Open Access Article
Open Access ArticleRapid generation of chemical combinations on a magnetic digital microfluidic array†
Yi Zhang *a and
Tza-Huei Wang*b
*a and
Tza-Huei Wang*b
aSchool of Mechanical and Aerospace Engineering, Nanyang Technological University, 50 Nanyang Avenue, Singapore 639798. E-mail: yi_zhang@ntu.edu.sg
bDepartment of Biomedical Engineering, Department of Mechanical Engineering, Johns Hopkins University, 3400 North Charles Street, Baltimore, MD 21218, USA. E-mail: thwang@jhu.edu.sg
First published on 12th July 2019
Abstract
Combinatorial screening is frequently used to identify chemicals with synergistic effects by measuring the response of biological entities exposed to various chemical-dose combinations. Conventional microwell-based combinatorial screening is resource-demanding, and the closed microfluidics-based screening requires sophisticated fluidic control systems. In this work, we present a novel combinatorial screening platform based on the surface energy trap (SET)-assisted magnetic digital microfluidics. This platform, known as FlipDrop, rapidly generates chemical combinations by coupling two droplet arrays with orthogonal chemical concentration gradients with a simple flip. We have illustrated the working principle of FlipDrop by generating combinations of quantum dots. We have also successfully demonstrated the screening of quantum dot fluorescence resonance energy transfer (QD-FRET) on the FlipDrop platform by measuring the FRET response. This report demonstrates that FlipDrop is capable of rapidly generating chemical combinations with unprecedented ease for combinatorial screening.
Introduction
Combinatorial screening is a method used to measure the response of biological entities to a combination of chemicals. One main application of combinatorial screening is to identify combinatorial therapy, a treatment regime using multiple drugs and adjuvants.1 It is evident that a combination of multiple compounds could be more effective than a single compound, hence reduces the required dosage and side effects and leads to more coveted clinical outcomes.1–5 In spite of its increasing popularity, many existing combinatorial therapies are empirical with limited evidence.6,7 To facilitate evidence-based combinatorial therapy, new combinatorial drugs ought to be screened systematically from a library of new or existing compounds using combinatorial screening.3,8Conventional microwell-based combinatorial screening platform is resource-demanding and time-consuming. Its large assay volume necessitates a large number of cells, and the generation of chemical combinations in the microwell plate requires complex liquid handling systems and is often laborious.
Microfluidics has long been proposed for screening.9–16 So far, many microfluidic systems have been developed to screen one single compound at a time. Only a limited number of examples have demonstrated the proof-of-concept of combinatorial screening. Kim et al. developed a programmable cell culture array that integrated a microfluidic concentration gradient generator with cell culture chambers for combinatorial drug screening.16 The drug combinations were generated by coupling two orthogonal concentration gradients with the cell culture chamber at the intersection. Each cell culture chamber on the device was individually addressable by the microfluidic network and was subjected to one unique drug combination. Jang et al. developed a similar device with two orthogonal concentration gradient generators.15 One of the concentration gradients was generated by the tree-like gradient generator, and the other one was generated by an active microfluidic mixer and dispenser. Eduati et al. demonstrated a discrete microfluidics-based combinatorial drug screening platform.14 The drug combinations were generated in aqueous plugs separated by the oil phase in a capillary tube. The drug composition in each aqueous plug was actively controlled by Braille valves. Cells were co-encapsulated and incubated with the drug combinations in the plug for screening. While these microfluidic platforms showed great promise in combinatorial screening, they were usually complex multi-layer devices that required sophisticated fluidic control systems. Another common limitation of these platforms was the difficulty in retrieving the cells for subsequent analysis because they were closed microfluidic systems. Therefore, the screening assay was often restricted to single-step homogeneous assays. In contrast to the conventional closed microfluidic systems, open microfluidic platforms control fluids on an open surface with no or only partial constrains. One example of open microfluidics is digital microfluidics.17,18 On a digital microfluidic platform, fluids in the form of droplets are manipulated using various mechanisms, such as magnetic force,19–27 electrowetting,28–36 surface acoustic wave.37–40 Among these manipulation methods, magnetic digital microfluidics has