Open Access Article
Open Access ArticleCloning, expression and characterization of a novel fructosyltransferase from Aspergillus niger and its application in the synthesis of fructooligosaccharides†
Shuhong Mao a,
Yanna Liub,
Juanjuan Yanga,
Xiaoyu Mab,
Fang Zengb,
Zhaohui Zhangb,
Shan Wangc,
Haichao Hana,
Hui-Min Qin
a,
Yanna Liub,
Juanjuan Yanga,
Xiaoyu Mab,
Fang Zengb,
Zhaohui Zhangb,
Shan Wangc,
Haichao Hana,
Hui-Min Qin *ac and
Fuping Lu
*ac and
Fuping Lu *ab
*ab
aKey Laboratory of Industrial Fermentation Microbiology, Ministry of Education, Tianjin University of Science & Technology, Tianjin 300457, China. E-mail: shuhongmao@tust.edu.cn; 13820601835@163.com; hanhaichao@qq.com; huiminqin@tust.edu.cn
bState Key Laboratory of Food Nutrition and Safety, Tianjin University of Science & Technology, Tianjin 300457, China. E-mail: yannaliu92@foxmail.com; 763454508@qq.com; 249653931@qq.com; 740929020@qq.com
cTianjin Key Laboratory of Industrial Microbiology, College of Biotechnology, Tianjin University of Science & Technology, No. 29, 13 Main Street, Tianjin Economic and Technological Development Zone, Tianjin 300457, China. E-mail: 2415825419@qq.com; lfp@tust.edu.cn; Fax: +86-22-60600810; Tel: +86-22-60600160
First published on 31st July 2019
Abstract
Fructosyltransferases have been used in the industrial production of fructooligosaccharides (FOS). However, it is still not possible to explain the difference in FOS production based on the variations observed in the FOS synthesizing enzymes of A. niger. In the present study, a novel fructosyltransferase (FT-A) from A. niger TCCC41686 with high FOS synthesis ability was characterized. The FT-A gene was obtained and expressed in Pichia pastoris. A homology model of FT-A showed that the changes of residues identified outside the conserved domains were found to have an effect on its characteristics and its FOS-synthesis capacity. The optimal activity of the recombinant FT-A was observed at 50 °C and pH 6.0, and it was stable below 50 °C and over the range of pH 3.0–11.0. The Km and Vmax values of FT-A were 151.13 g L−1 and 6.55 g L−1 min−1, respectively. The production of FOS using the recombinant FT-A remained above 60% during 50–80 min of synthesis based on sucrose as substrate. The novel fructosyltransferase (FT-A) investigated in this study can potentially be applied for the efficient industrial production of FOS. The results also provide more valuable information for explaining the relationship between the structure and function of the FT-A.
Introduction
Fructooligosaccharides (FOS) are widely used in the manufacture of low-calorie and functional foods owing to their prebiotic properties.1,2 For decades, many strategies have been developed to improve the production of FOS, and biotechnological approaches proved to be an efficient platform for FOS production.Many microorganisms, especially the genus Aspergillus that possesses fructosyltransferases with high transfructosylating activity, have been regarded as the most efficient producers of FOS. Whole cell catalysis using dried A. niger CGMCC 6640 mycelia containing β-fructofuranosidase achieved the production of 314.60 g L−1 FOS from sucrose.3 Ganaie et al. found that the average yield of FOS achieved by recycling A. niger cells was 59.71 ± 2.32% (w/w).4
Commercial production of FOS can also be carried out by fructosyltransferases (also called fructofuranosidase or invertase) using sucrose as substrate. A. niger proved to be the most suitable strain for fructosyltransferases production. The productivity of the enzyme from A. niger was very high. Nguyen et al. previously reported the production of FOS using β-fructofuranosidase from A. niger IMI303386.5 The enzyme was purified while the gene encoding it was not identified.6 The gene of fructofuranosidase from A. niger B60, also known as invertase, was first sequenced by Boddy et al. (1993) based on the amino-acid sequences of tryptic peptides (suc1: L06844).7 In subsequent years, more and more gene sequence of fructosyltransferase from A. niger were deciphered and deposited in the GenBank database (GeneBank accession no: L06844, AF029359, BAB67771, DQ233218, DQ233219, DQ233220, HQ450381, KF699529 and KT724055.7–15), in which three putative invertase genes (SucA, SucB, SucC) were reported based on the available complete genome sequence of A. niger CBS 513.88.10,11 SucA (DQ233218) was previously characterized as the secreted invertase encoded by gene suc1.11 SucB (DQ233219) was identified with transfructosylating activity by Goosen et al.16 Another A. niger strain, which was later reclassified as A. japonicus ATCC 20611, was also investigated for its increased fructofuranosidase activity.9,17,18 In addition, the characteristics of the fructosyltransferase (AjFT) derived from A. japonicus CB05 were determined and the AjFT was crystallized, revealing the active cavity and the residues that interact with the different substrates.19
Genetic engineering approaches were also employed for the development of efficient biocatalysts that can be applied in the industrial synthesis of FOS. The invertase gene (suc1) isolated from A. niger B60 was expressed in Trichoderma reesei, but the efficiency of the transformation was very low.20 P. pastoris has been developed as a host for the efficient production and secretion of foreign proteins.21 Several fructosyltransferases from A. niger were heterologously expressed in P. pastoris, and these recombinant fructosyltransferases showed better activity and higher FOS-synthesis ability than the corresponding native enzymes.14,15,22 A gene encoding an invertase from A. niger was isolated from xerophilic A. niger GH1, and the invertase was produced in the methylotrophic yeast P. pastoris with a specific activity of 3389 U mg−1 protein.12 However, data on FOS production using recombinant fructosyltransferase isolated from A. niger and expressed in P. pastoris were available only in three studies.
