Open Access Article
Open Access ArticleFactors affecting performance and functional stratification of membrane-aerated biofilms with a counter-diffusion configuration†
Tinggang Li *a and
Junxin Liu*b
*a and
Junxin Liu*b
aInstitute of Process Engineering, Chinese Academy of Sciences, Beijing 100190, People's Republic of China. E-mail: tgli@ipe.ac.cn; Fax: +86-10-82545078; Tel: +86-10-82545078
bResearch Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing 100085, People's Republic of China. E-mail: jxliu@rcees.ac.cn; Fax: +86-10-62849133; Tel: +86-10-62849133
First published on 17th September 2019
Abstract
Membrane-aerated biofilms (MABs) developed with a novel counter-diffusion configuration in oxygen and substrate supply were examined for the effect of biofilm thickness on the functional activity and microbial community structure of the biofilm with the simultaneous degradation of acetonitrile, and nitrification and denitrification. Results demonstrated that different biofilm thicknesses under different surface loading rates (SLRs) caused substantially varied profiles of the microbial activities with distinct functions in the biofilm. Both thick and thin MABs achieved high-rate performance in terms of acetonitrile removal (>99%), but the performance differed in the removal efficiencies of total nitrogen (TN), which was 1.3 times higher in the thick MAB (85%) than in the thin MAB (36.3%). The specific ammonia-oxidizing rate (SAOR) and the specific acetonitrile-degrading rate (SADR) exhibited similar declining and ascending trends in both the thin and thick MABs, respectively. In contrast, the specific denitrifying rate (SDNR) was relatively uniform at a concentration near the detection limit in the thin MAB but exhibited a hump-shaped variation with the highest rate occurring in an intermediate region in the thick MAB. Microbial community analysis revealed a dramatic shift in the dominant bacteria of the community composition with low diversity across the biofilm. This study suggests that the biofilm thickness developed under SLRs, which controls the mass transfer of oxygen and substrates into biofilms, is an important factor affecting the structural and functional stratification of bacterial populations in a single MAB treating organonitrile wastewater.
1. Introduction
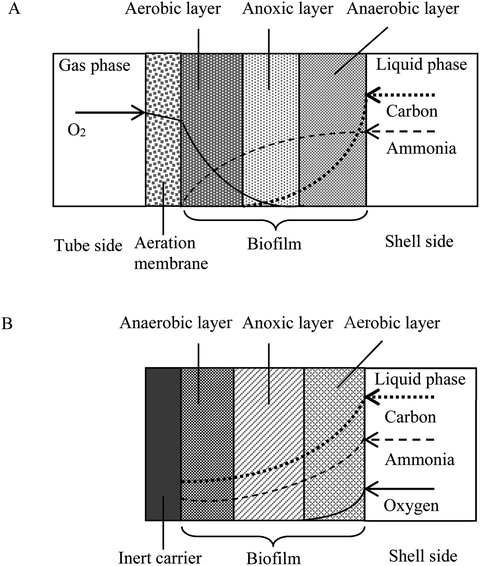
Adequate design and operation of bioreactors for wastewater treatment are of paramount importance in engineered systems. Bioreactor communities contain multiple interacting microbial populations, even when a single substrate is provided.1 Improved design and management of such communities greatly depend upon the formulation of experimentally validated ecological concepts. Not surprisingly, much of the engineering practice of bioreactors has been empirical, especially for an emerging bioreactor such as the membrane-aerated biofilm rector (MABR).2,3 In the MABR, membrane-aerated biofilms (MABs) immobilized on oxygen permeable membranes grow differently from conventional biofilms developed on inert surfaces because the oxygen and substrates required for microbial growth in MABs diffuse from opposite sides of the biofilm. The counter-diffusion of nutrients and oxygen result in a growth of environmental niche different from that of conventional biofilms which receive both oxygen and substrates from the same side. The counter-diffusion configuration allows development of a unique nutrient profile and, consequently, a microorganism population profile in the MAB, which can provide effective treatment for complex wastewater treatment in a single reactor unit (e.g. acetonitrile degradation, nitrification and denitrification processes).4–6 Due to the bubbleless aeration and high oxygen transfer efficiency, membrane aeration shows additional advantages such as lower operational cost and reduced stripping of odors and volatile pollutants from the wastewater being treated.7,8 Recent research attempts have been made toward MABR to advance treatment performance, prevention of secondary pollution and low energy consumption.4,9,10 The MABR biotechnology has been broadly used in the treatment of domestic sewage,11,12 ammonia-containing wastewater,11,13 simultaneously remove C and N pollutions,14,15 volatile pollutant,9,16 toxic xenobiotics in industrial wastewater.4,7,17–19 For example, a hybrid membrane-aerated bioreactor was used to treat acetonitrile (ACN) wastewater, and ACN removal rate, the total organic carbon TOC removal rate and total nitrogen (TN) removal rate reached 98.7, 82.4 and 44.1%, respectively.9Although the MABR is an attractive way for high-rate treatment of wastewater, some key problems still exist in engineering applications: (i) because of the limit to the biofilm thickness, the biomass amount of membrane-aerated biofilm may be constrained. (ii) Not all cultures successfully attach and grow to form biofilms on gas-permeable membranes. The startup of the MABR requires a more thorough understanding of formation behavior of biofilm consisting of oxic/anoxic/anaerobic zones that provide different functions for carbohydrate degradation, nitrification, and denitrification. (iii) There is still a need to develop an approach on immobilizing the desired bacterial community (e.g. nitrifying bacteria and volatile organic compounds degrading bacteria) in an MABR for resistance to environmental stresses.