unique advantages, such as simple fluidic operation and dual functionality of magnetic particles, compared to other actuation mechanisms.17 Droplets, which function as reaction chambers, are driven by the added magnetic particles to accomplish a range of fluidic operations required for biochemical assays. The substrate on which the droplets sit can be modified with additional features, such as the surface energy traps (SETs), to facilitate a full range of droplet manipulations for complex assays.19 Several open microfluidic platforms have been proposed for combinatorial screening. Ding et al. developed a print-to-screen platform that used a microfluidic manifold to dispense the chemical compounds into droplets through a dot-matrix printer head.41 Chemical combinations were generated by dispensing multiple droplets to the same location. A similar approach was demonstrated by Du et al.42 Instead of using a manifold, a 3-axis translational stage was used to guide the capillary printer head to designated locations on the open microfluidic platform. These approaches merely used the open microfluidic platform as a liquid holder. The generation of chemical combinations still relied on point-to-point dispensing. Rapid generation of droplet arrays was demonstrated on an open microfluidic platform by Wu et al. In that work, droplet array was generated by spreading the liquid over hydrophilic/hydrophobic patterns similar to SETs.43 Authors demonstrated the encapsulation of single cells in the droplet array.44 However, no strategy was proposed to generate chemical combinations for combinatorial screening using this approach. Kulesa et al. devised a rapid way of generating chemical combinations by coupling nanolitre-sized droplets on a droplet pairing array which trapped one pair of droplets in each microchamber.45 The paired droplets were merged by washing away the surfactant used for droplet stabilization. This approach generated chemical combinations with a great ease. However, because the droplet coupling was a stochastic process, the exact composition of the chemical combination was not designed a priori, but only unveiled posteriori by decoding the fluorescent coding dyes.
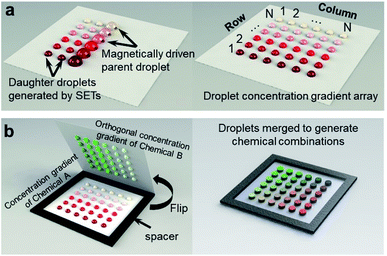
In this work, we present a SET-assisted magnetic digital microfluidic platform that generates chemical combinations rapidly with unprecedented ease (Fig. 1). This platform, which is referred to as FlipDrop, generates two droplet arrays with orthogonal concentration gradients using the SETs. Two chemicals, each of which has N different concentrations, are coupled into N2 number of chemical combinations with a simple flip. We demonstrate the rapid generation of chemical combinations using two different quantum dots with distinct fluorescent emission profiles. We also perform the screening of quantum dot fluorescent resonance energy transfer (QD-FRET)46–48 by measuring the FRET signals in response to various chemical combinations. To our best knowledge, FlipDrop is the first digital microfluidic platform using this novel mechanism for rapid generation of chemical combinations, which promises great potential for combinatorial screening.
Experimental
Device fabrication
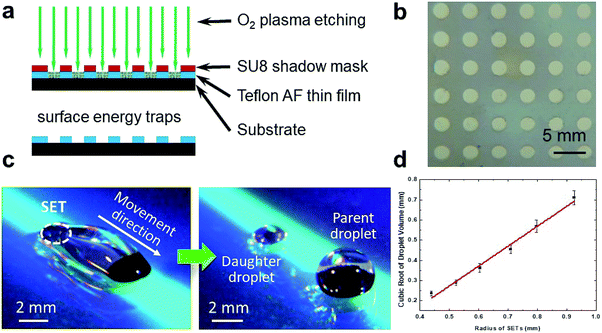
The SET-assisted magnetic digital microfluidic device was fabricated by oxygen plasma etching through an SU-8 (Microchem) shadow mask using a similar protocol we previously reported (Fig. 2).19,46 To briefly recap, the SU-8 shadow mask of ∼60 μm in thickness was fabricated with a lift-off process by using the Teflon AF (DuPond) as a sacrificial layer. This shadow mask was laminated on a glass substrate coated with a Teflon AF thin film of ∼700 nm thick. The SETs were created by etching the Teflon AF thin film with O2 plasma through the shadow mask. After etching, the Teflon AF thin film was annealed by heating the substrate above 200 °C for 30 s. The fabrication of the SET-assisted magnetic digital microfluidic device was accomplished within 3 minutes when using the ready-made shadow mask which could be reused.Droplet manipulation on magnetic digital microfluidic platform
The parent droplet of desired volume was dispensed onto the substrate with SETs. Micro magnetic particles (Qiagen) were washed and dried before being added to the droplet for actuation. A N52 grade cylindrical permanent magnet (1/8′′ in diameter and 1/2′′ in length) was placed beneath the substrate to control the movement of the magnetic particles. To move multiple droplets in parallel, a linear array of magnets was used. The spacing between the magnets matched that between the droplets.For the demonstration of droplet dispensing shown in Fig. 2, the diameter of the SET is 2.5 mm, the parent droplet volume is 20 μL, and the mass of magnetic particles is 0.36 mg. To create the droplet array for chemical combinations, the diameter of the SET is 1.6 mm, the parent droplet volume is 10 μL, and the mass of magnetic particles is 0.24 mg.