Fructosyltransferases belong to either the glycoside hydrolase family 32 (GH32) or GH68 which has three highly conserved domains allowing substrate binding and catalysis.19,23 The mutagenesis within the conserved domains usually negatively affects the activity of fructosyltransferases, while the mutation within the unconserved domains tend to increase the activity of fructosyltransferases.24,25 For instance, mutants D60A, D191A, and E292A located at the conserved domains of the fructosyltransferase (AjFT) from A. japonicus CB05 completely lost their activities,19 while the mutation A458V away from the conserved domains of β-fructofuranosidase (fopA GeneBank accession no: AB046383) significantly improved fructosyltransferase activity.26 Furthermore, Sergio et al. found that two fructosyltransferases, both derived from A. japonicus, had different total affinity energies of the ligands and FOS-synthesizing abilities, even though their sequence identity was as high as 96% and the TM-score for these two fructosyltransferases was 0.99, suggesting that their structures share the same overall fold.24 As such, changes of residues which are not directly involved in the protein–ligand interaction might result in improvement in FOS production. In this regard, it is still valuable to explore the diversity of fructosyltransferases present in A. niger and identify their characteristics to obtain valuable information for further understanding of the FOS synthesis process.
In this work, a fructosyltransferase (FT-A) with high FOS synthesis ability was identified in A. niger and its cDNA was obtained via RT-PCR, and overexpressed in P. pastoris. The molecular and biochemical characteristics of the recombinant FT-A were determined, and its application in FOS production was also investigated.
Experimental section
Strains, vectors and reagents
A. niger TCCC41686 was obtained from the Tianjin University of Science and Technology Culture Collection Center (TCCC; Tianjin, China), P. pastoris GS115 and plasmid pPIC9K were purchased from Invitrogen (Carlsbad, CA, USA). GF2, GF3, and GF4 were obtained from Wako Pure Chemical Industries, Ltd. (Osako, Japan). Other chemicals (e.g., sucrose) were obtained from Shanghai Sangon Biological Engineering Technology & Services Co. Ltd. (Shanghai, China). Unless noted otherwise, all enzymes for molecular manipulations of DNA used in this study were purchased from TaKaRa (Dalian, China) and all primers were from Beijing Genomics Institute (BGI, Beijing, China).Gene cloning of FT-A
The cDNA of FT-A from A. niger TCCC41686, without its introns and signal peptide,14 was synthesized using the HiFiScript cDNA Synthesis Kit (CoWin Biosciences, China), and the forward primer 5′-CCGGAATTCGCCTCGCCTTCCATGCAGAC-3′, and reverse primer 5′-ATAAGAATGCGGCCGCTTAGTGGTGATGGTGGTGATGAG ACTGACGATCCGGCCA-3′ was used. The primers contained the EcoR I (5′) and Not I (3′) restriction sites, respectively. The PCR was carried out with an initial denaturation step at 95 °C for 5 min; 30 cycles of 95 °C for 50 s, 58 °C for 1 min, and 72 °C for 2 min; and a final extension step at 72 °C for 10 min. The purified PCR product was sequenced by BGI (Beijing, China).Sequence analysis of FT-A
To compare FT-A with other fructosyltransferases from A. niger, we conducted a BLAST search with these sequences against the NCBI database (http://www.ncbi.nlm.nih.gov/). The three-dimensional (3D) molecular model of FT-A from A. niger TCCC41686 was generated using I-TASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/).27,28 The crystal structure of fructosyltransferase from A. japonicus CB05 (PDB ID: 3LF7, 62.40% sequence identity with FT-A) was chosen as the template.19 Homology modeling was performed using the auto-model command.Construction of the recombinant plasmid pPIC9K-FT-A
The cDNA encoding FT-A obtained by RT-PCR was digested with EcoR I and Not I, and cloned between the corresponding sites of pPIC9K. The plasmid pPIC9K-FT-A (Fig. S1†) was linearized and electroporated into P. pastoris GS115 at 1500 V, 5 ms. After growth on minimal dextrose medium (MD) agar plates at 30 °C for 3 d, the recombinant P. pastoris GS115 were selected on YPD agar plates with G418 (4.0 mg mL−1). The insertion of the FT-A segment in the resulting positive recombinant P. pastoris was confirmed by PCR.Expression of FT-A in P. pastoris
The selected recombinant P. pastoris cells (single colonies) were grown in 30 mL YPD medium at 30 °C and 220 rpm for 24 h. Then 1 mL of the seed broth was inoculated into 50 mL BMGY medium, and cultured at 30 °C and 220 rpm for about 17 h. Subsequently, the cells were collected by centrifugation at 4000 × g for 4 min at 4 °C, resuspended in BMMY medium (50 mL) and cultured at 30 °C and 220 rpm, and then 250 μL of methanol was added every 12 h to induce protein expression. After 96 h of induction, the fermented broth was centrifuged at 8000 × g for 15 min and the supernatant with crude FT-A was obtained.Purification of FT-A
To purify the FT-A with 6-His tag, the culture supernatant of recombinant P. pastoris was loaded onto a Ni-NTA Superflow column (QIAGEN, Germany) equilibrated with binding buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 7.4). The resin was washed twice with wash buffer (50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 7.4) and the target protein was eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 7.4). The purity and apparent molecular mass of FT-A were monitored by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE). The protein concentration was determined using a Bradford Protein Assay Kit (Solarbio, China) with BSA as standard.Enzyme activity assays
Fructosyltransferase activity of FT-A was measured via the content of GF2 produced from sucrose, which was determined by HPLC. The reaction solution contains appropriate amounts of fructosyltransferase and 10% sucrose (w/v) prepared in 50 mM phosphate buffer (pH 5.0). The reaction mixture (20 mL) was incubated at 50 °C with shaking at 200 rpm for 60 min and then heated at 85 °C for 10 min to stop the reaction. After cooling to room temperature, the solution was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 2 min and the supernatant was analyzed by HPLC. One unit of fructosyltransferase activity was defined as the amount of enzyme that produces 1 μmol of GF2 per min under the described assay conditions.