Understanding the microbial communities within the MABs' structures is a critical step towards improving engineering design and operational performance of MABRs. Depending on the biofilm thickness and other operating conditions (such as organic surface loading rate (SLR) to the biofilm on the membrane), the microbial species and activity through the thickness of the biofilm may vary. Several approaches have been applied to preferentially develop functional populations and MABR performance including controlling influent substrates,20 oxygen supply,21,22 alkalinity and pH,23 and the nature of the inoculum.24 While the bacterial communities, especially ammonia-oxidizing bacteria (AOB) and nitrifying bacteria, of MABs have been previously described,21,25 a gap in our understanding of the multiple microbial communities residing in MABs with acetonitrile-degrading, nitrification and denitrification still remains. Recognizing the community structure and the links within the key functional groups in a single biofilm system can lead to better ways to optimize treatment functions as well as to improve process stability.26,27 Additional research is therefore needed to understand the multifunctional stratification of the microbial community as a function of biofilm as well as the interrelated factors (e.g. surface loading rate (SLR), recirculation rate, etc.) that control the development of multiple functional populations within a single MAB.
In our previous study, we investigated rapid start-up and formation property of biofilm in the MABR and showed microbial adaptation for biodegradation of acetonitrile.16,28,29 The aim of this work was to investigate the integrated performance of MABR under a specific oxygen supply and liquid recirculation rate but two different SLRs (5.66 and 10.54 g m−2 d−1). Thin and thick MABs were developed under the above defined conditions to elucidate the stratification of MABs with respect to ecological shifts and function of MAB communities.
2. Materials and methods
2.1. MABR design and operational conditions
Two identical MABRs in a cylindrical form with a 650 mm height and 55 mm diameter were set up. The MABRs were constructed from Pyrex glass and the working volume of each MABR was 1.42 L. Each of the MABRs contained a membrane module (30 mm diameter) consisting of microporous polypropylene hollow fibers (320 μm o.d. and 200 μm i.d.) (Zenon, Singapore) in a dead-end configuration. The membrane pore size is about 0.07 μm and the specific surface area of the reactor from the contained hollow fibers was 84.5 m2 m−3 as previously described.5 The MABRs were inoculated with a microbial consortium acclimated to biodegrading acetonitrile as the initial carbon and nitrogen source in a synthetic mineral salt (SMS) medium with the trace-element composition as described in details elsewhere.28 In each biofilm growth experiment, the membrane module was supplied with oxygen at 8.6 mL min−1 (from the lumen side of the fibers) under a trans-membrane pressure of 2 psi and the reactor was operated at a HRT of 30 h (Table 1). A centrifugal pump was used to recirculate the liquid in the reactor (at a rate of 3.5 L min−1) to achieve a mean upflow fluid velocity of 12 cm s−1. MABs were developed for a period of over 10 months in two separate reactors at an average SLR of 5.66 g m−2 d−1 (SLR1) or 10.54 g m−2 d−1 (SLR2), respectively. The reactor was operated at 25 ± 1 °C in a temperature controlled room.| Stage | Incubation time (day) | Trans-membrane pressure (psi) | Recirculation rate (L min−1) | Upflow fluid velocity (cm s−1) | HRT (h) | SLR (g m−2 d−1) | Biofilm characteristics |
|---|---|---|---|---|---|---|---|
| S1 | 1–15 | 2 | 3.5 | 12 | 30 | 3.41 ± 0.07 | Formation of biofilm |
| S2 | 16–166 | 2 | 3.5 | 12 | 30 | 5.66 ± 0.09 | Thin biofilm |
| S3 | 167–317 | 2 | 3.5 | 12 | 30 | 10.54 ± 0.13 | Thick biofilm |
2.2. General analysis
Mixed liquor suspended solid (MLSS), volatile suspended solid (VSS), NH4+–N and NO3−–N were measured in accordance with the standard methods.30 Total organic carbon (TOC) and total nitrogen (TN) were determined through a TOC-VCSH plus nitrogen analyzer (Shimadzu, Japan). Ammonia, nitrite, and nitrate were measured using the Hach test kits together with a UV-vis spectrophotometer (DR5000, Hach, USA). Acetonitrile, acetamide, and acetic acid concentrations were measured through a gas chromatograph (6890N, Agilent, USA) as described previously.28 Biofilm thickness was determined through a noninvasive method,31 and the images were captured through a VH-Z75 microscope (Keyence, Japan) via a charge-coupled device that was connected to a computer. Dissolved oxygen (DO) concentrations were determined using a Clark-type microelectrode (Unisense, Aarhus, Denmark) with a tip diameter of 10 μm and a response time of less than 5 s.5 Measurements of biofilm thicknesses and DO concentrations were repeated three times at each location, and the average values were reported in this paper.Biofilm samples were harvested from the reactors after over 6 months development during the steady-state operation under a constant upflow fluid velocity of 12 cm s−1 but with different surface loading rates (SLR1 and SLR2). Sample preparation was performed according to the previous protocol.20,21 In each experiment, the biofilm attached to the membrane at different locations were excised with a razor blade and immediately frozen at −15 °C. Then a series of biofilm slices of a 200 μm thickness were prepared from the frozen biofilm sample using a combined cryostat/microtome (CM 3050, Germany) at −20 °C. The biofilm mass from each slice was placed in a sterile centrifuge tube and stored at −20 °C for further analysis.
