacterial group was incubated separately with control groups and RF@AgNCs (0.125 μg mL−1, on the basis of Ag atoms) under continuous orbital shaking at 200 rpm at 37 °C for 3 h. Then the treated bacteria suspension of each group was diluted by 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 folds, and 50 μL of the dilutions were added into the agar plates to let the bacteria grow at 37 °C. Finally, the inactivation of bacterial cells was evaluated by counting the number of colony forming units (CFU) on the agar plate after 48 h incubation. Each assay was carried out in triplicate.
000 folds, and 50 μL of the dilutions were added into the agar plates to let the bacteria grow at 37 °C. Finally, the inactivation of bacterial cells was evaluated by counting the number of colony forming units (CFU) on the agar plate after 48 h incubation. Each assay was carried out in triplicate.
2.4. Fluorescent microscopy study
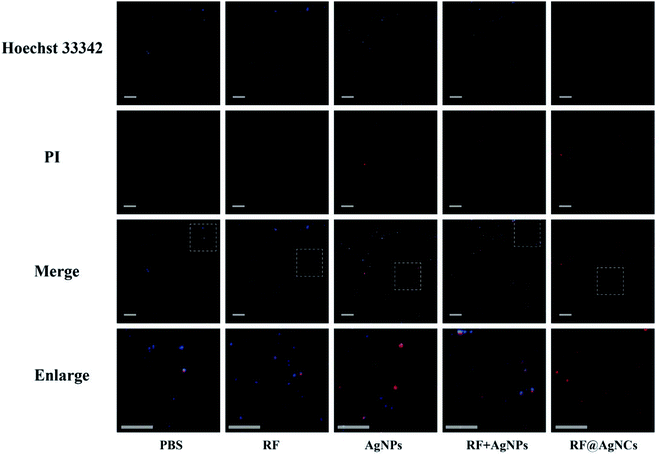
The bacterial cells (1 × 107 CFU mL−1) were incubated separately with RF@AgNCs and control groups at 37 °C for 3 h. Then 1 μg mL−1 Hoechst 33342 and 5 μg mL−1 PI were added to the treated bacteria solutions respectively and the solutions were kept in dark for 15 min. After rinsed with PBS for 3 times, the new suspension (5 μL) was dropped on a glass slide for fluorescent microscopy imaging (Nikon ECLIPSE Ti, Japan).2.5. TEM images of bacteria cells
The bacteria solutions (1 × 107 CFU mL−1, 500 μL) were mixed with RF@AgNCs solution (2 μg mL−1, 500 μL) at room temperature, respectively. After incubation for 2 h, 2 μL of the mixture were dropped on a carbon-coated Cu grid (400 Mesh) for TEM observation.2.6. ROS measurement
DCFH-DA fluorescent probe was used for the evaluation of the level of intracellular reactive oxygen species (ROS) when exposed to NPs. The bacteria cells were incubated with RF@AgNCs and control groups for 2 h respectively. The treated bacteria solution was incubated with 5 μM DCFH-DA in an orbital shaker (200 rpm) at 37 °C for 30 min. Then the bacteria suspension was centrifuged, washed and re-suspended in ultrapure water to remove the redundant DCFH-DA outside cells. The fluorescent signal (λex/λem = 484/525 nm) was measured on a fluorescence microscope (Nikon ECLIPSE Ti, Tokyo, Japan) and Microplate Reader (Tecan Infinite M200 Pro) to evaluate ROS production in bacteria cells. Each sample was taken for three replicates.2.7. Hemolysis assay
Hemolysis assay was performed with red blood cells (RBCs). The erythrocytes were collected from fresh human blood (5 mL) by centrifugation at 2000 rpm for 5 min, and washed three times with PBS. Then 0.5 mL of RBC solution was mixed with 0.5 mL of RF@AgNCs. After incubation at 37 °C (150 rpm) for 30 min, the mixture was centrifuged at 4000 rpm for 5 min. At last, the absorbance of the supernatant was measured at 570 nm. RBCs incubated with PBS and water were treated as negative and positive control, respectively. All samples were prepared in triplicate at one test. The percent hemolysis was calculated by the following equation:2.8. MTT assay