We calibrated the volume of the daughter droplets as a function of the SET diameter. The volume of the daughter droplet was too small to be measured directly. We therefore estimated the droplet volume by fluorescence. 10 μL of water was added to each daughter droplet. The volume of the daughter droplet Vd could be calculated according to
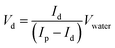 | (1) |
Generation of chemical combinations with FlipDrop
The 6 × 6 SET array was used to create the droplet array. The diameter of each SET was 1.6 mm, and the spacing between SETs was 5 mm. The glass substrate was cut into a 35 mm × 35 mm square. The coupling of the two arrays were done manually by aligning the edge of the square glass substrate.We demonstrated the generation of chemical combinations with two types of quantum dots. A twofold serial dilution of Qdot 525 (Invitrogen) with 6 different concentrations ranging from 0.625 nM to 20 nM was dispensed onto the SET-assisted magnetic digital microfluidic device in the form of droplets. The volume of each parent droplet was 10 μL, and 0.24 mg of magnetic particles were added to each droplet. All 6 parent droplets were dragged over the SETs in parallel by the permanent magnets. As the parent droplets moved over the SETs, daughter droplets were created by the passive dispensing mechanism illustrated in Fig. 1. After the parent droplets passed through the SETs, a droplet array of Qdot 525 concentration gradient was created. The droplet array of QD 605 (Invitrogen) concentration gradient was created in the same way. To generate the chemical combination, the Qdot 605 droplet array was flipped and coupled with the Qdot 525 array in such an orientation that the two concentration gradients were orthogonal to each other. A polydimethylsiloxane (PDMS) spacer of 1 mm thick was placed between the two substrates to prevent the droplets from being squashed. The fluorescent image was acquired using the Typhoon™ laser scanner (GE Healthcare) with a 488 nm laser excitation and proper emission filters for Qdot 525 and Qdot 605. The total intensities of Qdot 525 and Qdot 605 of each coupled droplet were extracted from the image using ImageQuant and normalized between 0 and 255. For all fluorescent images presented in this report, only the areas of interest, i.e. the merged droplets, were shown.
To screen the QD-FRET response at various quantum dot and Cy5 combinations, a droplet array of Qdot 605 concentration gradient and a droplet array of Cy5 concentration gradient were created on the SET-assisted magnetic digital microfluidic platform. The Qdot 605 was conjugated with streptavidin (Invitrogen), and the Cy5 was conjugated to the 3′ terminal of a 20-base DNA oligonucleotide. The 5′ terminal of the DNA oligonucleotide was conjugated with biotin. Once mixed, the quantum dot and the DNA oligonucleotide would self-assemble into a nanocomplex, bringing Cy5 to the close proximity of the quantum dot and promoting the energy transfer.