2.3. Microbial activity analysis
Batch studies were performed to determine the specific activities of ammonia-oxidizing bacteria (AOB), denitrifying bacteria (DNB) and acetonitrile-degrading bacteria (ADB) in the MABs. Specific ammonia oxidizing rate (SAOR), specific denitrification rate (SDNR) and specific acetonitrile-degrading rate (SADR) were determined based on maximum rates of substrate utilization per biomass unit for AOB, DNB and ADB for biofilm samples, expressed as mg NH4+–N per g VSS per h, mg NO3−–N per g VSS per h and mg acetonitrile per g VSS per h. A 1 mL portion of biofilm sample from each slice were inoculated to 49 mL SMS in 100 mL flasks. The content was incubated at 28 ± 1 °C and was shaken at 180 rpm on a rotary shaker. The initial substrate concentrations were 50 mg L−1 of NH4+–N (in term of 191.1 mg L−1 of NH4Cl) and 200 mg CaCO3 L−1 of alkalinity (in terms of 168 mg L−1 of NaCO3) for SAOR, or 50 mg L−1 of NO3−–N (as 303.6 mg L−1 of NaNO3) and 400 mg L−1 of COD (supplied as CH3COONa) for SDNR, or 200 mg L−1 of acetonitrile for SADR. Any DO in the denitrifying medium was first removed by purging the medium in the flask with nitrogen gas. The flasks for the determination of SADR were sealed using aluminum caps with PTFE/silicone septum to prevent acetonitrile volatilization loss and was supplied with the same DO concentration as that for the biofilm in MABRs. Samples were withdrawn at an interval of 30 min, and substrate concentration (NH4+–N, NO3−–N and acetonitrile) were determined according to the methods described earlier.2.4. Microbiological analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and chloroform. The amount of extracted DNA was quantified using a UV-vis spectrophotometer (Jasco V-550, Japan) at the wavelength of 260 nm. The extracted DNA was precipitated overnight with a sodium acetate–ethanol mix at −20 °C and dissolved in sterile deionized water. Extracted DNA samples were stored in a −20 °C freezer before analysis.
1), and chloroform. The amount of extracted DNA was quantified using a UV-vis spectrophotometer (Jasco V-550, Japan) at the wavelength of 260 nm. The extracted DNA was precipitated overnight with a sodium acetate–ethanol mix at −20 °C and dissolved in sterile deionized water. Extracted DNA samples were stored in a −20 °C freezer before analysis.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stock solution) in 5× TAE buffer was used. A 45 μL of PCR amplicons from DNA of biofilm mass slices was loaded in each gel well. Electrophoresis was conducted in 1× TAE buffer solution at 85 V and 60 °C for 15 h. After electrophoresis, the gel was stained with ethidium bromide for 15 min and then destained for 1 h with 1× TAE buffer solution, and visualized by a SynGene Bio Imaging system (GeneGenius, UK). SynGene Gene-Tools software was used for DGGE band pattern analysis.
1 stock solution) in 5× TAE buffer was used. A 45 μL of PCR amplicons from DNA of biofilm mass slices was loaded in each gel well. Electrophoresis was conducted in 1× TAE buffer solution at 85 V and 60 °C for 15 h. After electrophoresis, the gel was stained with ethidium bromide for 15 min and then destained for 1 h with 1× TAE buffer solution, and visualized by a SynGene Bio Imaging system (GeneGenius, UK). SynGene Gene-Tools software was used for DGGE band pattern analysis.2.5. Statistical methods
Principle component analysis (PCA) of the DGGE data was performed by program of Statistical Package for the Social Sciences (SPSS). The DGGE-banding patterns were scored by a SynGene Bio Imaging system with SynGene Gene-Tools software (GeneGenius, UK) described early, and a binary matrix was made based on the presence (1) or absence (0) of the bands. The binary data representing the banding patterns were used to PCA. The two-dimensional PCA graphs were used to visualize and interpret relative spatial and temporal changes in the microbial community structure between groups.3. Results
3.1. Characteristics of MABRs and MABs
Both MABRs achieved high-rate performance at SLR1 and SLR2 in terms of acetonitrile removal, but the performance differed in the removal efficiencies of TOC and TN (Table 2). TN removal efficiency was much higher in the MAB grown at SLR2 than that in the MAB grown at SLR1. The performance results of greater than 99%, 96.2, and 85% of ACN removal rate, TOC removal rate and TN removal rate in the thick MAB are comparable with results from other study.9,38 The MAB grown at SLR1 developed an average VSS of 47 g m−2 and a final biofilm thickness of 950 μm (thin biofilm), in contrast to an average VSS of 88 g m−2 and a final biofilm thickness of 1600 μm (thick biofilm) for the MAB grown at SLR2. The average ratio of dissolved oxygen penetration was about 100% and 56% at SLR1 and SLR2, respectively (Table 2). DO concentrations in the biofilms were at a similar level at near the membrane/biofilm interface and then declined with the biofilm thickness for the MABs developed at the two SLRs (Fig. S1†), similar to that described in our previous study.5 The DO in the bulk liquid and more than one third of the biofilm on the membrane side was around zero for the thick MAB grown at SLR2, indicating that anoxic or anaerobic region was obtained in the MAB at the higher SLR. The aerobic/anoxic or aerobic/anoxic/anaerobic zones were successfully formed within the MABs under different SLRs.| MABs | Acetonitrile removal efficiency (%) | TOC removal efficiency (%) | TN removal efficiency (%) | NH4+–N effluent concentration (mg L−1) | NO2−–N effluent concentration (mg L−1) | NO3−–N effluent concentration (mg L−1) | Final thickness (μm) | VSS (g m−2) | O2 penetration (μm) |