Two types of cells (HK-2 cells and HUVE cells) were used in MTT assay to evaluate the effects of RF@AgNCs treatment on cell proliferation behavior. In brief, cells were maintained in low-glucose Dulbecco's Modified Eagle's Medium with 10% fetal bovine serum (FBS) and 1% antibiotic (100 U mL−1 penicillin and 100 μg mL−1 streptomycin). Cells were cultured at 37 °C under a humidified atmosphere of 5% CO2 (96-well plate, 4000 cells per well). Media was removed and cells were washed with PBS. Cells were treated with RF@AgNC at various concentrations (0.125–5 μg mL−1) for 24 and 48 h, respectively. Then cell samples were incubated with MTT solution for another 4 h to determine living cells. The culture medium containing MTT was removed and 150 μL dimethyl sulfoxide (DMSO) was added into each well to dissolve the formazan crystals. The OD570 nm value was measured by a Microplate Reader. Cells without any treatment were normalized as 100%. Each experiment was performed in triplicate.3. Results and discussion
3.1. Characterization of RF@AgNCs
Riboflavin (vitamin B2) has two forms of oxidation and reduction types because there is a conjugated double bond of isoalloxazine ring in its structure. In this study, riboflavin (RF) was used as reducing agent to synthesize RF-protected silver nanoclusters (RF@AgNCs). Synthesized RF@AgNCs were characterized by UV-vis absorption spectrum, TEM, and zeta potential measurements. In UV-vis absorption spectrum, the peaks decrease of RF and the appearance of the peak at 535 nm indicates the formation of RF@AgNCs (Fig. 2a). TEM images show that synthesized RF@AgNCs are well-dispersed and quasi-spherical (Fig. 2b). The lattice fringes of the RF@AgNCs are consistent with metallic Ag which exhibits a discerned lattice spacing of 2.07 Å (inset in Fig. 2b). The size distribution of RF@AgNCs was in concordance with normal distribution curve and the average size was 2.4 ± 1.2 nm (Fig. S1†). The zeta potential of RF@AgNCs was +0.283 mV, indicating a neutral surface of RF@AgNCs. Moreover, AgNPs as control group were also characterized by UV-vis absorption spectrum and zeta potential measurements (Fig. S2†).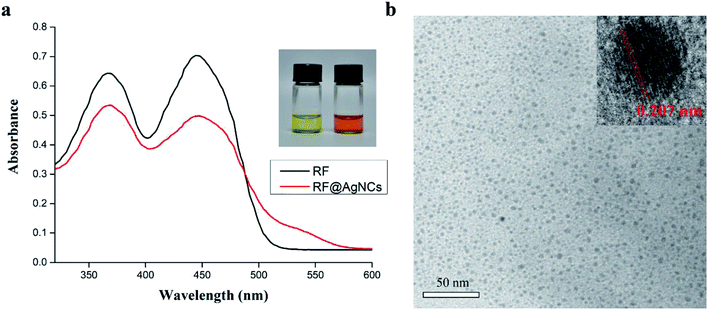 | ||
| Fig. 2 Characteristics of synthesized RF@AgNCs. (a) UV-vis absorption spectrum of RF and RF@AgNCs. (b) TEM image of RF@AgNCs. | ||
3.2. In vitro antibacterial activity of RF@AgNCs
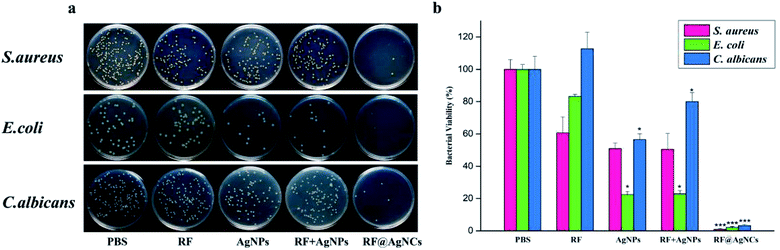
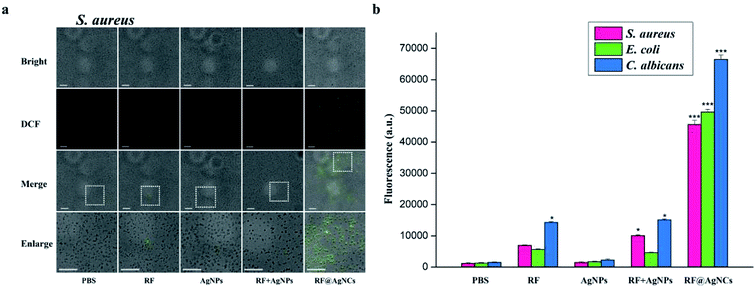
