Open Access Article
Open Access ArticleProteomic analysis reveals the potential neuroprotective effects of tetramethylpyrazine dimer in neuro2a/APPswe cells†
Xiaoyi Linab,
Benhong Xub,
Zaijun Zhangd,
Ying Yange,
Gongping Liue,
Feiqi Zhuf,
Xiaohu Renb,
Jianjun Liub,
Shupeng Li*c,
Xianfeng Huang*ab and
Xifei Yang*b
aCollege of Pharmaceutical Engineering and Life Sciences, Changzhou University, No. 21, Gehu Middle Road, Wujin District, Changzhou, China 213000. E-mail: huangxf@cczu.edu.cn; Fax: +86 13914325607; Tel: +86 13914325607
bKey Laboratory of Modern Toxicology of Shenzhen, Shenzhen Center for Disease Control and Prevention, No. 8, Longyuan Road, Nanshan District, Shenzhen, China 518055. E-mail: xifeiyang@gmail.com; Fax: +86 75525508584; Tel: +86 75525601914
cState Key Laboratory of Oncogenomic, School of Chemical Biology and Biotechnology, Peking University Shenzhen Graduate School, Shenzhen 518055, China. E-mail: lisp@pkusz.edu.cn; Fax: +86 75526032325; Tel: +86 75526032325
dInstitute of New Drug Research and Guangzhou, Key Laboratory of Innovative Chemical Drug Research in Cardio-Cerebrovascular Diseases, Jinan University College of Pharmacy, Guangzhou, 510632, China
eDepartment of Pathophysiology, School of Basic Medicine and the Collaborative Innovation Center for Brain Science, Key Laboratory of Ministry of Education of China for Neurological Disorders, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China
fCognitive Impairment Ward of Neurology Department, The 3rd Affiliated Hospital of Shenzhen University, China
First published on 14th June 2019
Abstract
Alzheimer's disease (AD) is a common neurodegenerative disease characterized by pathological processes, including abnormal amyloid deposits and filament tangles, oxidative stress, neuroinflammation, and neurotrophic insufficiency, leading to chronic and prolonged neuronal loss and cognitive deficits. Tetramethylpyrazine (TMP) is one of the main active components of Ligusticum wallichii, a traditional Chinese medicine widely used for brain related disease. Here, we synthesized the TMP derivative tetramethylpyrazine dimer (DTMP), and evaluated the potential mechanisms underlying its potential neuroprotective effects using the murine neuron-like cells (N2a) transfected with the human “Swedish” mutant amyloid precursor protein (N2aAPP). ELISA results indicated that DTMP reduced the levels of Aβ1–40 and Aβ1–42 in N2aAPP. Then through proteomic analysis we identified a total of 208 differentially expressed proteins in N2aAPP cells compared to the wild-type N2a cells (N2aWT), including 144 increased and 64 decreased proteins. 449 differentially expressed proteins were revealed in N2aAPP cells on DTMP treatment with 69 increased and 380 decreased proteins. Bioinformatic analysis suggested that these proteins are enriched in mitochondrial function, the electronic transmission chain, ATP binding, oxidative phosphorylation, GTPase function, the transcriptional translation process, amino acid metabolism, nucleotide binding and others. Given the vital role of mitochondria in the pathogenesis of AD, we selected the electron transport chain pathway-related molecules to further validate these findings. Western-blot analysis demonstrated that DTMP significantly increased the levels of complex I (NDUAA), complex II (SDHB), complex III (UCRI), complex IV (COX5A) and complex V (ATP5A) in N2aAPP cells. The modulation of dysregulated proteins implicated in AD pathogenesis implies the pharmacological mechanisms of DTMP and its potential as a novel therapeutic choice in AD.
1. Introduction
Alzheimer's disease (AD) is one of the most common neurodegenerative diseases.1,2 Approximately 46.8 million people suffer from AD worldwide and the number is expected to reach 74.7 million by 2030 as the population ages.3,4 Characterized pathological changes of AD mainly include the formation of β-amyloid (Aβ), neuronal synaptic loss, neurofibrillary tangles (NFT) and mitochondrial impairment.5,6 However, the efficient therapeutic strategies are limited.2,3,5,6-Tetramethylpyrazine (TMP) is one of the main active ingredients of Ligusticum wallichii, a traditional Chinese medicine.7 TMP has been reported to inhibit platelet aggregation and thrombolysis, reduce Ca2+ overload, inhibit apoptosis and scavenge free radicals.8,9 In the whole brain and focal cerebral ischemia model, TMP exhibited strong neuroprotective effects in both global and focal cerebral ischemic injury models.10 TMP can also elevate learning and memory ability, and alleviate cholinergic dysfunction of D-galactose injury in mice.11 Some studies showed TMP can reduce oxidative stress induced neuronal death, protect against toxicity induced by kainite excitability, and benefit mitochondrial biogenesis.12–14 Similarly, diallyl disulfide (DADS), a compound in garlic has shown cardioprotective and neuroprotective effects.15,16 It was found that the extract containing DADS can prevent APP processing and tau phosphorylation in an AD transgenic mouse model.17–19 Both TMP and DADS showed therapeutic potentials in AD, but their pharmacological mechanisms are yet to be defined. In this study, we designed and synthesized a combined molecular of TMP and DADS, named tetramethylpyrazine dimer (DTMP), and evaluated it neuroprotective effects. The potential mechanism of DTMP neuroprotection was assessed using N2a cells. Proteomic and bioinformatic strategies were used to identify differentially expressed proteins in cells with or without DTMP-treated N2aAPP cells. Our results revealed that DTMP had a potential therapeutic effect on AD by modulating the dysregulated proteins involved in the pathogenesis of AD, such as mitochondria-associated molecules.