|---|---|---|---|---|---|---|---|---|---|
| a Thin biofilm grown at SLR1.b Thick biofilm grown at SLR2. | |||||||||
| Thin biofilma | 100 ± 8.3 | 99.1 ± 5.6 | 36.3 ± 1.46 | 6.2 ± 0.31 | 28.6 ± 0.25 | 91.9 ± 2.85 | 950 ± 165 | 47 ± 3.21 | 950 ± 45 |
| Thick biofilmb | 99.5 ± 6.1 | 96.2 ± 8.4 | 85 ± 2.15 | 47.2 ± 2.25 | 2.7 ± 0.16 | 2.6 ± 0.13 | 1600 ± 270 | 88 ± 5.46 | 900 ± 35 |
3.2. Distribution and functional stratification of microbial activity
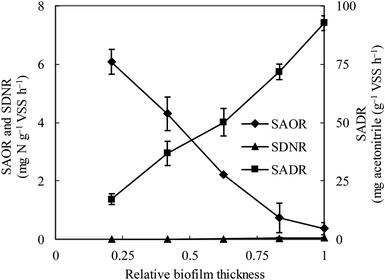
The specific ammonia-oxidizing rate (SAOR), specific denitrifying rate (SDNR), and specific acetonitrile-degrading rate (SADR) were analyzed for both the thin and thick MABs. In the thin MAB grown at SLR1, the SAOR decreased from the membrane side towards the outer region of the biofilm, a wide range of SADR was observed throughout the biofilm whilst no SDNR was detected (see Fig. 1). In contrast, the functional microbial activity profiles in the thick MAB grown at SLR2 (see Fig. 2) were substantially different from those in the thin MABs grown at SLR1 (Fig. 1). Moreover, in the thick MAB grown at SLR2, the highest SAOR of 7.53 mg N per g VSS per h took place on the membrane side (rich oxygen), and the SAOR reduced along the biofilm thickness from the membrane surface and eventually became zero on the bulk liquid side. The SDNR exhibited a hump-shaped variation as a function of biofilm depth, from the lowest in the biofilm on the membrane side and increasing to the highest of 6.74 mg N per g VSS per h at about relative biofilm thickness of 0.625. The SDNR gradually reduced with further increase in the biofilm thickness toward the outer surface of the biofilm on the bulk liquid side. Although the highly anaerobic level in the MAB on the bulk liquid side facilitate denitrification, low concentrations of nitrified products were detected in this zone, certainly due to the unique mass transfer pattern in MABs (Fig. S1†). Consequently, the maximum SDNR was obtained within the biofilm rather than at the outermost zone of the MAB on the bulk liquid side. The SADR in the thick biofilm exhibited similar distribution pattern to that of the thin biofilm increasing from the lowest to the highest from the oxic zone, through the anoxic zone, and to the anaerobic zone in the thick biofilm, which was consistent with the observation in the previous study.16 Result in Fig. 2 provided evidence confirming that the stratified MAB grown at higher SLR2 led to simultaneous functions including acetonitrile degradation, nitrification, and denitrification in a single MABR.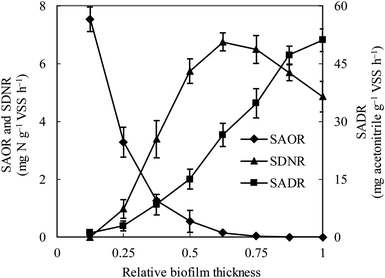 | ||
| Fig. 2 Variations of ammonia-oxidizing rate (SAOR), denitrifying rate (SDNR) and acetonitrile-degrading rate (SADR) in the thick biofilm at SLR2. ♦, SAOR; ▲, SDNR; ■, SADR. Vertical bars represent the standard deviation calculated from five replications. The definition of relative biofilm thickness is consistent with that in Fig. 1. | ||
3.3. Functional bacterial community profiles
The bacterial community structure was investigated as a function of depth of the thick biofilm grown at SLR2 of 10.54 g m−2 d−1 by DGGE of 16S rRNA gene fragments (Fig. 3). DGGE profiles of 16S rRNA were spatially varied and showed a substantial shift in community structure as a function of biofilm depth from the membrane to the outer anaerobic zone of the biofilm. New dominant bands appeared, and some initially important bands disappeared as a function of biofilm depth. Diversity, assessed by the number of clear DGGE bands, varied markedly and was reasonably low throughout the biofilm. In general, the intensity of a band represents the relative abundance of the corresponding microbial species. Several dominant bands (the brightest bands in the lane) are specific to the different zones of the biofilm, for example, two bright bands specific for the inner aerobic zone (IAZ, near <400 μm from the membrane; e.g., bands 1 and 2); two bright bands specific for the middle anoxic zone (MAnZ, from 600 to 1000 μm; e.g., bands 3 and 4); four bright bands specific for the outer anaerobic zone (OAnZ, >1200 μm; e.g., bands 5–8); whereas minor dim bands were detected throughout the entire depth of the biofilm. It is noted that the common bands were found in the zone interfaces within biofilm at 500 and 1100 μm.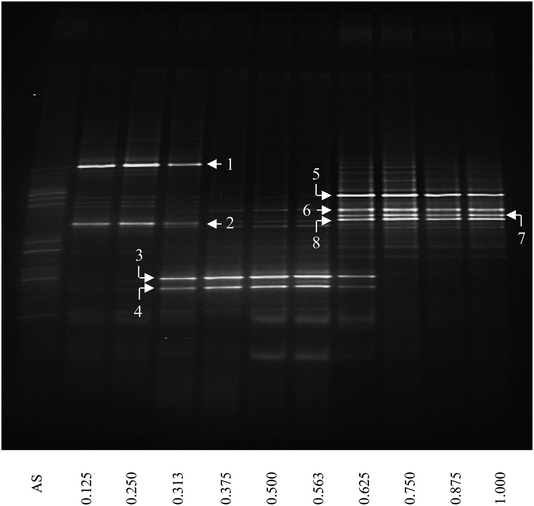 | ||
| Fig. 3 DGGE image of PCR-amplified 16S rRNA gene fragments of the thick MAB grown at SLR2. AS below the first lane refers to inoculum of activated sludge and the number below other lanes stands for the relative biofilm thickness as a function of distance from membrane. Numbers and arrows indicate the specific DGGE bands selected for cloning and sequencing. The definition of relative biofilm thickness is consistent with that in Fig. 1. | ||
The above results were further confirmed by principal component analysis (PCA) of the samples. PCA indicated that differences of community structures were obvious as a function of biofilm depth (Fig. 4). PCA1 explained 37.6% of the observed variation, and PCA2 explained 21.0% of the variation. ANOVA tests showed that there are significant differences in PCA1 and PCA2 between the zones (p < 0.001 in both zones). Community structure had three clear groups as a function of biofilm depth, suggesting that functional dominant bacteria stratified at different zones of the biofilm and established an ecological niche to support the activity and growth of microorganisms.