000 × g for 2 min and the supernatant was analyzed by HPLC. One unit of fructosyltransferase activity was defined as the amount of enzyme that produces 1 μmol of GF2 per min under the described assay conditions.
Enzyme activity (X) was calculated according to formula (1), and expressed as U mL−1.
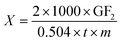 | (1) |
In formula (1) GF2 represents the percent of GF2 (%); t represents the reaction time (min); m represents the amount of enzyme (mL).
Effects of temperature on FT-A activity
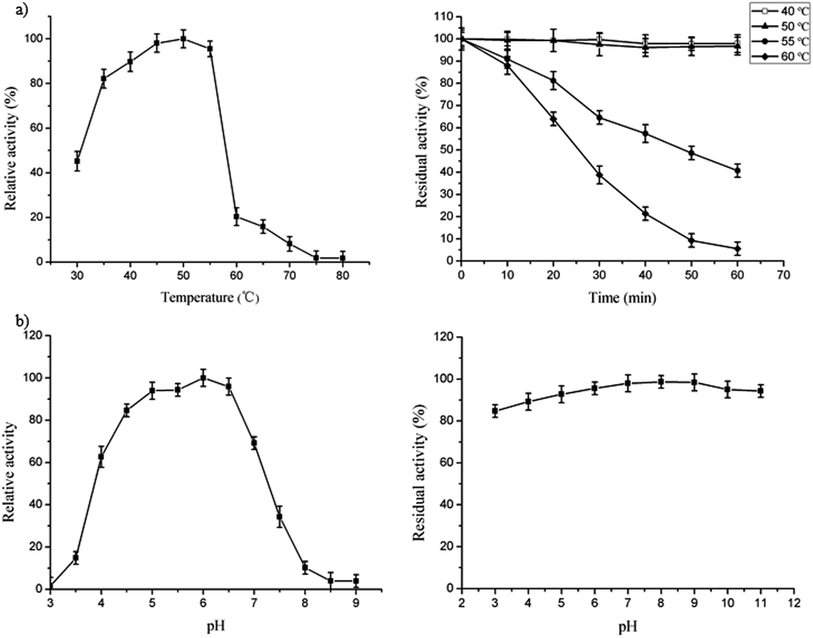
The optimum temperature of FT-A was determined by measuring the FT-A activity at different reaction temperatures (ranging from 30 °C to 80 °C) with the other reaction conditions and the assay method kept unchanged.15,22 The highest activity measured under different temperature conditions is defined as 100%. The thermal stability of FT-A was determined by incubating the enzyme at different temperatures (40 °C, 50 °C, 55 °C and 60 °C), and the residual activities at various incubation times were measured by the method mentioned in “Enzyme activity assays”. The activity of FT-A without incubation in different temperatures was taken as 100%.Effects of pH on FT-A activity
To determine the optimal pH of FT-A, the activity assay was performed at 50 °C using different phosphate buffers (pH from 3 to 9),15,22 and the other reaction conditions and activity assay method are as described in “Enzyme activity assays”. The highest activity measured under different pH conditions is defined as 100%. To investigate the effect of pH on the stability of FT-A, FT-A was pre-incubated in various phosphate buffers (pH from 3.0 to 11.0) at 4 °C for 24 h, and the residual activities were measured as described in “Enzyme activity assays”. The activity of FT-A without incubation in phosphate buffers with different pH values was taken as 100%.Effect of metal ions on FT-A activity
To analyze the effect of metal ions on FT-A activity, 0.5 mM and 5.0 mM metal ions (K+, Li+, Ca2+, Mg2+, Fe2+, Zn2+, Mn2+, Ba2+, Cu2+, Ni2+, and Al3+) and 0.5 mM and 5.0 mM ethylene diamine tetraacetic acid (EDTA) were added in the reaction solution, respectively.22,29 The enzyme activities were assayed as described in “Enzyme activity assays” at 50 °C and pH 6.0. The activity obtained without the addition of any ions was defined as 100%.Determination of kinetic parameters of FT-A
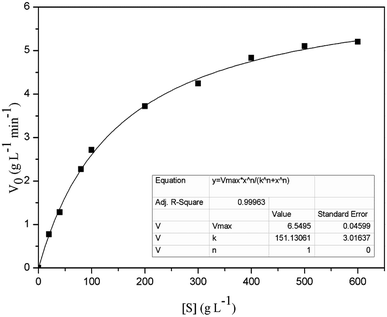
To determine the kinetic parameters of FT-A, the activity assay was performed using different concentrations of sucrose (from 20 to 600 g L−1), with the other reaction conditions and the assay method conducted as described in “Enzyme activity assays”. The Km and Vmax of FT-A were obtained using a non-linear fit to the Michaelis Menten equation in Origin 8.0 software (https://www.originlab.com/).30Synthesis of FOS
In the reaction system, 10% (w/v) sucrose solution was used as substrate in a 20 mL reaction system and different amounts of FT-A (40 U, 80 U, 160 U, 400 U) were added. The transformations were carried out for 6 h at 50 °C, pH 6.0 (phosphate buffer, 50 mM) and samples withdrawn at different reaction times were analyzed by HPLC.HPLC analysis