The fluorescent images of QD-FRET were acquired using the Typhoon™ laser scanner with a 488 nm laser excitation and proper emission filters for Qdot 605 and Cy5. The intensities of Qdot 605 and Cy5 through FRET were extracted from the images, and the FRET ratio was calculated according to
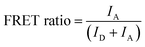 | (2) |
Results and discussion
Principle of FlipDrop
FlipDrop comprises of two droplet arrays created using SET-assisted magnetic digital microfluidics (Fig. 1). First, N number of parent droplets of Chemical A are dispensed onto the magnetic digital microfluidic platform, and magnetic particles are added into the droplets. The concentrations of the parent droplets follow a concentration gradient. As the magnetic particles drag a parent droplet over a SET, the surface tension splits a portion of the liquid from the parent droplet and holds it in the SET in the form of a daughter droplet (Fig. 1a, ESI Video 1†). A total of N daughter droplets are created from each parent droplet. In the end, an N × N droplet array is created. The concentration gradient is along the column direction with N different concentrations, and each concentration is made into N number of droplets in a row. Next, Chemical B is also made into an N × N droplet array in the same way. The concentration gradient of the Chemical B array is orthogonal to the Chemical A array, which means that the concentration gradient is along the row and each concentration is dispensed into N droplets in a column (Fig. 1b). Chemical combinations are generated by flipping the Chemical B array 180° and coupling it to the Chemical A array. The droplets on the two arrays are paired and merged as the two arrays are brought together. A 1 mm thick spacer is placed between the two arrays to prevent the droplets from being squashed. In the end, a sandwiched N × N droplet array with N2 number of distinct chemical combinations is generated.Droplet dispensing on the magnetic digital microfluidic array
The concentration gradient array on FlipDrop is created by dispensing daughter droplets using SETs on the magnetic digital microfluidic platform.19 The SETs are high-surface-energy areas patterned on the low-surface-energy substrate. SETs are fabricated by oxygen plasma etching that removes the Teflon AF thin film through a shadow mask (Fig. 2a). The regions with the Teflon AF thin film have a low surface energy. The regions where the Teflon AF is removed exposes the underlying substrate and have a high surface energy (Fig. 2b). We have previously demonstrated a full range of droplet manipulation on the SET-assisted magnetic digital microfluidic platform.19 In this study, we mainly use SETs for liquid dispensing, one operation that is unattainable on the traditional magnetic digital microfluidic platform. The mechanism of the SET-assisted liquid dispensing has been discussed in our previous work.19,20 To briefly recap, as the magnetic particles drag the parent droplet over the SET, the surface tension provided by the SET and the magnetic force pull the droplet from both sides and form two necking points, one of which is around the magnetic particle cluster and the other is around the SET (Fig. 2c). Depending on the size of the SET and the mass of the magnetic particles, one of the necking points would break. The breakage of the necking point around the SET would lead to the liquid dispensing. The volume of the dispensed daughter droplet is determined by the size of the SET. We discover that the cubic root of the daughter droplet volume is proportional with the diameter of the SET (Fig. 2d). This finding provides a guideline for SET design. The desired volume of the dispensed daughter droplet can be easily estimated based on the size of the SET.Generation chemical combinations using FlipDrop
We first demonstrate the creation of droplet arrays on the SET-assisted magnetic digital microfluidic platform using the approach described above. A representative picture of a droplet array created on the SETs is presented in Fig. 3a. The droplets show reasonably good uniformity. Our previous study indicates that the intra-chip variation of the daughter droplet volume is ∼3% and the chip-to-chip variation is ∼10%.19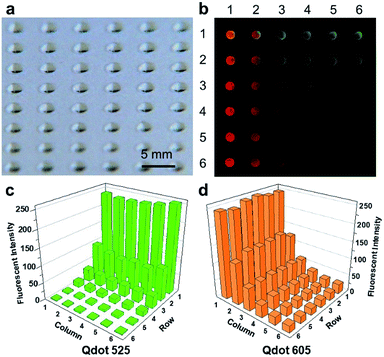 | ||
| Fig. 3 Chemical combinations generated by FlipDrop. (a) Droplet array generated by SET-assisted magnetic digital microfluidics. (b) Fluorescent image of FlipDrop array. Each droplet contains a unique combination of Qdot 525 and Qdot 605. (c) The fluorescent intensity Qdot 525. (d) The fluorescent intensity of Qdot 605. The raw data are included in the ESI.† | ||
We used a 6 × 6 SET array and two types of quantum dots to demonstrate the generation of chemical combinations on the FlipDrop platform (Fig. 3b). First, 6 parent droplets of Qdot 525 were dispensed on one SET array. The concentration of the 6 parent droplets followed a twofold serial dilution ranging from 0.63 nM to 20 nM. The 6 parent droplets were moved horizontally over the SET array so that the daughter droplets in each row had the same concentration. Next, 6 parent droplets of Qdot 605 were dispensed on the other SET array. The concentrations of the Qdot 605 droplets were the same as the Qdot 525. The Qdot 605 parent droplets were moved vertically over the SETs so that the daughter droplets generated in each column had the same concentration. As a result, the two droplet concentration gradient arrays were orthogonal to each other. The Qdot 605 array was then flipped and coupled with the QD 525 array. The flipping resulted in 36 merged droplets with distinct chemical combinations, which could be applied to combinatorial screening. With a simple flip, chemicals with a total of 2N number of concentrations were converted into an array with N2 number of chemical combinations.