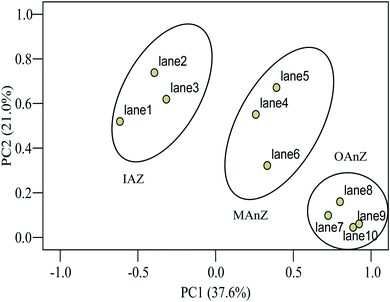 | ||
| Fig. 4 Principal component analysis of DGGE profiles of 16S rRNA from the thick membrane-aerated biofilm grown at SLR2. | ||
Eight prominent bands from different vertical band positions were excised from three different lanes of the DGGE gel (i.e. relative biofilm thickness of 0.125, 0.5 and 1), amplified, cloned and sequenced (Fig. 3). The cloning results showed that a band often was made up of more than one unique sequence. Hence, the total number of strains was more than the number of bands. These bands contained 16 unique DNA sequences, or true operational taxonomic units (OTUs), determined by using 99% minimum similarity as the threshold.24 The phylogenetic distributions from the biofilm using a bootstrap neighbour-joining method are shown in Fig. 5. The sequences of DGGE bands (8 OTUs) fall into 3 putative main phylogenetic divisions, respectively. Dominant bacteria could be divided into two groups: Bacteroidetes and β-Proteobacteria families. Strains of β-Proteobacteria existed in the most communities and appeared to be key members.
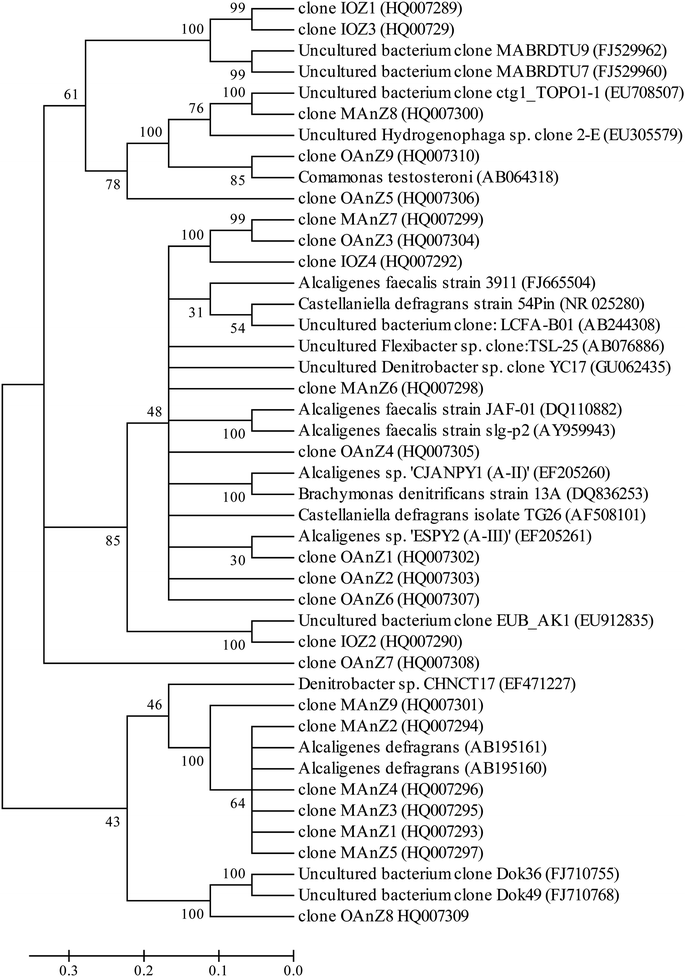 | ||
| Fig. 5 Phylogenetic neighbor-joining tree derived from V3 region of 16S rRNA sequences of DGGE bands and clone libraries and sequences from the database. GenBank accession numbers are given in parentheses. Bootstrap values of 30% for neighbor joining are shown (percentages of 1000 resamplings). Bar indicates 0.1 divergences. The definition of a clone abbreviation for the location from which they were detected is consistent with that in Table 3. | ||
For aerobic zone of biofilm nearest the membrane, the specific OTUs were bands 1 and 2 (Fig. 3). These specific populations were affiliated with bacteria such as uncultured Bacteroidetes and uncultured Hydrogenophaga sp. (Table 3 and Fig. 5). The result has therefore supported the observed high SAOR at the location nearest the membrane. For anoxic zone in the middle of biofilm, the prominent OTUs were bands 3 and 4 (Fig. 3). These populations were affiliated with Alcaligenes defragrans, Alcaligenes sp. and Denitrobacter sp., respectively (Table 3 and Fig. 5). These populations certainly played an important role in denitrification (see Fig. 2). For the outer region of MAB at the biofilm–liquid interface, the specific OTUs were bands 5, 6, 7 and 8 (Fig. 3). These dominant populations were affiliated with acetonitrile-degrading bacteria such as, Comamonas testosteroni, Alicycliphilus sp., Flavobacterium sp. and Brachymonas denitrificans, (Table 3 and Fig. 5). Although there were substantial differences between these three bacterial communities, the common OTUs were detected as a function of biofilm depth with weak and insignificant in functional reaction activity.