The samples were analyzed by HPLC on a Agilent 1260 Infinity instrument (Agilent Technologies, Germany) equipped with a Prevail Carbohydrate ES column (4.6 × 250 mm × 5 μm; Grace, America) and a ELSD 2000ES evaporative light-Scattering detector (ELSD; Alltech, America). The drift tube temperature of the ELSD was 90 °C and the ventilatory capacity was 2.2 L min−1. The mobile phase comprised acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (65
water (65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35, v/v) at 30 °C, and the flow rate was 1.0 mL min−1. Samples comprising 5 μL were injected, and the total elution time of 20 min. Three independent measurements were performed for each sample.
35, v/v) at 30 °C, and the flow rate was 1.0 mL min−1. Samples comprising 5 μL were injected, and the total elution time of 20 min. Three independent measurements were performed for each sample.
Statistical analysis
All experiments, except the construction of the recombinant strain for efficient FT-A expression, were performed in triplicates, and the results are expressed in mean values and standard deviation values as shown in figures. Data analysis was performed using origin statistical program (Origin Software, MA, USA).Results and discussion
Structural analysis of the FT-A homology model
Sequence analysis of the FT-A gene from A. niger TCCC41686 revealed that it was 1842 bp (Fig. S2†) and encoded a protein of 613 amino acids with a theoretical molecular weight of 66 kDa. It was also observed the presence of three specific amino acids in FT-A, two aspartic acids (Asp; D) and one glutamic acid (Glu; E) at the conserved domains A (HVLPPNGQIG![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) PCL), domain D (VTAFR
PCL), domain D (VTAFR![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) PYVFQ), and domain E (AFNF
PYVFQ), and domain E (AFNF![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) TGN), respectively (Fig. S3†).19,23 These domains allow substrate binding and catalysis, and residues Asp49 (49D), Asp179 (179D) and Glu256 (256E) of FT-A (Fig. 1) homologous to Asp60, Asp191 and Glu292 of AjFT, which were described as the putative catalytic nucleophile, transition-state stabilizer, and general acid/base catalyst, respectively.19,31
TGN), respectively (Fig. S3†).19,23 These domains allow substrate binding and catalysis, and residues Asp49 (49D), Asp179 (179D) and Glu256 (256E) of FT-A (Fig. 1) homologous to Asp60, Asp191 and Glu292 of AjFT, which were described as the putative catalytic nucleophile, transition-state stabilizer, and general acid/base catalyst, respectively.19,31
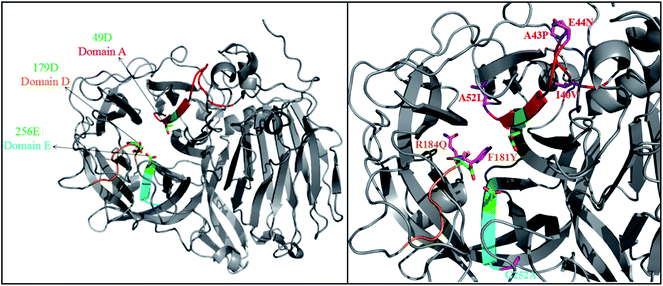 | ||
| Fig. 1 3D structures of FT-A. The crystal structure of fructosyltransferase from A. japonicus CB05 (PDB ID: 3LF7) was chosen as the template. | ||
A. niger SG610 fructosyltransferase (FruSG, GenBank accession no. KT724055),15 A. niger YZ59 fructosyltransferase (fwt),22 A. niger QU10 fructosyltransferase (fts1, GenBank accession no. AHC54391)14 with FT-A in this study shared an identity between 96 and 99%, and the residues inside the domains A, D and E are completely conserved, while there are variable outside the conserved domains (Fig. S3†). Previous reports have suggested that some mutations indirectly involved in the interacting with substrates are responsible for specific characteristics of fructosyltransferases and could be used to improve the production of FOS.19,24–26 In this sense, the changes of residues in FT-A outside the conserved domains may also affect its specific characteristics and its capacity to synthesize FOS.