2. Materials and methods
2.1 Reagents
TMP and di-dodecyl dimethylammonium bromide were bought from Aladdin Chemical Reagent Co., Ltd (Shanghai, China); dichloromethane, chloroform, petroleum ether and ethyl acetate were bought from Shanghai Lingfeng Chemical Reagent Co., Ltd (Shanghai, China); sodium sulfide were bought from Wuxi Yasheng Chemical Co., Ltd (Wuxi, China); carbon tetrachloride were bought from Jiangsu Yongfeng Chemical Reagent Co., Ltd (Danyang, China); benzoyl peroxide were bought from Jiangsu Runjie Chemical Reagent Co., Ltd (Nanjing, China); N-bromo succinimide were bought from Jiangsu Qiangsheng Functional Chemical Co., Ltd (Changshu, China); ascending Sulfur were bought from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China) and dimethyl sulfoxide were purchased from bought from Thermo Fisher Scientific (Waltham, MA, United States). DMSO for preparing DTMP (50 mM) and used as a stock solution.2.2 Synthesis of DTMP
The lead compound was synthesized as illustrated in Fig. 1. First, ligustrazine (5.0 g, 36.76 mmol) and benzoyl peroxide (0.84 g, 3.47 mmol) were dissolved in carbon tetrachloride, and N-bromo succinimide (6.74 g, 3.45 mmol) were added in three portions at 45 °C. Then the temperature was raised to 70 °C for 10 hours and then stopped. It was filtered with hot saturated sodium bicarbonate solution, extracted with dichloromethane.In the second step, sublimed sulfur (122.76 mg, 3.84 mmol) and sodium sulfide (1.815 g, 23.27 mmol) were dissolved in 82.5 ml of water and reacted at 50 °C for 30 minutes, taken out and cooled to room temperature. First, all of the compound 2 was added, then dodecyl dimethylammonium bromide (2640 mg, 5.70 mmol), 82.5 ml of chloroform was added and reacted at room temperature. 4 h later, an appropriate amount of water and dichloromethane were added for extraction. The organic layer was taken, dried over anhydrous sodium sulfate, and evaporated, and then purified by column chromatography to give the desired product of ligustrazine dimer (3.24 g, 52.77%) [Mp: 64.3–65.5 °C, 1H NMR (300 MHz, CDCl3) δ 3.99 (s, 4H), 2.55 (s, 6H), 2.50 (s, 12H)].
2.3 Cell culture
The method was referenced to a previous report.202.4 ELISA
After 24 hours of DTMP treatment, cells were washed twice with 1× PBS. 200 μL of IP lysis buffer was added to each 25 cm2 flask for 30 minutes on ice. Cells were scraped from the flask and collected into a 1.5 mL tube, and centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C for 30 min. The resulting supernatant was diluted prior to assay 20 times. The levels of Aβ1–40 and Aβ1–42 in each fraction were determined using the ELISA kits (R&D Systems, Minneapolis, MN, USA) according to the kit instructions.
000g at 4 °C for 30 min. The resulting supernatant was diluted prior to assay 20 times. The levels of Aβ1–40 and Aβ1–42 in each fraction were determined using the ELISA kits (R&D Systems, Minneapolis, MN, USA) according to the kit instructions.
2.5 Sample preparation
After 24 hours of 1 μM DTMP treatment, cells were washed twice with 1× PBS. 400 μL of urea lysis buffer was added to each 75 cm2 flask for 30 minutes on ice. Collect cells scraped from the culture flask and sonicated for 2 minutes (6 seconds on and four seconds off) with Fisher 550 Sonic Dismemberer (Pittsburgh, Pennsylvania) in 45% of the power until the samples was transparent. Then, the samples were then cleaved on ice for 30 min and centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min at 4 °C. Protein concentration was measured using Nanodrop2000 (Thermo Scientific, NJ) according to the manufacturer's instruction. The 1 mg from six individual samples (per group) of the pooled protein 10 mM dithiothreitol (DTT) at 55 °C incubated for 30 min and then incubated at room temperature (dark environment) with 25 mM iodoacetamide (IAA) for 1 h. Samples were diluted to a final concentration of 1.0 M urea with PBS (pH 8.0) and digested with trypsin (1
000g for 30 min at 4 °C. Protein concentration was measured using Nanodrop2000 (Thermo Scientific, NJ) according to the manufacturer's instruction. The 1 mg from six individual samples (per group) of the pooled protein 10 mM dithiothreitol (DTT) at 55 °C incubated for 30 min and then incubated at room temperature (dark environment) with 25 mM iodoacetamide (IAA) for 1 h. Samples were diluted to a final concentration of 1.0 M urea with PBS (pH 8.0) and digested with trypsin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 w/w) (Promega, WI, USA) for 14 hours at 37 °C. After digestion, peptides were adjusted to a pH value of 1–2 with 1% formic acid (FA) and centrifuged at 12
100 w/w) (Promega, WI, USA) for 14 hours at 37 °C. After digestion, peptides were adjusted to a pH value of 1–2 with 1% formic acid (FA) and centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min to collect the supernatant, and then reversed phase column (Oasis HLB, Waters, USA) desalted and dried in a vacuum centrifuge and then dissolved in 50 μL triethylammonium bicarbonate buffer (TEAB, 200 mM, pH 8.5).