The chemical composition of the combinatorial droplet array was examined by measuring the fluorescent intensities emitted by the two quantum dots. The Qdot 525 is represented by green and the Qdot 605 is represented by red in the combined false color image (Fig. 3b). Each spot illustrates a merged droplet that contains a unique combination of Qdot 525 and Qdot 605. The fluorescent intensities of the two quantum dots in each droplet is shown in Fig. 3c and d. The droplet is denoted by Eij where i stands for the row number and j stands for the column number. Droplet E11 contains high concentrations of both Qdot 525 and Qdot 605, hence appears bright orange in the false colored image. Droplet E61 contains a high concentration of Qdot 605 but a low concentration of Qdot 525, hence appears red. In contrast, droplet E16 contains a high concentration of Qdot 525 but a low concentration of Qdot 605, hence appears green. The concentrations of Qdot 525 and Qdot 605 are both low in droplet E66, therefore it is barely visible.
Screening of QD-FRET response using FlipDrop
As a proof-of-concept, we demonstrate the screening of QD-FRET at various combinations of Qdot 605 and Cy5 (Fig. 4). QD-FRET is a type of nanosensor widely used in many biosensing applications.46,48–50 The model QD-FRET system consists of a streptavidin-conjugated Qdot 605 and a DNA oligonucleotide labeled with biotin and Cy5 at its two termini (Fig. 4a). The quantum dot and the DNA self-assemble into a nanocomplex via the biotin–streptavidin interaction. When excited by the 488 nm laser, the quantum dot fluoresces with a maximum at 605 nm. The emission of the quantum dot in turn excites and transfers the energy to Cy5, leading to the Cy5 emission with a maximum at 650 nm. The FRET ratio is a parameter indicating the degree of energy transfer between the FRET donor (Qdot 605) and the FRET acceptor (Cy5). QD-FRET is a multi-acceptor-single-donor system whose FRET ratio is influenced by the acceptor to donor ratio. The higher the Cy5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qdot ratio, which means the more Cy5-labeled DNA oligonucleotides bind to each quantum dot, the higher the FRET ratio becomes.
Qdot ratio, which means the more Cy5-labeled DNA oligonucleotides bind to each quantum dot, the higher the FRET ratio becomes.
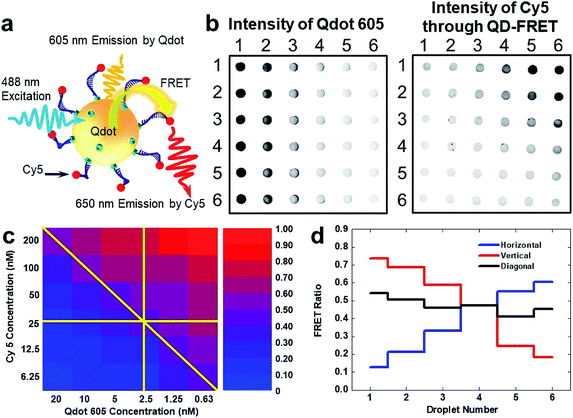 | ||
| Fig. 4 Screening of QD-FRET response at various Qdot 605 and Cy5 combinations on FlipDrop. (a) Schematic illustration of QD-FRET. The streptavidin-labelled quantum dots and the biotin-Cy5 dually labelled oligonucleotide self-assemble into a nanocomplex. The nanocomplex is excited by a 488 nm source, which causes the fluorescent emission of the Qdot 605. A portion of the 605 nm emission from the Qdot in turn excites Cy5 through FRET, causing another fluorescent emission peaked at 670 nm. Both emissions centered at 605 nm and 670 nm are recorded to evaluate FRET efficiency. (b) Qdot 605 fluorescent image of the FlipDrop array. Cy5 fluorescent image of the FlipDrop array acquired with 488 nm excitation. The Cy5 emission is due to FRET. (c) The FRET ratio map of the FlipDrop array. The raw data are shown in the ESI.† (d) The FRET ratio along the directions indicated in (c). | ||