A homology model of FT-A was generated using fructosyltransferase (AjFT, PDB ID: 3LF7) from A. japonicus CB05 as the template, with which FT-A shared 62.40% sequence identify (Fig. 1). Compared with AjFT, most of the residues were conserved at the conserved domains, but some of them are variable (Fig. 1). In particular, I40V, A43P, E44N and A52L were located at domain A. F181Y and R184Q were surrounded by D179 of domain D. G252A was near to E256 of domain E. Most of the residues were hydrophobic amino acids and located at the peripheral of the active site. They were speculated to form hydrophobic interaction with the surrounding residues resulting in the change of hydrophobic environment at the active site. Interestingly, 184Q and 44N were located at the surface of FT-A. They might form electrostatic interaction and change the molecular surface charge, which may in turn result in the different performance for FT-A catalyzing FOS production.
General properties of FT-A expressed in P. pastoris
The P. pastoris GS115 strain containing only the plasmid pPIC9K was used as the control and no fructosyltransferase activity was observed. The fructosyltransferase activity (124.5 U mL−1) was detected in the supernatant of the P. pastoris harboring plasmid pPIC9K-FT-A, which was up to 100 fold higher than the activity of fructosyltransferase from A. niger TCCC41686. In addition, the specific activity of the purified FT-A reached 6448.92 U mg−1, which was higher when compared with fructosyltransferase from A. niger SG610 (2284.90 U mg−1).15 The recombinant FT-A purified by His-tag affinity chromatography was analyzed by SDS-PAGE (Fig. S4†), and the apparent molecular mass of FT-A without its signal peptide was 116 kDa, almost two times the calculated molecular weight (66 kDa). This difference could be attributed to the glycosylation of the protein, which is typical for fructosyltransferase expression in P. pastoris.13,14 For instance, 12 potential N-glycosylation sites (increase in mass of about 30 kDa) in fructosyltransferase (calculated molecular weight: 67 kDa) of A. niger QU10 may result in the molecular mass of approximately 100 kDa.14Effect of temperature on FT-A activity
The optimal temperature for FT-A was 50 °C (Fig. 2a, left), which was similar to what was observed for the fructosyltransferase from A. niger YZ59 (55 °C) or A. niger SG610 (50 °C).15,22 However, the activity began to decrease dramatically when the temperature was over 55 °C (Fig. 2a, left). FT-A showed a better thermostability when it was incubated at 40 °C and 50 °C for 60 min, with no loss of its initial activity and over 80% residual activity after incubation for 24 h at the same temperature (Fig. 2a, right). However, the enzyme only retained 64% of its initial activity after incubation at 60 °C for 20 min (Fig. 2a, right). Thus, FT-A was stable below 50 °C, which was more stable than all the other reported fructosyltransferases from A. niger.15,22Effects of pH on FT-A activity
A recombinant fructosyltransferase with high transfructosylation activity under optimum reaction conditions is desirable for scaling up the scale of FOS production. The optimal pH of FT-A was 6.0 (Fig. 2b, left) which was slightly more alkaline than the optimum pH of other fructosyltransferases from A. niger (pH 5.0 and 5.5).15,22 The FT-A cloned in this study retained more than 90% of its initial activity after incubation at 4 °C for 24 h between pH 4.0 and 11.0 (Fig. 2b, right), and also retained more than 80% of its initial activity even at pH 3.0, which was more stable over a wide range of pH spectrum than the fructosyltransferase from A. niger SG610 and A. niger YZ59.15,22Effect of metal ions on FT-A activity
Metal ions have an important influence on the folding and catalytic ability of enzymes.32 The effects of different metal ions on FT-A activity was also determined (Table 1). FT-A activity was enhanced by 5.0 mM K+, Ni2+, Mg2+ and Mn2+, while other metal ions had slightly inhibitory effects on FT-A activity. EDTA exhibited an obvious negative effect on enzyme activity when FT-A was treated with 5.0 mM EDTA, indicating that the FT-A activity might be dependent on divalent ions.22 This was consistent with previous reports that fructosyltransferase from A. niger YZ59 was activated by Ni2+ and Mg2+ (5.0 mM) and fructosyltransferase from A. niger SG610 was significantly activated by 5.0 mM Mg2+ and 1.0 mM Fe2+.15,22| Metal ions | Concentration (mM) | |
|---|---|---|
| 0.5 | 5.0 | |
| Relative activity (%) | ||