000g for 15 min to collect the supernatant, and then reversed phase column (Oasis HLB, Waters, USA) desalted and dried in a vacuum centrifuge and then dissolved in 50 μL triethylammonium bicarbonate buffer (TEAB, 200 mM, pH 8.5).
2.6 TMT labeling
The TMT reagent (dissolved in 40 μL of acetonitrile 0.8 mg TMT) was added to the peptide solution incubated for 1 h at room temperature, and then reacted by adding 5% hydroxylamine at RT for 15 minutes to terminate the reaction. The peptide samples were labeled as follows: TMT-126, N2aWT; TMT-127, N2aAPP; TMT-128, DTMP. The labeled from each group were mixed together, as were the labeled total peptides from each group, then the mixtures were desalted and dried for fractionation. The TMT-labeled peptide mixtures were individually fractionated using a high-pH reversed-phase peptide fractionation kit (Thermo Scientific, NJ, USA) and divided into eight different fractions. Per fraction was dried by using vacuum centrifuge, then dissolved in 20 μL 0.1% FA for liquid chromatography-tandem mass spectrometry analysis.2.7 LC-MS/MS analysis and bioinformatics analysis
The method was referenced to a previous report.21,222.8 Western-blot
Reference experimental method 2.4 protein sample preparation. Protein samples were combined with loading buffer (Thermo Fisher Scientific) and heated at 96 °C for 8 minutes, then separated on 10% SDS-PAGE and transferred to PVDF membrane. After blocking for 1 h at room temperature, it was incubated with primary antibody in 1× TBST overnight at 4 °C. The primary antibodies used in our study are anti-β-actin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000, Santa Cruz, sc-47778), anti-α-tubulin (1
3000, Santa Cruz, sc-47778), anti-α-tubulin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000, Santa Cruz, sc-73242), anti-NDUAA (1
3000, Santa Cruz, sc-73242), anti-NDUAA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Abcam, ab103026), anti-SDHB (1
1000, Abcam, ab103026), anti-SDHB (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Abcam, ab14714), anti-UCRI (1
1000, Abcam, ab14714), anti-UCRI (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Abcam, ab131152), anti-COX5A (1
1000, Abcam, ab131152), anti-COX5A (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000, Santa Cruz, sc-376907), and anti-ATP5A (1
3000, Santa Cruz, sc-376907), and anti-ATP5A (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Abcam, ab14748). The membranes were washed with TBST and incubated with anti-rabbit or anti-mouse IgG HRP secondary antibody for 1 hour. Thereafter, the membrane was washed with TBST three times and visualized using the chemiluminescent reagent from ECL kit (Thermo Scientific Pierce ECL, USA) color. Quantitative densitometry analysis was performed using quantity one software.
1000, Abcam, ab14748). The membranes were washed with TBST and incubated with anti-rabbit or anti-mouse IgG HRP secondary antibody for 1 hour. Thereafter, the membrane was washed with TBST three times and visualized using the chemiluminescent reagent from ECL kit (Thermo Scientific Pierce ECL, USA) color. Quantitative densitometry analysis was performed using quantity one software.