We screened the QD-FRET using FlipDrop by measuring the FRET ratio. Two droplet arrays with orthogonal concentration gradients of Qdot 605 and Cy5 were generated using the SET-assisted magnetic digital microfluidics. Both Qdot 605 and Cy5 had 6 concentrations in a twofold serial dilution. The concentration of Qdot 605 ranged from 0.63 nM to 20 nM, and the concentration of Cy5 ranged from 6.25 nM to 200 nM. Combinations of Qdot 605 and Cy5 were generated by flipping the Cy5 array and coupling it with the Qdot array, which resulted in 36 merged droplets with distinct chemical compositions. The combined droplets were incubated at room temperature in a humid chamber for 10 min before measurement. The fluorescent emission intensities of Qdot 605 and Cy5, which were the FRET donor and the FRET acceptor respectively, were measured using a fluorescent scanner (Fig. 4b). The droplet with a high Cy5 intensity (excited by 488 nm, emission through FRET) and a low Qdot 605 intensity had a high FRET ratio, and vice versa. The FRET ratios of all droplets on FlipDrop were calculated according to eqn (2) and plotted in a heatmap (Fig. 4c). The FRET ratio showed an increasing trend from the bottom left to the top right. As mentioned above, the FRET ratio was influenced by the Cy5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qdot 605 ratio. Droplet E61 at the bottom left corner had the lowest Cy5
Qdot 605 ratio. Droplet E61 at the bottom left corner had the lowest Cy5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qdot 605 ratio of 0.31, and droplet E16 at the top right corner had the highest Cy5
Qdot 605 ratio of 0.31, and droplet E16 at the top right corner had the highest Cy5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qdot 605 ratio of 320. Therefore, droplet E61 had the smallest FRET ratio, and the droplet E16 had the largest FRET ratio in the heatmap. If we followed the heatmap in the horizontal, vertical and diagonal directions as indicated in Fig. 4c, we could observe the trend. In row 4, the Cy5 concentration was kept at 25 nM (before merging) while the Qdot 605 concentration decreased from 20 nM to 0.63 nM from left to right. As a result, the FRET ratio increased from droplet E41 to E46 (Fig. 4d). Similarly, in column 4, the Qdot 605 concentration was kept at 2.5 nM while the Cy5 concentration decreased from 200 nM to 6.25 nM. Consequently, the FRET ratio decreased from droplet E14 to E64. In the diagonal direction from droplet E11 at the top left corner to E66 at the bottom right corner, the Cy5
Qdot 605 ratio of 320. Therefore, droplet E61 had the smallest FRET ratio, and the droplet E16 had the largest FRET ratio in the heatmap. If we followed the heatmap in the horizontal, vertical and diagonal directions as indicated in Fig. 4c, we could observe the trend. In row 4, the Cy5 concentration was kept at 25 nM (before merging) while the Qdot 605 concentration decreased from 20 nM to 0.63 nM from left to right. As a result, the FRET ratio increased from droplet E41 to E46 (Fig. 4d). Similarly, in column 4, the Qdot 605 concentration was kept at 2.5 nM while the Cy5 concentration decreased from 200 nM to 6.25 nM. Consequently, the FRET ratio decreased from droplet E14 to E64. In the diagonal direction from droplet E11 at the top left corner to E66 at the bottom right corner, the Cy5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qdot 605 ratio was the same in all these droplets despite the fact that none of them had the same dose combination of Qdot 605 and Cy5. The results indicated that the QD-FRET ratio was determined by the Cy5
Qdot 605 ratio was the same in all these droplets despite the fact that none of them had the same dose combination of Qdot 605 and Cy5. The results indicated that the QD-FRET ratio was determined by the Cy5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qdot 605 ratio but not the concentration of the two components.
Qdot 605 ratio but not the concentration of the two components.