| Control group (none) | 100.0 | 100.0 |
| K+ | 92.7 ± 2.5 | 107.6 ± 1.9 |
| Li+ | 94.7 ± 1.5 | 96.4 ± 0.8 |
| Ca2+ | 89.8 ± 1.5 | 94.3 ± 1.6 |
| Mg2+ | 86.3 ± 1.0 | 104.8 ± 2.4 |
| Fe2+ | 97.3 ± 0.9 | 98.9 ± 2.2 |
| Zn2+ | 83.7 ± 1.2 | 90.5 ± 1.7 |
| Mn2+ | 91.4 ± 2.1 | 103.4 ± 1.4 |
| Ba2+ | 87.4 ± 0.6 | 93.4 ± 1.1 |
| Cu2+ | 78.3 ± 1.6 | 87.4 ± 2.1 |
| Ni2+ | 94.7 ± 0.9 | 110.5 ± 1.2 |
| Al3+ | 94.5 ± 1.5 | 96.7 ± 1.8 |
| EDTA | 88.3 ± 1.5 | 5.4 ± 0.2 |
Kinetic parameters of FT-A
The Km and Vmax values of FT-A with sucrose as substrate were 151.13 g L−1 and 6.55 g L−1 min−1, respectively (Fig. 3). The turnover number (kcat) and catalytic efficiency (kcat/Km) values of FT-A were 3.02 × 103 s−1 and 20.00 L g−1 s−1, respectively. In a previous study, the kinetic constants of a fructosyltransferase from A. aculeatus were determined, and the Km value for sucrose was 183.10 g L−1.33 Yang et al. determined the properties of the fructosyltransferase from A. niger YZ59, and found that the Km and Vmax values for sucrose were 159.80 g L−1 and 0.66 g L−1 min−1.22 Km is a very important kinetic parameter of enzyme. The smaller Km means the greater the affinity of the enzyme to the substrate, and the faster the enzymatic reaction. The results indicated that the substrate affinity of the fructosyltransferase from this work was higher than that of the fructosyltransferases from A. aculeatus and A. niger YZ59.FOS synthesis using FT-A
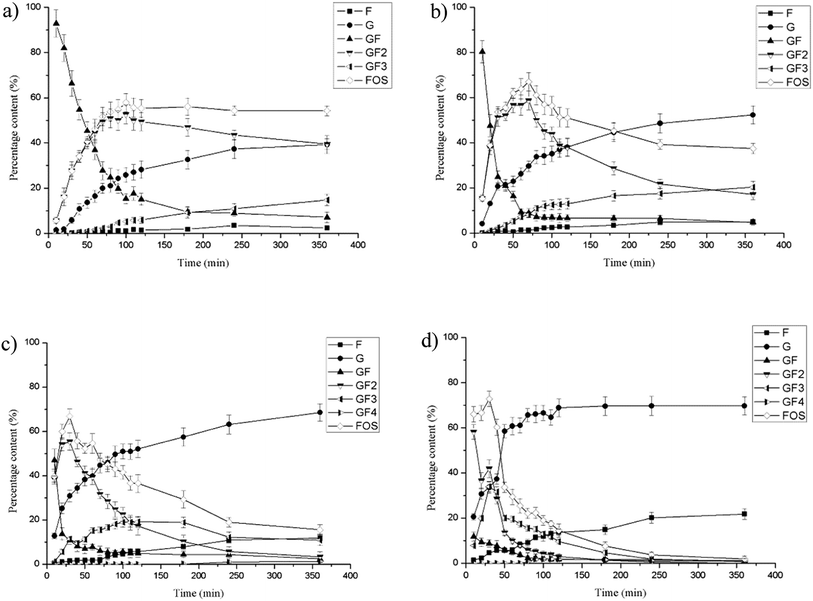
Fructosyltransferases exhibit both hydrolytic and transfructosylating activities.13,34,35 The extent of the two activities depends not only on the origin of the enzymes, but also on the concentration of sucrose. At high sucrose concentrations, increased transfructosylation activity has been observed, while for lower sucrose concentrations, hydrolytic activity is favored.16,35–37 In this study, the effect of different amounts of FT-A on FOS production at 100 g L−1 sucrose solutions was investigated. The carbohydrate concentration profiles showed that the biotransformation proceeded from sucrose to 1-kestose (GF2) and further to nystose (GF3). The highest FOS production (52.70% of GF2 and 5.13% of GF3) was obtained at about 100 min, and remained basically constant thereafter when lower amounts of FT-A (40 U) was used (Fig. 4a). As the addition of FT-A increased to 80 U, the maximum production of FOS increased to 67.05 ± 3.12% (58.82% of GF2 and 8.23% of GF3) at about 70 min, and stayed above 60% from 50 min to 80 min (Fig. 4b and S5†), suggesting the potential to achieve higher yield of FOS in industrial production. As can be seen in Fig. 4c and d, the further addition of FT-A to the reaction resulted in higher FOS production efficiency, and the concentration of FOS reached maximum quickly (30 min). Subsequently, the glucose and fructose concentrations increased along with the rapidly decreasing FOS concentration, indicating that the transfructosylating activity of FT-A is dominant at first, which is helpful for FOS production, and the reaction should be stopped at this point to obtain the maximum product yield. In the later reaction phase, the hydrolytic activity became more pronounced, and the FOS initially produced was degraded by the enzyme. Nevertheless, no additional sucrose appeared during this period, and the observed hydrolysis of the product is not associated with the ping-pong mechanism reported for fructosyltransferases, in which sucrose would be produced from FOS.38,39 Therefore, the amount of enzyme as well as the reaction time was very important control parameters that should be optimized to obtain the maximum yield of FOS.The fructosyltransferase (fts1) gene from A. niger QU10 was functionally expressed in P. pastoris GS115, and FOS yield reached 58% based on the total sugars present in the reaction mixture after 4 h of reaction.14 The highest yield of FOS with a purified fructosyltransferase (fwt) from A. niger YZ59 (CICIM F0901) reached approximately 57% at 2 h.22 Guo et al. achieved maximum FOS production of 64.5% using the fructosyltransferase (FruSG) isolated from A. niger SG610 (CICIM F0902) after 18 h of reaction.15 To our best knowledge, the maximum FOS production yield obtained with recombinant FT-A was the highest among those with all the other reported fructosyltransferases, and also shorter durations were required to achieve the maximum FOS yield. The difference in the amino acids outside the conserved domains may have a significant impact on the catalytic properties of FT-A as well as the synthesis of FOS.