The plate counting method was used to evaluate the antimicrobial activity of RF@AgNCs against Gram-positive S. aureus, Gram-negative E. coli, and fungi C. albicans. Fig. 3a shows the antibacterial photographs after the bacteria were treated with RF@AgNCs (0.125 μg mL−1, on the basis of Ag atoms) and the control groups including PBS, RF (0.28 μg mL−1), AgNPs (0.125 μg mL−1), and the mixed RF & AgNPs (RF + AgNPs) for 2 h, respectively. Apparently, RF@AgNCs efficiently inhibited the growth of three kinds of bacteria because the colonies were hardly observed on the Petri dish. Although AgNPs and mixed RF & AgNPs showed antibacterial effects against E. coli, the colony number was a bit more than that observed in the sample treated by RF@AgNCs. The relative viabilities of S. aureus, E. coli and C. albicans after treated with RF@AgNCs were 0.83, 2.08, 3.24%, respectively (Fig. 3b). The results indicate that RF@AgNCs are more effective in inhibiting the proliferation of the bacteria and fungi than control groups (RF, AgNPs, and RF + AgNPs), exhibiting superior antibacterial efficacy.3.3. Study on the mechanisms about antibacterial activity of RF@AgNCs
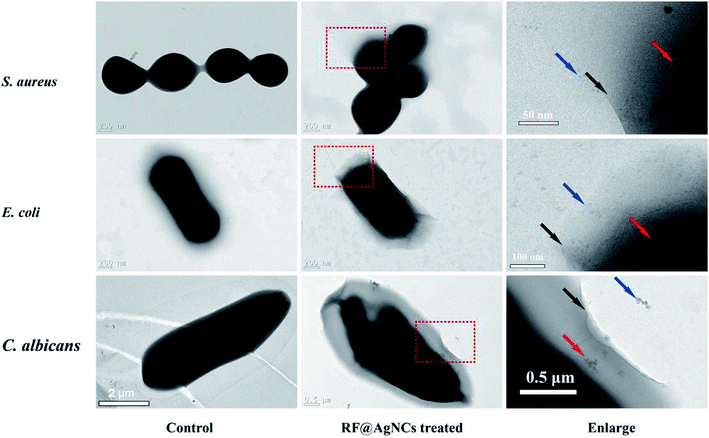
We also used TEM to directly observe the change of bacterial cell membranes integrity. Morones J. R. reported that AgNPs (1–10 nm) could enter into bacterial cells and cause further damage by possibly interacting with sulfur- and phosphorus containing compounds.20 Fig. 5 shows TEM images of S. aureus, E. coli, and C. albicans untreated and treated with RF@AgNCs for 2 h. In the absence of RF@AgNCs, three bacterial cells exhibit the typical smooth structure, while the morphology and membrane integrity were strongly changed after the treatment with RF@AgNCs. We can see that RF@AgNCs penetrate bacteria cell easily and are aggregated or accumulated inside cells, causing structural changes in bacterial cell walls or membranes. It can even be observed that some bacterial cells were broken (Fig. S5†). These morphological changes demonstrate the beneficial antibacterial activities of RF@AgNCs against Gram-positive S. aureus, Gram-negative E. coli, and fungi C. albicans.
3.4. Hemolysis and cytotoxicity assays
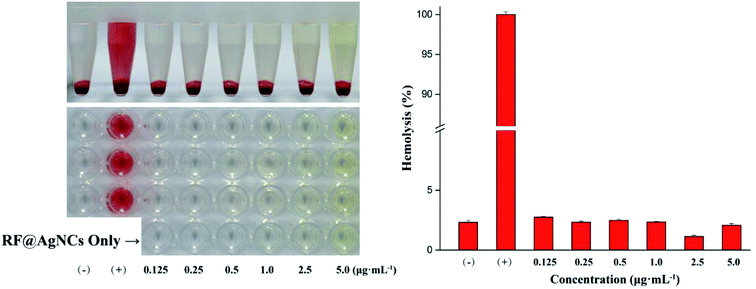
To evaluate the biocompatibility or cytotoxicity of RF@AgNCs, hemolysis assay on human red blood cells (RBCs) and MTT assay on mammalian cells of HUVE cells and HK-2 cells were carried out. As shown in Fig. 7, the treatment of RBCs with RF@AgNCs at various concentrations (0.125–5 μg mL−1) show no observable hemolytic activity, indicating the biocompatibility of RF ligands with human blood cells. MTT assay were used to assess the proliferation of HUVE cells and HK-2 cells after exposed to RF@AgNCs at various concentrations (0.125–5 μg mL−1). As shown in Fig. 8, RF@AgNCs do not show obvious influence on the proliferation of either HUVE cells or HK-2 cells after 24 h and 48 h incubations, respectively. Therefore, the results demonstrate that RF@AgNCs have good biocompatibility with human blood cells and mammalian cells.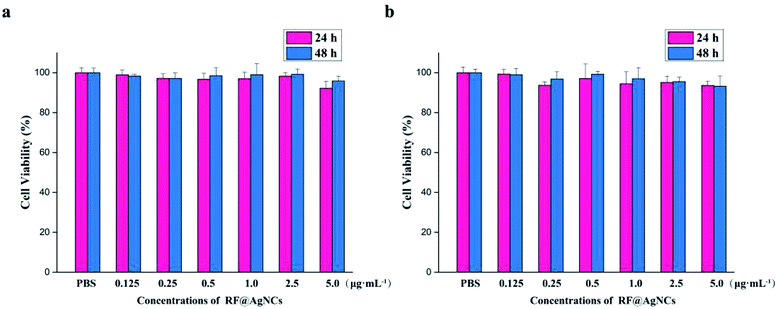 | ||
| Fig. 8 Cell viability assay of RF@AgNCs at different concentrations. (a) HUVE cells and (b) HK-2 cells. | ||