| Clones (DGGE band) | Most closely related sequence (accession no.) | Taxonomic affiliation | 16S rRNA gene sequence identity (%) | Sequence length (bases) |
|---|---|---|---|---|
| a DGGE band numbers identify specific bands that were excised from the DGGE gel (see Fig. 3). Clones given in parentheses are identified by an abbreviation for the location from which they were detected (IAZ = inner aerobic zone; MAnZ = middle anoxic zone; OAnZ = outer anaerobic zone) and a number. | ||||
| Band 1 (clone IAZ1) | Uncultured bacterium clone MABRDTU9 (FJ529962) | Bacteriodetes | 97.9 | 295 |
| Band 2 (clone IAZ4) | Uncultured Hydrogenophaga sp. clone 2-E (EU305579) | β-Proteobacteria | 99.7 | 325 |
| Band 3 (clone MAnZ1) | Alcaligenes sp. CJANPY1 (A-II) (EF205260) | β-Proteobacteria | 99.9 | 693 |
| Band 4 (clone MAnZ3) | Denitrobacter sp. CHNCT17 (EF471227) | β-Proteobacteria | 97.5 | 635 |
| Band 5 (clone OAnZ1) | Comamonas testosteroni (AB064318) | β-Proteobacteria | 98.0 | 295 |
| Band 6 (clone OAnZ2) | Alcaligenes faecalis strain JAF-01 (DQ110882) | β-Proteobacteria | 100 | 371 |
| Band 7 (clone OAnZ10) | Uncultured bacterium clone EUB_AK1 (EU912835) | Bacteroidetes | 99.4 | 350 |
| Band 8 (clone OAnZ11) | Brachymonas denitrificans strain 13A (DQ836253) | β-Proteobacteria | 100 | 344 |
4. Discussion
An understanding of the factors controlling the performance and stratification of activity and community structure in MABs is significant to optimize their application for biological wastewater treatment and management. For the thick biofilm grown at SLR2, the biofilm was stratified into oxic/anoxic/anaerobic zones (Fig. 6 and S1†), and able to achieve acetonitrile removal as well as nitrification and denitrification for nitrogen removal (Fig. 2 and 3). The MAB examined in this work is complex, with the region nearest the membrane being rich in oxygen but low in substrate concentration, and the outer region of the biofilm being void of oxygen but rich in substrate. As the biofilm thickness increased (as a result of the increased SLR), diffusion rates of oxygen and the nutrient (e.g. acetonitrile, metabolic products of ammonia and possibly nitrate) in the biofilm decreased, resulting in the stratification of active regions in the MABs (Fig. 2, 3 and 6). Therefore, the region near the membrane would be highly aerobic while the region next to the bulk liquid (wastewater) could be fully anaerobic in the thick biofilm grown at SLR2. These changing growth conditions within the biofilm promote various microbial activities, resulting in development of stratified MABs (Fig. 2–4).DGGE profiles and cloning of 16S rRNA genes also provided further evidence for the heterogeneous stratification and distribution of activity and functional population in MABs versus the relative biofilm thickness (Fig. 1–3 and Table 3). Ammonia-oxidizing bacteria were found in the biofilm near the membrane side, whereas acetonitrile-degrading bacteria proliferated mainly in the middle and outer regions of biofilms; denitrifying bacteria were found in the anoxic and anaerobic outer regions of the MAB grown at high SLR2 (see Fig. 2 and Table 3). However, such stratification was not observed in the thin MAB grown at the low SLR1 (data not shown). Our results differ from previous efforts to characterize community structure in MABs which reported that denitrifying bacteria were in outer biofilm.21,39 The enrichment of denitrifying bacteria in the middle region of MAB as observed in this study could be explained by the fact that denitrifying bacteria can easily utilize nitrate (metabolic product of AOB) diffusing from inner oxic zone of MAB, while shielding away from toxicity or inhibition of acetonitrile and/or its biodegradation intermediates in the bulk liquid. Consequently, these results suggest that the difference in microbial community or dominant populations can be due to differences in the composition of the inoculums, operation conditions and substrate types. This study found that using the same inoculum and substrate, development of microbial communities can be controlled by the MABR operating conditions. Differences in community structure undoubtedly reflect the differences in reaction activity, which can be controlled by operational parameters such as air or oxygen supply and flow velocity.20,22 The identification of key microbial community members for the treatment of organonitrile-containing wastewater can help effective treatment while maintaining a stable microbial community in the reactor.