In this proof-of-concept experiment, we primarily focus on the rapid generation of the chemical combinations on the FlipDrop platform. Therefore, we do not emphasize the generation of the concentration gradient. The concentration gradient of the parent droplets is created off-chip. FlipDrop is fully compatible with the SET-assisted magnetic digital microfluidic serial dilution platform we reported earlier.20 The serial dilution platform utilizes SET to meter desired amounts of liquids from the stock solution droplet and merges them with the dilution buffer droplets to create a serial dilution. This operation would generate a concentration gradient in the form of droplets that can be directly transferred to FlipDrop. Droplets on both the serial dilution platform and FlipDrop are manipulated by an array of permanent magnet, and both use SETs to passively dispense daughter droplets. By combining these two platforms, we would be able to rapidly create concentration gradients from the stock solution and generate a large number of chemical combinations with unprecedented ease. It takes around 2 minutes to generate the chemical combination on the FlipDrop platform. Compared to the manual preparation of chemical combinations in microwell platform, FlipDrop significantly shortens the preparation time and reduces the number of pipetting operations. The magnetic digital microfluidics-based serial dilution takes about 1 minute. The overall time required to prepare desired chemical combinations from the raw stock solutions is on a par with the automated liquid handling platform.
Unlike conventional microfluidic systems, the SET-assisted magnetic digital microfluidics, which forms the foundation of FlipDrop and the serial dilution platform, does not require sophisticated fluidic control systems. All the fluidic operations are accomplished in the form of droplets, and all the control actions can be done by a translational stage. Therefore, we believe FlipDrop would greatly simplify the preparation of chemical combinations for combinatorial screening.
Application of FlipDrop to drug screening
This work mainly focuses on the generation of combinatorial arrays of multiple chemicals using FlipDrop. The droplet is well-suited for suspension cell culture. Drug screening can be performed by culturing the cells in the droplets together with the drug combinations. We have earlier demonstrated the bacteria suspension culture in droplets on a magnetic digital microfluidic platform for antimicrobial susceptibility testing.51 In that work, the bacteria were incubated together with the antibiotics in droplets kept in a humid environment to prevent the evaporation. Bacterial cells showed normal growth in the no-drug control.A similar workflow can be applied to combinatorial drug screening. On one of the arrays, the cell suspension is added to the parent droplets and split into daughter droplets together with one of the drugs. After being coupled with the other array, cell suspensions could be cultured in the combined droplets. The droplets contain the same initial number of cells but different drug combinations. The FlipDrop platform is then incubated at 37 °C in a humid chamber. The cell's response to the drug combinations could be monitored by observing the turbidity of the culture or measuring the fluorescent reporter.52
Conclusions
In conclusion, we have demonstrated a novel platform known as FlipDrop that rapidly generates chemical combinations for potential combinatorial screening. FlipDrop use SET-assisted magnetic digital microfluidics to create two orthogonal concentration gradients in the form of droplet arrays. The droplet arrays are then coupled together by a simple flip and generate a large number of chemical combinations for combinatorial screening. The working principle of FlipDrop is demonstrated with two quantum dots. We also demonstrate the screening of QD-FRET at various combinations of Cy5 and Qdot 605 on FlipDrop. The future work involves integrating FlipDrop with our magnetic digital microfluidic serial dilution platform. Once integrated, FlipDrop has the potential to change the paradigm of microfluidics-based combinatorial screening, making it more efficient and less resource demanding.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Yi Zhang would like to thank the funding support by the Start-Up Grant from the School of Mechanical and Aerospace Engineering at Nanyang Technological University and Tier 1 Grant (RG49/17) from Singapore Ministry of Education. Tza-Huei Wang would like to acknowledge the funding support from National Institutes of Health (R01AI117032, R01AI137272).Notes and references
- G. Cottarel and J. Wierzbowski, Trends Biotechnol., 2007, 25, 547–555 CrossRef CAS.
- C. Walsh, Nat. Rev. Microbiol., 2003, 1, 65 CrossRef CAS.
- M. Tschöp and R. DiMarchi, Diabetes, 2017, 66, 1766–1769 CrossRef.
- D. Leigh, K. Bradnock and J. Marriner, J. Antimicrob. Chemother., 1981, 7, 229–236 CrossRef CAS.
- B. Al-Lazikani, U. Banerji and P. Workman, Nat. Biotechnol., 2012, 30, 679 CrossRef CAS.
- L. Ejim, M. A. Farha, S. B. Falconer, J. Wildenhain, B. K. Coombes, M. Tyers, E. D. Brown and G. D. Wright, Nat. Chem. Biol., 2011, 7, 348 CrossRef CAS.