4. Conclusions
In summary, we used riboflavin as biocompatible surface ligand to synthesize ultrasmall silver nanoclusters (RF@AgNCs). The RF@AgNCs exhibited very good antibacterial properties against S. aureus, E. coli, and C. albicans. The antibacterial mechanism studies showed that the integrity of bacteria cell membranes was disrupted and a large amount of intracellular ROS was produced after exposure to RF@AgNCs. Significantly, RF@AgNCs maintained intrinsic good biocompatibility with red blood cells and mammalian cells, making these systems promising for antimicrobial therapeutic use. With the efficient antimicrobial effect and good or acceptable biocompatibility, RF@AgNCs would have great potential in biomedical applications as well as other fields such as textile industry.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 21175103 and 21125525), and the Key Program of Natural Science Foundation of Shaanxi Province (Grant No. 2018JZ2002).References
- M. Rai, A. Yadav and A. Gade, Biotechnol. Adv., 2009, 27, 76–83 CrossRef CAS PubMed.
- W. R. Li, X. B. Xie, Q. S. Shi, H. Y. Zeng, Y. S. Ou-Yang and Y. B. Chen, Appl. Microbiol. Biotechnol., 2010, 85, 1115–1122 CrossRef CAS PubMed.
- V. K. Sharma, R. A. Yngard and Y. Lin, Adv. Colloid Interface Sci., 2009, 145, 83–96 CrossRef CAS PubMed.
- G. Franci, A. Falanga, S. Galdiero, L. Palomba, M. Rai, G. Morelli and M. Galdiero, Molecules, 2015, 20, 8856–8874 CrossRef CAS PubMed.
- I. V. Vrcek, I. Zuntar, R. Petlevski, I. Pavicic, M. D. Sikiric, M. Curlin and W. Goessler, Environ. Toxicol., 2016, 31, 679–692 CrossRef CAS PubMed.
- S. Chernousova and M. Epple, Angew. Chem., Int. Ed., 2013, 52, 1636–1653 CrossRef CAS PubMed.
- M. Ovais, I. Ahmad, A. T. Khalil, S. Mukherjee, R. Javed, M. Ayaz, A. Raza and Z. K. Shinwari, Appl. Microbiol. Biotechnol., 2018, 102, 4305–4318 CrossRef CAS PubMed.
- S. B. Seo, S. H. S. Dananjaya, C. Nikapitiya, B. K. Park, R. Gooneratne, T. Y. Kim, J. Lee, C. H. Kim and M. De Zoysa, Fish Shellfish Immunol., 2017, 68, 536–545 CrossRef CAS PubMed.
- J. Tian, K. K. Y. Wong, C. M. Ho, C. N. Lok, W. Y. Yu, C. M. Che, J. F. Chiu and P. K. H. Tam, ChemMedChem, 2007, 2, 129–136 CrossRef CAS PubMed.
- M. K. Rai, S. D. Deshmukh, A. P. Ingle and A. K. Gade, J. Appl. Microbiol., 2012, 112, 841–852 CrossRef CAS PubMed.
- R. K. Kunkalekar, M. M. Naik, S. K. Dubey and A. V. Salker, J. Chem. Technol. Biotechnol., 2013, 88, 873–877 CrossRef CAS.
- T. C. Dakal, A. Kumar, R. S. Majumdar and V. Yadav, Front. Microbiol., 2016, 7, 17 Search PubMed.
- A. Panacek, L. Kvitek, R. Prucek, M. Kolar, R. Vecerova, N. Pizurova, V. K. Sharma, T. Nevecna and R. Zboril, J. Phys. Chem. B, 2006, 110, 16248–16253 CrossRef CAS PubMed.
- C. Carlson, S. M. Hussain, A. M. Schrand, L. K. Braydich-Stolle, K. L. Hess, R. L. Jones and J. J. Schlager, J. Phys. Chem. B, 2008, 112, 13608–13619 CrossRef CAS PubMed.