2.9 Statistical analysis
Results are expressed as mean ± SEM. One-way analysis of variance was used for statistical analysis (GraphPad Prism 7.0, http://www.graphpad.com/). P < 0.05 were considered statistically significant.3. Results
3.1 DTMP synthesis
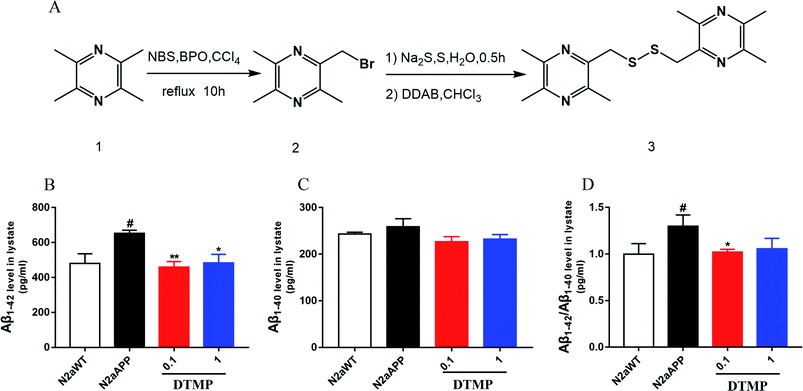
Tetramethylpyrazine1 was used as a raw material and carbon tetrachloride as a solvent. They were heated to 70 °C degrees under reflux with N-bromo succinimide (NBS) and the catalysis of benzoyl peroxide (BPO), which carry out a free radical reaction to obtain 2-bromomethyl-3,5,6-trimethylpyrazine.2 In the second step, sulfur and sodium sulfide were first heated in water to form polysulfide in the aqueous phase. The polysulfide then reacted with dodecyl dimethylammonium bromide (DDAB), and the product entered the organic phase to undergo phase transfer catalyzed reaction with compound 2 to obtain the desired product. The structure of the compound was confirmed by 1H NMR (Fig. S1†), and its molecular weight was 334.5.3.2 DTMP reduces Aβ accumulation
The effect of DTMP on the expression of Aβ1–42 and Aβ1–40 protein in N2aAPP cells was evaluated by ELISA. N2aAPP significantly increased the level of Aβ1–42, while DTMP significantly decreased the level of Aβ1–42 as shown in Fig. 1B. N2aAPP and DTMP did not significantly reduce the level of Aβ1–40 as shown in Fig. 1C. N2aAPP significantly increased the relative expression of Aβ1–42/Aβ1–40, while DTMP significantly decreased the relative expression of Aβ1–42/Aβ1–40 as shown in Fig. 1D.This result indicates that DTMP reduces the levels of Aβ1–40 and Aβ1–42 in N2aAPP.3.3 Proteomic profiles on DTMP treatment
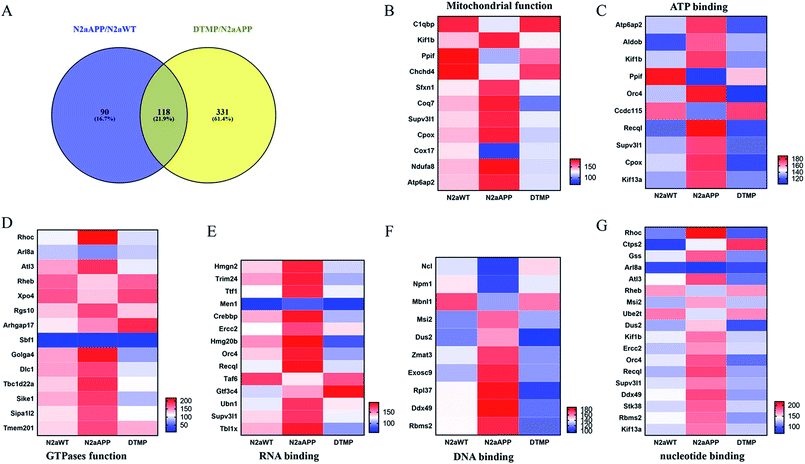
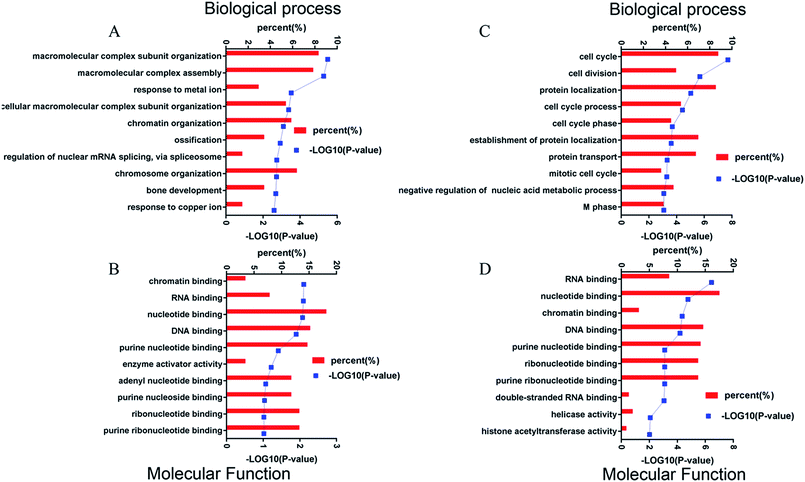
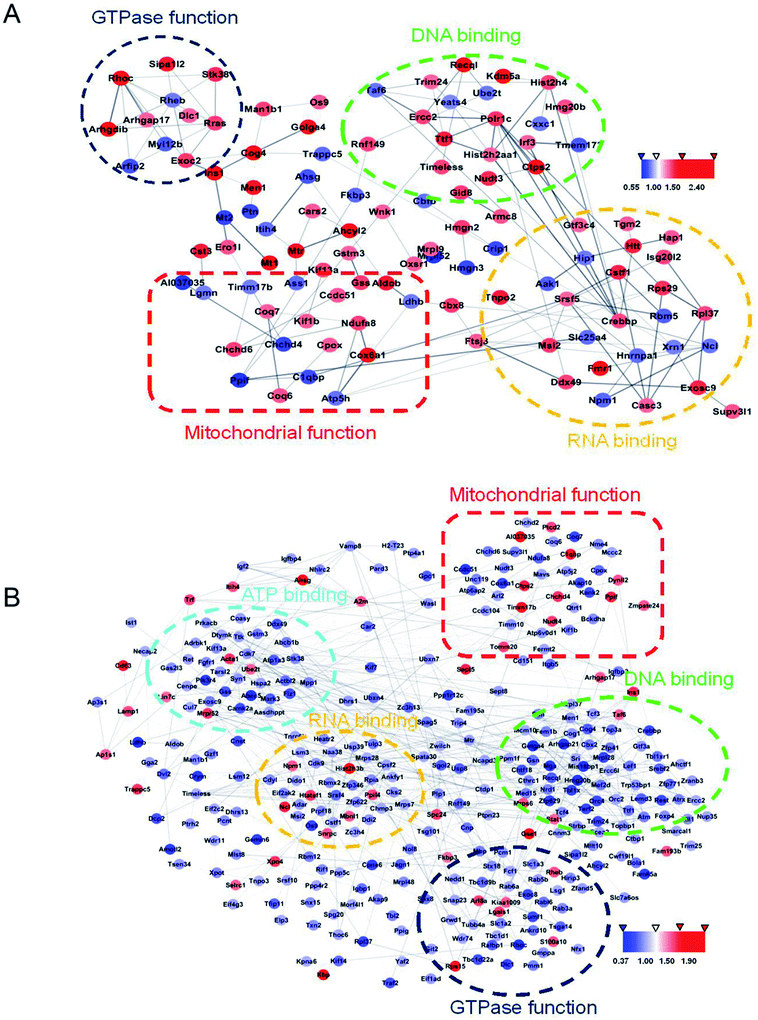
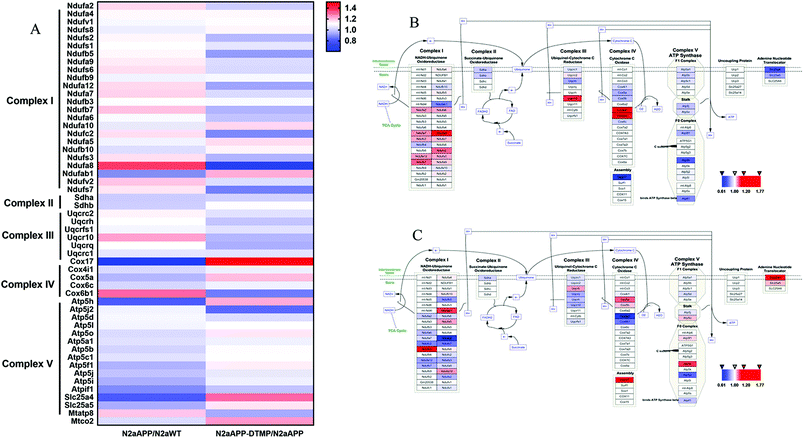
Protein changes of N2aWT cells, N2aAPP cells, and N2aAPP cells treated with 1 μM DTMP were analyzed by TMT marker proteomics. 3490 proteins were identified with at least one unique peptide with a false discovery rate (FDR) of less than 1%. The relative protein expression ratios were compared by calculating the relative abundance of the proteins identified in each group and then used to determine the increase or decrease in protein in the two groups [i.e., (N2aAPP)/(N2aWT) and (DTMP)/(N2aAPP)]. The standard ratios e set as ≥1.25 or <0.8. N2aAPP significantly altered 208 proteins (including 144 increased and 64 decreased), and DTMP significantly changed 449 proteins (including 69 increased and 380 decreased). The number of differential proteins in the two groups were analyzed with Venny in Fig. 2A. (N2aAPP)/(N2aWT) accounted for 16.7%; (DTMP)/(N2a/APP) accounted for 61.4%; these two together accounted for 21.9%. 