Conclusions
In this work, a novel fructosyltransferase (FT-A) from A. niger TCCC41686 was isolated, and its cDNA was successfully expressed in P. pastoris GS115. The recombinant FT-A has high substrate affinity, stability and synthetic ability to produce FOS. The FOS yield obtained using recombinant FT-A reached 67.05 ± 3.12%, indicating the enzyme's potential for use in industrial production of FOS. These results significantly contribute to the understanding of the FOS synthesis process, and may have a significant impact on the rational design of fungal fructosyltransferases for improved FOS production.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by National Key R&D Program of China (2017YF0400304), National Natural Science Foundation of China (21878233, 31771911), Applied Basic Research Programs of Science and Technology Commission Foundation of Tianjin (No. 18JCYBJC91400). We also thank Professor Xiaoguang Liu for editing the English text of this manuscript.References
- A. E. Maiorano, R. M. Piccoli, E. S. Da Silva and M. F. de Andrade Rodrigues, Biotechnol. Lett., 2008, 30, 1867–1877 CrossRef CAS PubMed.
- R. Simmering and M. Blaut, Appl. Microbiol. Biotechnol., 2001, 55, 19–28 CrossRef CAS PubMed.
- X. Zeng, K. Zhou, D. Liu, C. S. Brennan, M. Brennan, J. Zhou and S. Yu, LWT--Food Sci. Technol., 2016, 65, 1072–1079 CrossRef CAS.
- M. A. Ganaie and U. S. Gupta, Carbohydr. Polym., 2014, 110, 253–258 CrossRef CAS PubMed.
- Q. D. Nguyen, F. Mattes, Á. Hoschke, J. Rezessy-szabó and M. K. Bhat, Biotechnol. Lett., 1999, 21, 183–186 CrossRef CAS.
- Q. D. Nguyen, J. M. Rezessy-Szabó, M. K. Bhat and Á. Hoschke, Process Biochem., 2005, 40, 2461–2466 CrossRef CAS.
- L. M. Boddy, T. Berges, C. Barreau, M. H. Vainstein, M. J. Dobson, D. J. Ballance and J. F. Peberdy, Curr. Genet., 1993, 24, 60–66 CrossRef CAS PubMed.
- R. I. Somiari, H. Brzeski, R. Tate, S. Bieleck and J. Polak, Biotechnol. Lett., 1997, 19, 1243–1247 CrossRef CAS.
- K. Yanai, A. Nakane, A. Kawate and M. Hirayama, Biosci., Biotechnol., Biochem., 2001, 65, 766–773 CrossRef CAS PubMed.
- H. J. Pel, J. H. de Winde, D. B. Archer, P. S. Dyer, G. Hofmann, P. J. Schaap, G. Turner, R. P. de Vries, R. Albang, K. Albermann, M. R. Andersen, J. D. Bendtsen, J. A. Benen, M. van den Berg, S. Breestraat, M. X. Caddick, R. Contreras, M. Cornell, P. M. Coutinho, E. G. Danchin, A. J. Debets, P. Dekker, P. W. van Dijck, A. van Dijk, L. Dijkhuizen, A. J. Driessen, C. D'Enfert, S. Geysens, C. Goosen, G. S. Groot, P. W. de Groot, T. Guillemette, B. Henrissat, M. Herweijer, J. P. van den Hombergh, C. A. van den Hondel, R. T. van der Heijden, R. M. van der Kaaij, F. M. Klis, H. J. Kools, C. P. Kubicek, P. A. van Kuyk, J. Lauber, X. Lu, M. J. van der Maarel, R. Meulenberg, H. Menke, M. A. Mortimer, J. Nielsen, S. G. Oliver, M. Olsthoorn, K. Pal, N. N. van Peij, A. F. Ram, U. Rinas, J. A. Roubos, C. M. Sagt, M. Schmoll, J. Sun, D. Ussery, J. Varga, W. Vervecken, P. J. van de Vondervoort, H. Wedler, H. A. Wosten, A. P. Zeng, A. J. van Ooyen, J. Visser and H. Stam, Nat. Biotechnol., 2007, 25, 221–231 CrossRef PubMed.
- X. Yuan, C. Goosen, H. Kools, M. J. E. C. van der Maare, C. A. M. J. van den Hondel, L. Dijkhuizen and A. F. J. Ram, Microbiol-Sgm, 2006, 152, 3061–3073 CrossRef CAS PubMed.
- F. Veana, J. A. Fuentes-Garibay, C. N. Aguilar, R. Rodríguez-Herrera and M. Guerrero-Olazarán, Enzyme Microb. Technol., 2014, 63, 28–33 CrossRef CAS PubMed.