DGGE analysis revealed three functional reaction regions within the biofilms with low bacterial diversity (Fig. 3). This is a result of the selection pressure exerted by high SLR operation, which would favor the enrichment of certain bacterial species to the detriment of others. Furthermore, the toxicity or recalcitrance of acetonitrile as the biofilm thickness increased contributed to the limited diversity observed, as only a handful of isolates have so far been characterized and known to utilize acetonitrile as a sole carbon and energy source.40 Many mechanisms could be involved in the enhancement of metabolic efficiency in the adapted biofilms.38,41,42 First, this enhancement could be the result of syntrophic interactions between the functional populations in the biofilms. Second, metabolic activity enhancement could result from an increase of bacterial diversity through the exchange of genetic material among the bacteria within the biofilms in the ecological niche. Interestingly, there is an evidence to suggest that genes involved in the degradation of ether fuels might be transferred horizontally between bacteria belonging to the Rhodococcus group that is known to be capable of degrading acetonitrile.43,44 Although the extent to which each of the adaptive mechanisms contributed to the development of acetonitrile-degrading biofilm has yet to be determined, such adaptation selected individual or groups of microorganisms that best suited to tolerate and degrade acetonitrile, and these microorganisms eventually swept over and dominated the microbial community, suggesting that an ecological niche established to support the survival and growth of differential functional microorganisms.
Collectively, our results demonstrated that the functional model of the microbial community and its activity stratification within the MABs fed with nitrogen-containing acetonitrile versus the biofilm thickness (Fig. 2–4 and S1†). All these microbial groups cooperate with each other to ensure the formation and stability of biofilm exposed to high acetonitrile concentrations in the bulk milieu. Within the biofilms under appropriate operation, the bacteria will distribute and organize themselves to best meet the needs of each other and the community in the particular microenvironment.45 Therefore, bacteria with high acetonitrile biodegradation ability and high growth rates arranged near the biofilm surface as part of the overall community's scheme to counter toxicity due to high acetonitrile concentrations. Concurrently, slow-growing species (e.g. AOB) or nutrient competitive populations (denitrifying bacteria) can take refuge and accumulate inside or middle the biofilm (Fig. 3 and Table 2). In fact, the structural adaptations and interrelationships that exist in highly structured biofilms have caused them to be regarded as multicellular organisms with developed internal interdependencies and coordinated activities.46
5. Conclusions
In this study, we developed two types of MABs with a thin or a thick biofilm under different SLRs for the simultaneous degradation of acetonitrile, and nitrification and denitrification in a single reactor. Different biofilm thicknesses caused substantially different profiles of the microbial activities with different functions in the biofilm. Results showed that ammonia-oxidizing bacteria were found in the biofilm near the membrane side, whereas acetonitrile-degrading bacteria proliferated mainly in the middle and outer regions of biofilms; denitrifying bacteria were found in the anoxic and anaerobic outer regions of the MAB grown at high SLR2. However, such stratification was not observed in the thin MAB grown at the low SLR1. This study demonstrated that the biofilm thickness developed under SLRs is an important factor affecting the structural and functional stratification of bacterial populations in a single MABR treating volatile nitrogen-containing organonitrile wastewater. It suggested the potential of using the MABR for high-rate volatile organic pollutant removal, organic carbonaceous pollutant biodegradation, and nitrification/denitrification processes.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant 50978246 and 51138009).References
- A. S. Fernandez, S. A. Hashsham, S. L. Dollhopf, L. Raskin, O. Glagoleva, F. B. Dazzo, R. F. Hickey, C. S. Criddle and J. M. Tiedje, Appl. Environ. Microbiol., 2000, 66, 4058–4067 CrossRef CAS PubMed.
- E. Syron and E. Casey, Environ. Sci. Technol., 2008, 42, 1833–1844 CrossRef CAS PubMed.
- R. Nerenberg, Curr. Opin. Biotechnol., 2016, 38, 131–136 CrossRef CAS PubMed.
- A. Celik, E. Casey and H. Hasar, J. Hazard. Mater., 2018, 356, 26–33 CrossRef CAS PubMed.
- T. G. Li, R. B. Bai and J. X. Liu, J. Biotechnol., 2008, 135, 52–57 CrossRef CAS PubMed.
- N. Landes, A. Morse and W. A. Jackson, Environ. Eng. Sci., 2013, 30, 606–616 CrossRef CAS.
- B. Heffernan, C. D. Murphy, E. Syron and E. Casey, Environ. Sci. Technol., 2009, 43, 6776–6785 CrossRef CAS PubMed.
- D. G. Ohandja and D. C. Stuckey, J. Chem. Technol. Biotechnol., 2010, 85, 294–301 CAS.
- X. Mei, Y. Chen, C. Fang, L. Xu, J. Li, S. Bi, J. Liu, Y. Wang, P. Li, Z. Guo, H. Qin, J. Gu, Y. Xiao, X. Yang, B. Zhou and Z. Zhang, Bioresour. Technol., 2019, 289, 121754 CrossRef PubMed.
- B. Taşkan, E. Casey and H. Hasar, Sci. Total Environ., 2019, 682, 553–560 CrossRef PubMed.
- L. Sun, Z. Wang, X. Wei, P. Li, H. Zhang, M. Li, B. Li and S. Wang, Chem. Eng. Sci., 2015, 135, 559–565 CrossRef CAS.
- J. Lin, P. Zhang, G. Li, J. Yin, J. Li and X. Zhao, Int. Biodeterior. Biodegrad., 2016, 113, 74–79 CrossRef CAS.
- Y. Ma, C. Domingo-Félez, B. G. Plósz and B. F. Smets, Environ. Sci. Technol., 2017, 51, 6146–6155 CrossRef CAS.
- C. T. Kinh, T. Suenaga, T. Hori, S. Riya, M. Hosomi, B. F. Smets and A. Terada, Water Res., 2017, 124, 363–371 CrossRef CAS PubMed.