118 differentially expressed proteins shared by these two and their classification were shown in Fig. 2B–G, Tables S1 and S2,† mainly related to mitochondrial function and ATP binding, GTPase function related, RNA binding, DNA binding, nucleotide binding, and others.Using Gene Ontology (GO) annotation, we analyzed the biological characteristics (biological processes and molecular functions) of differentially expressed proteins. Gene ontology enrichment analysis identified the top ten enrichment of biological processes and molecular functions of 208 differential proteins in N2aAPP cells (Fig. 3A, B, Tables S3 and S4†), biological processes include macromolecular complex subunit organization, response to metal ion, macromolecular complex assembly, and ossification; molecular functions include: chromatin binding, RNA binding, purine nucleotide binding, and nucleotide binding. While DTMP-treated N2aAPP cells were enriched in the top ten classes of biological processes and molecular functions of 449 differentially expressed proteins (Fig. 3C, D, Tables S5 and S6†), biological processes include cell cycle, cell division, protein localization, cell cycle process and cell cycle phase; molecular function include: RNA binding, nucleotide binding, purine nucleotide binding, and chromatin binding. To illustrate protein–protein interactions from N2aAPP and DTMP differential proteins, we used the STRING database to construct a protein network. The network was visualized by using Cystoscope 3.6.0 to determine the potential relationship as shown in Fig. 4. Many interacting proteins are involved in AD processes, such as mitochondrial function and electron transport chain, RNA binding, DNA binding, ATP binding and GTPase function, which are highly compatible with the pathogenesis of AD. These characteristics were further reflected by using the web-based Gene Set Analysis Toolkit (Web Gestalt: http://bioinfo.vanderbilt.edu/webgestalt/) for 118 differentially expressed proteins shared by the two groups shown in Table 1. In view of the important role of mitochondria in the pathogenesis of AD, we selected electron transport chain pathway-related molecules to further demonstrate the potential neuroprotective effects of DTMP.