- F. Veana, D. G. Rodríguez-Reyna, E. T. Aréchiga-Carvajal, C. N. Aguilar and R. Rodríguez-Herrera, Mycopathologia, 2014, 12, 77–82 Search PubMed.
- G. Zhang, J. Yang, J. Shi, S. Qian and Y. Chao, Shengwu Gongcheng Xuebao, 2015, 31, 512–522 CAS.
- W. Guo, H. Yang, S. Qiang, Y. Fan, W. Shen and X. Chen, Eur. Food Res. Technol., 2016, 242, 1159–1168 CrossRef CAS.
- C. Goosen, X. L. Yuan, J. M. van Munster, A. F. Ram, M. J. van der Maarel and L. Dijkhuizen, Eukaryotic Cell, 2007, 6, 674–681 CrossRef CAS PubMed.
- M. Hirayama, N. Sumi and H. Hidaka, Agric. Biol. Chem., 1989, 53, 667–673 CAS.
- H. Hidaka, M. Hirayama and N. Sumi, Agric. Biol. Chem., 1988, 52, 1181–1187 CAS.
- P. Chuankhayan, C. Y. Hsieh, Y. C. Huang, Y. Y. Hsieh, H. H. Guan, Y. C. Hsieh, Y. C. Tien, C. D. Chen, C. M. Chiang and C. J. Chen, J. Biol. Chem., 2010, 285, 23251–23264 CrossRef CAS PubMed.
- T. Bergès, C. Barreau, J. F. Peberdy and L. M. Boddy, Curr. Genet., 1993, 24, 53–59 CrossRef.
- J. M. Cregg, J. L. Cereghino, J. Shi and D. R. Higgins, Mol. Biotechnol., 2000, 16, 23–52 CrossRef CAS PubMed.
- H. Yang, Y. Wang, L. Zhang and W. Shen, New Biotechnol., 2016, 33, 164–170 CrossRef CAS PubMed.
- C. J. Alméciga-Díaz, Á. M. Gutierrez, I. Bahamon, A. Rodríguez, M. A. Rodríguez and O. F. Sánchez, Gene, 2011, 484, 26–34 CrossRef PubMed.
- S. Olarte-Avellaneda, A. Rodríguez-López, J. D. Patiño, C. J. Alméciga-Díaz and O. F. Sánchez, Interdiscip. Sci.: Comput. Life Sci., 2018, 10, 53–67 CrossRef CAS PubMed.
- M. Alvaro-Benito, M. de Abreu, F. Portillo, J. Sanz-Aparicio and M. Fernandez-Lobato, Appl. Environ. Microbiol., 2010, 76, 7491–7499 CrossRef CAS PubMed.
- K. M. Trollope, H. H. Nieuwoudt, J. F. Görgens and H. Volschenk, Appl. Microbiol. Biotechnol., 2014, 98, 4063–4073 CrossRef CAS PubMed.
- J. Yang, R. Yan, A. Roy, D. Xu, J. Poisson and Y. Zhang, Nat. Methods, 2015, 12, 7–8 CrossRef CAS PubMed.
- A. Roy, A. Kucukural and Y. Zhang, Nat. Protoc., 2010, 5, 725–738 CrossRef CAS PubMed.
- H. Driouch, A. Roth, P. Dersch and C. Wittmann, Appl. Microbiol. Biotechnol., 2010, 87, 2011–2024 CrossRef CAS PubMed.
- J. G. Moberly, M. T. Bernards and K. V. Waynant, J. Cheminf., 2018, 10, 5–6 Search PubMed.
- W. Lammens, K. Le Roy, L. Schroeven, A. Van Laere, A. Rabijns and W. Van den Ende, J. Exp. Bot., 2009, 60, 727–740 CrossRef CAS PubMed.
- H. Yang, L. Liu, J. Li, G. Du and J. Chen, Ann. Microbiol., 2012, 62, 1219–1226 CrossRef CAS.
- I. Ghazi, L. Fernandez-Arrojo, H. Garcia-Arellano, M. Ferrer, A. Ballesteros and F. J. Plou, J. Biotechnol., 2007, 128, 204–211 CrossRef CAS PubMed.
- M. Kurakake, R. Masumoto, K. Maguma, A. Kamata, E. Saito, N. Ukita and T. Komaki, J. Agric. Food Chem., 2010, 58, 488–492 CrossRef CAS PubMed.
- O. F. Sánchez, A. M. Rodriguez, E. Silva and L. A. Caicedo, Food Bioprocess Technol., 2010, 3, 662–673 CrossRef.
- R. C. Fernández, B. G. Maresma, A. Juárez and J. Martínez, J. Chem. Technol. Biotechnol., 2004, 79, 268–272 CrossRef.
- L. L'Hocine, Z. Wang, B. Jiang and S. Xu, J. Biotechnol., 2000, 81, 73–84 CrossRef.
- R. G. Crittenden and M. J. Playne, Trends Food Sci. Technol., 1996, 7, 353–361 CrossRef CAS.
- J. W. Yun, Enzyme Microb. Technol., 1996, 19, 107–117 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra02520k |
| This journal is © The Royal Society of Chemistry 2019 |