- S. Pal, Y. K. Tak and J. M. Song, Appl. Environ. Microbiol., 2007, 73, 1712–1720 CrossRef CAS PubMed.
- A. M. El Badawy, R. G. Silva, B. Morris, K. G. Scheckel, M. T. Suidan and T. M. Tolaymat, Environ. Sci. Technol., 2011, 45, 283–287 CrossRef CAS PubMed.
- S. H. Tang and J. Zheng, Adv. Healthcare Mater., 2018, 7, 10 Search PubMed.
- G. A. Martinez-Castanon, N. Nino-Martinez, F. Martinez-Gutierrez, J. R. Martinez-Mendoza and F. Ruiz, J. Nanopart. Res., 2008, 10, 1343–1348 CrossRef CAS.
- K. Y. Zheng, M. I. Setyawati, T. P. Lim, D. T. Leong and J. P. Xie, ACS Nano, 2016, 10, 7934–7942 CrossRef CAS PubMed.
- J. C. Jin, X. J. Wu, J. Xu, B. B. Wang, F. L. Jiang and Y. Liu, Biomater. Sci., 2017, 5, 247–257 RSC.
- K. W. H. Kwok, W. Dong, S. M. Marinakos, J. Liu, A. Chilkoti, M. R. Wiesner, M. Chernick and D. E. Hinton, Nanotoxicology, 2016, 10, 1306–1317 CrossRef CAS PubMed.
- X. Y. Yang, A. P. Gondikas, S. M. Marinakos, M. Auffan, J. Liu, H. Hsu-Kim and J. N. Meyer, Environ. Sci. Technol., 2012, 46, 1119–1127 CrossRef CAS PubMed.
- A. K. Suresh, D. A. Pelletier, W. Wang, J. W. Moon, B. H. Gu, N. P. Mortensen, D. P. Allison, D. C. Joy, T. J. Phelps and M. J. Doktycz, Environ. Sci. Technol., 2010, 44, 5210–5215 CrossRef CAS PubMed.
- Y. M. Long, L. G. Hu, X. T. Yan, X. C. Zhao, Q. F. Zhou, Y. Cai and G. B. Jiang, Int. J. Nanomed., 2017, 12, 3193–3206 CrossRef CAS PubMed.
- W. T. Lu, D. Senapati, S. G. Wang, O. Tovmachenko, A. K. Singh, H. T. Yu and P. C. Ray, Chem. Phys. Lett., 2010, 487, 92–96 CrossRef CAS PubMed.
- F. Liu, F. F. Wei, L. Wang, H. Liu, X. P. Zhu and Y. C. Liang, Physiol. Mol. Plant Pathol., 2010, 74, 330–336 CrossRef CAS.
- H. Boubakri, J. Chong, A. Poutaraud, C. Schmitt, C. Bertsch, A. Mliki, J. E. Masson and I. Soustre-Gacougnolle, Eur. J. Plant Pathol., 2013, 136, 837–855 CrossRef CAS.
- R. P. Goodrich, Vox Sang., 2000, 78, 211–215 CAS.
- K. Makdoumi, A. Backman, J. Mortensen and S. Crafoord, Graefes Arch. Clin. Exp. Ophthalmol., 2010, 248, 207–212 CrossRef CAS PubMed.
- B. Roy, P. Bairi and A. K. Nandi, Analyst, 2011, 136, 3605–3607 RSC.
- M. Svecova, P. Ulbrich, M. Dendisova and P. Matejka, Spectrochim. Acta, Part A, 2018, 195, 236–245 CrossRef CAS PubMed.
- Y. Q. Ll, Y. X. Zhu and Y. Zhang, Chem. J. Chin. Univ., 2008, 29, 669–672 Search PubMed.
- O. Choi, K. K. Deng, N. J. Kim, L. Ross, R. Y. Surampalli and Z. Q. Hu, Water Res., 2008, 42, 3066–3074 CrossRef CAS PubMed.
- G. Applerot, A. Lipovsky, R. Dror, N. Perkas, Y. Nitzan, R. Lubart and A. Gedanken, Adv. Funct. Mater., 2009, 19, 842–852 CrossRef CAS.
- L. S. A. de Melo, A. S. L. Gomes and S. Saska, J. Fluoresc., 2012, 22, 1633–1638 CrossRef PubMed.
- R. Aiello, D. Castrogiovanni and J. Parisi, Photochem. Photobiol., 2018, 94, 1159–1166 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra02079a |
| This journal is © The Royal Society of Chemistry 2019 |