| ID | Name | #Gene | FDR |
|---|---|---|---|
| WP730 | Glutathione and one carbon metabolism | 3 | 1.59E−01 |
| WP435 | One carbon metabolism | 2 | 1.00E+00 |
| WP310 | mRNA processing | 7 | 1.00E+00 |
| WP567 | Eukaryotic transcription initiation | 2 | 1.00E+00 |
| WP157 | Glycolysis and gluconeogenesis | 2 | 1.00E+00 |
| WP1770 | One carbon metabolism and related pathways | 2 | 1.00E+00 |
| WP18 | Heme biosynthesis | 1 | 1.00E+00 |
| WP1248 | Electron transport chain | 2 | 1.00E+00 |
| WP1244 | Estrogen signaling | 2 | 1.00E+00 |
| WP578 | Leptin insulin overlap | 1 | 1.00E+00 |
The electron transport chain pathway-associated proteins were hierarchically clustered from the 3490 proteins (Fig. 5). The majority of proteins involved in ETC were changed, and these proteins (NDUFB1, NDUFA5, NDUFA10, COX5A, ATP5A and SLC25A4) were up-regulated in DTMP-treated N2aAPP cells.
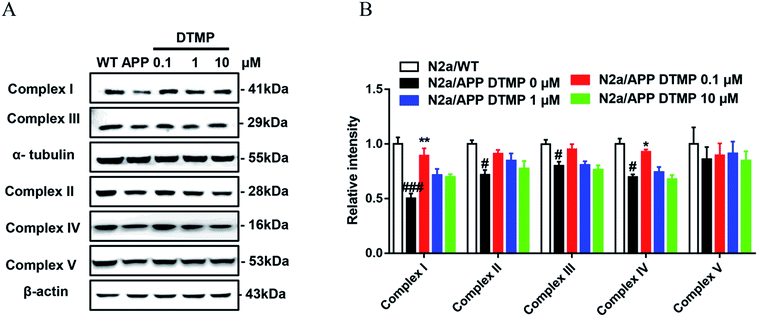
3.4 Confirmation of differential protein expression by Western-blot analysis
To confirm proteomic data, we verified the expression of differential proteins by Western blot analysis. Five related proteins complex I (NDUAA), complex II (SDHB), complex III (UCRI), complex IV (COX5A) and complex V (ATP5A) in the electron transport chain were selected for validation. The expression levels of NDUAA, SDHB, UCRI and COX5A were decreased in N2a/APP cells, while the treatment of DTMP increased the levels of NDUAA and COX5A in N2aAPP cells (Fig. 6). Overall, the Western blot results (NDUAA, UCRI and COX5A) are consistent with the proteomic data.4. Discussion
In this study, our methods of proteomics research DTMP neuroprotective effect on neuroblastoma N2a cells in AD-related injuries, using human Swedish mutant amyloid precursor gene transfected mouse neural element N2a cells as an AD model. The results of ELISA showed that DTMP has neuroprotective effect on N2aAPP cells. Proteomic analysis of proteins expressed by N2aAPP 208 cells and proteins differentially DTMP 449 differentially. Bioinformatics analysis indicated that these proteins are mainly involved in mitochondrial function, electron transport chain, ATP binding, oxidative phosphorylation, GTPase function, and nucleotide binding, suggesting that multiple molecular processes may be involved in the neuroprotective effects of DTMP on neuroblastoma N2a cells.Mitochondria are an organelle in eukaryotic cells, in addition to being responsible for energy supply, as a regulator of cell death and a major feature of neurodegeneration.23 Due to mitochondrial dysfunction, both increased ROS production and oxidative damage may lead to neurodegenerative diseases.24 There is increasing evidence that mitochondrial metabolism, especially defects in the electron transport chain, may play a role in the pathogenesis of AD.25 In our research, many proteins in the electron transport chain have changed. Next, we focus on these differentially expressed proteins that are closely related to mitochondrial function and electron transport chain.