- M. J. Semmens, K. Dahm, J. Shanahan and A. Christianson, Water Res., 2003, 37, 4343–4350 CrossRef CAS PubMed.
- T. G. Li, J. X. Liu, R. B. Bai and F.-S. Wong, Environ. Sci. Technol., 2008, 42, 2099–2104 CrossRef CAS PubMed.
- X. Mei, J. Liu, Z. Guo, P. Li, S. Bi, Y. Wang, W. Shen, Y. Wang, Y. Xiao, X. Yang, B. Zhou, H. Liu and S. Wu, J. Hazard. Mater., 2019, 363, 99–108 CrossRef CAS PubMed.
- H. Tian, Y. Hu, X. Xu, M. Hui, Y. Hu, W. Qi, H. Xu and B. Li, Bioresour. Technol., 2019, 289, 121649 CrossRef CAS PubMed.
- M. Lan, M. Li, J. Liu, X. Quan, Y. Li and B. Li, Bioresour. Technol., 2018, 270, 120–128 CrossRef CAS PubMed.
- T. M. LaPara, A. C. Cole, J. W. Shanahan and M. J. Semmens, J. Ind. Microbiol. Biotechnol., 2006, 33, 315–323 CrossRef CAS PubMed.
- A. C. Cole, M. J. Semmens and T. M. LaPara, Appl. Environ. Microbiol., 2004, 70, 1982–1989 CrossRef CAS PubMed.
- L. S. Downing and R. Nerenberg, Biotechnol. Bioeng., 2008, 101, 1193–1204 CrossRef CAS PubMed.
- J. W. Shanahan and M. J. Semmens, Water Res., 2015, 74, 10–22 CrossRef CAS PubMed.
- A. Terada, S. Lackner, K. Kristensen and B. F. Smets, Environ. Microbiol., 2010, 12, 2858–2872 CAS.
- K. Hibiya, A. Terada, S. Tsuneda and A. Hirata, J. Biotechnol., 2003, 100, 23–32 CrossRef CAS PubMed.
- F. D. Hulot, G. Lacroix, F. Lescher-Moutoue and M. Loreau, Nature, 2000, 405, 340–343 CrossRef CAS PubMed.
- H. L. Jiang, J. H. Tay, A. M. Maszenan and S. T. L. Tay, Appl. Environ. Microbiol., 2004, 70, 6767–6775 CrossRef CAS PubMed.
- T. G. Li and J. X. Liu, Biochem. Eng. J., 2017, 121, 156–162 CrossRef CAS.
- T. G. Li, R. B. Bai, D.-G. Ohandja and J. X. Liu, Biodegradation, 2009, 20, 569–580 CrossRef CAS PubMed.
- APHA, Standard methods for the examination of water and wastewater, American Public Health Association, Washington, DC, 20th edn, 1998 Search PubMed.
- L. M. Freitas dos Santos and A. G. Livingston, Biotechnol. Bioeng., 1995, 47, 82–89 CrossRef CAS PubMed.
- G. A. Kowalchuk, J. R. Stephen, W. DeBoer, J. I. Prosser, T. M. Embley and J. W. Woldendorp, Appl. Environ. Microbiol., 1997, 63, 1489–1497 CAS.
- G. Muyzer, E. C. Dewaal and A. G. Uitterlinden, Appl. Environ. Microbiol., 1993, 59, 695–700 CAS.
- K. Watanabe, M. Teramoto, H. Futamata and S. Harayama, Appl. Environ. Microbiol., 1998, 64, 4396–4402 CAS.
- P. D. Schloss and J. Handelsman, Appl. Environ. Microbiol., 2005, 71, 1501–1506 CrossRef CAS PubMed.
- T. A. Hall, Nucleic Acids Symp. Ser., 1999, 41, 95–98 CAS.
- S. Kumar, K. Tamura, I. B. Jakobsen and M. Nei, Bioinformatics, 2001, 17, 1244–1245 CrossRef CAS PubMed.
- R. Muñoz, M. Jacinto, B. Guieysse and B. Mattiasson, Appl. Microbiol. Biotechnol., 2005, 67, 699–707 CrossRef PubMed.
- A. Terada, K. Hibiya, J. Nagai, S. Tsuneda and A. Hirata, J. Biosci. Bioeng., 2003, 95, 170–178 CrossRef CAS.
- S.-L. Sun, T.-Q. Lu, W.-L. Yang, J.-J. Guo, X. Rui, S.-Y. Mao, L.-Y. Zhou and Y.-J. Dai, RSC Adv., 2016, 6, 15501 RSC.
- K. Håkansson, U. Welander and B. Mattiasson, Water Res., 2005, 39, 648–654 CrossRef PubMed.
- T. Manolov, H. Kristina and G. Benoit, Appl. Microbiol. Biotechnol., 2005, 66, 567–574 CAS.
- M. Kobayashi, N. Yanaka, T. Nagasawa and H. Yamada, J. Bacteriol., 1990, 172, 4807–4815 CrossRef CAS PubMed.
- B. R. Langdanhl, P. Bisp and K. Ingvorse, Microbiology, 1996, 142, 145–154 CrossRef.
- P. Watnick and R. Kolter, J. Bacteriol., 2000, 182, 2675–2679 CrossRef CAS PubMed.
- P. Stoodley, K. Sauer, D. G. Davies and J. W. Costerton, Annu. Rev. Microbiol., 2002, 56, 187–209 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra03128f |
| This journal is © The Royal Society of Chemistry 2019 |