It has been reported that dysfunction of the mitochondrial electron transport chain will lead to ATP synthesis, free radical production and oxidative damage leading to neuronal dysfunction.26,27 10 aberrantly expressed proteins in N2aAPP cells are involved in the mitochondrial electron transport chain, including: complex I in NDUFS5, NDUFAB1; complex II in SDHA, SDHB; complex III in UCRB; complex IV in COX5A, COX17 and complex V in ATP5I, ATP5H and ATP5J.28 The results of the experiment indicated that these proteins have been down-regulated, supporting previous studies that mitochondrial disorders will lead to neurodegenerative diseases.29,30 Also of interest, seven differentially expressed proteins in DTMP-treated N2aAPP cells are involved in the mitochondrial electron transport chain, including: complex I in NDUFAB1, NDUFA10; complex III in UCRB; complex IV in COX17 and complex V in ATP5O, ATP5H and ATP5F1.28 The results showed that proteins from these increases have taken place, so DTMP may have a role in the treatment of neurodegenerative diseases.
5. Conclusion
In summary, these data indicate that DTMP may be used as a potential drug candidate to ameliorate the symptoms of AD by modulating the mitochondrial electron transport chain involved in the expression of the relevant protein.Conflicts of interest
The authors declare that they have no conflict of interest to disclose.Acknowledgements
This work was supported by National Natural Science Foundation of China (81673134, 81401570), Shenzhen Special Fund Project on Strategic Emerging Industry Development (JCYJ20160428143433768, JCYJ20170818111012390, JCYJ20150529164656093, JCYJ20150529153646078, JCYJ20160422143433757, JCYJ20150529112551484 and JCYJ20170810163329510) and Sanming Project of Medicine in Shenzhen (SZSM201611090).References
- A. Sona, K. A. Ellis and D. Ames, Rapid cognitive decline in Alzheimer's disease: a literature review, Int. Rev. Psychiatry, 2013, 25(6), 650–658 CrossRef PubMed.
- T. B. Robins Wahlin and G. J. Byrne, Personality changes in Alzheimer's disease: a systematic review, Int. J. Geriatr. Psychiatry, 2011, 26(10), 1019–1029 CrossRef PubMed.
- L. Nie, G. Wei, S. Peng, Z. Qu, Y. Yang and Q. Yang, et al., Melatonin ameliorates anxiety and depression-like behaviors and modulates proteomic changes in triple transgenic mice of Alzheimer's disease, BioFactors, 2017, 43(4), 593–611 CrossRef CAS PubMed.
- X. Du, X. Wang and M. Geng, Alzheimer's disease hypothesis and related therapies, Transl. Neurodegener., 2018, 7, 2 CrossRef PubMed.
- K. Iqbal and I. Grundke-Iqbal, Alzheimer's disease, a multifactorial disorder seeking multitherapies, Alzheimer's Dementia, 2010, 6(5), 420–424 CrossRef CAS PubMed.
- A. Kumar and A. Singh, A review on mitochondrial restorative mechanism of antioxidants in Alzheimer's disease and other neurological conditions, Front. Pharmacol., 2015, 6, 206 Search PubMed.
- J. B. Lin, C. J. Zheng, X. Zhang, J. Chen, W. J. Liao and Q. Wan, Effects of Tetramethylpyrazine on Functional Recovery and Neuronal Dendritic Plasticity After Experimental Stroke, Evid.-Based Complementary Altern. Med., 2015, 2015, 394926 Search PubMed.
- H. Y. Chen, D. P. Xu, G. L. Tan, W. Cai, G. X. Zhang and W. Cui, et al., A Potent Multi-Functional Neuroprotective Derivative of Tetramethylpyrazine, J. Mol. Neurosci., 2015, 56(4), 977–987 CrossRef CAS.
- W. Qian, X. Xiong, Z. Fang, H. Lu and Z. Wang, Protective effect of tetramethylpyrazine on myocardial ischemia-reperfusion injury, Evid.-Based Complementary Altern. Med., 2014, 2014, 107501 Search PubMed.
- Y. Chang, G. Hsiao, S. H. Chen, Y. C. Chen, J. H. Lin and K. H. Lin, et al., Tetramethylpyrazine suppresses HIF-1alpha, TNF-alpha, and activated caspase-3 expression in middle cerebral artery occlusion-induced brain ischemia in rats, Acta Pharmacol. Sin., 2007, 28(3), 327–333 CrossRef CAS PubMed.
- C. Zhang, S. Z. Wang, P. P. Zuo, X. Cui and J. Cai, Protective effect of tetramethylpyrazine on learning and memory function in D-galactose-lesioned mice, Chin. Med. Sci. J., 2004, 19(3), 180–184 CAS.
- S. Y. Li, Y. H. Jia, W. G. Sun, Y. Tang, G. S. An and J. H. Ni, et al., Stabilization of mitochondrial function by tetramethylpyrazine protects against kainate-induced oxidative lesions in the rat hippocampus, Free Radical Biol. Med., 2010, 48(4), 597–608 CrossRef CAS PubMed.
- H. T. Liu, Y. G. Du, J. L. He, W. J. Chen, W. M. Li and Z. Yang, et al., Tetramethylpyrazine inhibits production of nitric oxide and inducible nitric oxide synthase in lipopolysaccharide-induced N9 microglial cells through blockade of MAPK and PI3K/Akt signaling pathways, and suppression of intracellular reactive oxygen species, J. Ethnopharmacol., 2010, 129(3), 335–343 CrossRef CAS PubMed.
- Z. Tan, Neural protection by naturopathic compounds-an example of tetramethylpyrazine from retina to brain, J. Ocul. Biol. Dis. Infor., 2009, 2(2), 57–64 CrossRef PubMed.
- H. Y. Park, N. D. Kim, G. Y. Kim, H. J. Hwang, B. W. Kim and W. J. Kim, et al., Inhibitory effects of diallyl disulfide on the production of inflammatory mediators and cytokines in lipopolysaccharide-activated BV2 microglia, Toxicol. Appl. Pharmacol., 2012, 262(2), 177–184 CrossRef CAS.
- V. B. Gupta, S. S. Indi and K. S. Rao, Garlic extract exhibits antiamyloidogenic activity on amyloid-beta fibrillogenesis: relevance to Alzheimer's disease, Phytother. Res., 2009, 23(1), 111–115 CrossRef PubMed.
- N. B. Chauhan, Anti-amyloidogenic effect of Allium sativum in Alzheimer's transgenic model Tg2576, J. Herb. Pharmacother., 2003, 3(1), 95–107 CrossRef CAS.
- N. B. Chauhan, Effect of aged garlic extract on APP processing and tau phosphorylation in Alzheimer's transgenic model Tg2576, J. Ethnopharmacol., 2006, 108(3), 385–394 CrossRef PubMed.
- N. B. Chauhan and J. Sandoval, Amelioration of early cognitive deficits by aged garlic extract in Alzheimer's transgenic mice, Phytother. Res., 2007, 21(7), 629–640 CrossRef CAS PubMed.
- X. Huang, J. Wang, X. Chen, P. Liu, S. Wang and F. Song, et al., The Prenylflavonoid Xanthohumol Reduces Alzheimer-Like Changes and Modulates Multiple Pathogenic Molecular Pathways in the Neuro2a/APPswe Cell Model of AD, Front. Pharmacol., 2018, 9, 199 CrossRef PubMed.
- B. Xu, R. Tian, X. Wang, S. Zhan, R. Wang and Y. Guo, et al., Protein profile changes in the frontotemporal lobes in human severe traumatic brain injury, Brain Res., 2016, 1642, 344–352 CrossRef CAS PubMed.
- X. Zhou, W. Xiao, Z. Su, J. Cheng, C. Zheng and Z. Zhang, et al., Hippocampal proteomic alteration in triple transgenic mouse model of Alzheimer's disease and implication of PINK 1 regulation in donepezil treatment, J. Proteome Res., 2019, 18, 1542–1552 CrossRef CAS PubMed.
- M. T. Lin and M. F. Beal, Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases, Nature, 2006, 443(7113), 787–795 CrossRef CAS PubMed.
- J. Hroudova, N. Singh and Z. Fisar, Mitochondrial dysfunctions in neurodegenerative diseases: relevance to Alzheimer's disease, BioMed Res. Int., 2014, 2014, 175062 Search PubMed.
- V. Rhein, G. Baysang, S. Rao, F. Meier, A. Bonert and F. Muller-Spahn, et al., Amyloid-beta leads to impaired cellular respiration, energy production and mitochondrial electron chain complex activities in human neuroblastoma cells, Cell. Mol. Neurobiol., 2009, 29(6–7), 1063–1071 CrossRef CAS.
- H. Yu, X. Lin, D. Wang, Z. Zhang, Y. Guo and X. Ren, et al., Mitochondrial Molecular Abnormalities Revealed by Proteomic Analysis of Hippocampal Organelles of Mice Triple Transgenic for Alzheimer Disease, Front. Mol. Neurosci., 2018, 11, 74 CrossRef PubMed.
- D. C. Chan, Mitochondria: dynamic organelles in disease, aging, and development, Cell, 2006, 125(7), 1241–1252 CrossRef CAS PubMed.
- W. J. Koopman, F. Distelmaier, J. A. Smeitink and P. H. Willems, OXPHOS mutations and neurodegeneration, EMBO J., 2013, 32(1), 9–29 CrossRef CAS PubMed.
- W. E. Muller, A. Eckert, C. Kurz, G. P. Eckert and K. Leuner, Mitochondrial dysfunction: common final pathway in brain aging and Alzheimer's disease-therapeutic aspects, Mol. Neurobiol., 2010, 41(2–3), 159–171 CrossRef PubMed.
- M. J. Calkins, M. Manczak, P. Mao, U. Shirendeb and P. H. Reddy, Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer's disease, Hum. Mol. Genet., 2011, 20(23), 4515–4529 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra03054a |
| This journal is © The Royal Society of Chemistry 2019 |