- K. Bush, Synergistic Antibiotic Combinations, Springer, 2017 Search PubMed.
- S. Malo and A. Geuna, Scientometrics, 2000, 47, 303–321 CrossRef CAS.
- Y. Ding, J. Choo and A. J. deMello, Microfluid. Nanofluid., 2017, 21, 58 CrossRef.
- C.-M. Huang, Y. Zhu, D.-Q. Jin, R. T. Kelly and Q. Fang, Anal. Chem., 2017, 89, 9009–9016 CrossRef CAS.
- J. Jeon, N. Choi, H. Chen, J.-I. Moon, L. Chen and J. Choo, Lab Chip, 2019, 19, 674–681 RSC.
- A. B. MacConnell, A. K. Price and B. M. Paegel, ACS Comb. Sci., 2017, 19, 181–192 CrossRef CAS.
- G. Wang, C. Lim, L. Chen, H. Chon, J. Choo, J. Hong and A. J. deMello, Anal. Bioanal. Chem., 2009, 394, 1827–1832 CrossRef CAS.
- F. Eduati, R. Utharala, D. Madhavan, U. P. Neumann, T. Longerich, T. Cramer, J. Saez-Rodriguez and C. A. Merten, Nat. Commun., 2018, 9, 2434 CrossRef.
- Y.-H. Jang, M. J. Hancock, S. B. Kim, Š. Selimović, W. Y. Sim, H. Bae and A. Khademhosseini, Lab Chip, 2011, 11, 3277–3286 RSC.
- J. Kim, D. Taylor, N. Agrawal, H. Wang, H. Kim, A. Han, K. Rege and A. Jayaraman, Lab Chip, 2012, 12, 1813–1822 RSC.
- Y. Zhang and N.-T. Nguyen, Lab Chip, 2017, 17, 994–1008 RSC.
- K. Choi, A. H. Ng, R. Fobel and A. R. Wheeler, Annu. Rev. Anal. Chem., 2012, 5, 413–440 CrossRef CAS.
- Y. Zhang and T. H. Wang, Adv. Mater., 2013, 25, 2903–2908 CrossRef CAS.
- Y. Zhang, D. J. Shin and T. H. Wang, Lab Chip, 2013, 13, 4827–4831 RSC.
- Y. Zhang, S. Park, K. Liu, J. Tsuan, S. Yang and T. H. Wang, Lab Chip, 2011, 11, 398–406 RSC.
- D. J. Shin, Y. Zhang and T.-H. Wang, Microfluid. Nanofluid., 2014, 17, 425–430 CrossRef CAS.
- J. Pipper, Y. Zhang, P. Neuzil and T. M. Hsieh, Angew. Chem., Int. Ed., 2008, 47, 3900–3904 CrossRef CAS.
- J. Pipper, M. Inoue, L. F. P. Ng, P. Neuzil, Y. Zhang and L. Novak, Nat. Med., 2007, 13, 1259–1263 CrossRef.
- C. H. Chiou, D. J. Shin, Y. Zhang and T. H. Wang, Biosens. Bioelectron., 2013, 50, 91–99 CrossRef CAS PubMed.
- D. Shin, P. Athamanolap, L. Chen, J. Hardick, M. Lewis, Y.-H. Hsieh, R. Rothman, C. A. Gaydos and T. Wang, Sci. Rep., 2017, 7, 4495 CrossRef CAS.
- D. J. Shin, A. Y. Trick, Y.-H. Hsieh, D. L. Thomas and T.-H. Wang, Sci. Rep., 2018, 8, 9793 CrossRef.
- M. Abdelgawad, S. L. S. Freire, H. Yang and A. R. Wheeler, Lab Chip, 2008, 8, 672–677 RSC.
- K. Choi, A. H. C. Ng, R. Fobel and A. R. Wheeler, Annu. Rev. Anal. Chem., 2012, 5, 413–440 CrossRef CAS.
- E. M. Miller and A. R. Wheeler, Anal. Chem., 2008, 80, 1614–1619 CrossRef CAS.
- A. H. C. Ng, M. D. Chamberlain, H. Situ, V. Lee and A. R. Wheeler, Nat. Commun., 2015, 6, 7513 CrossRef CAS.
- A. H. C. Ng, K. Choi, R. P. Luoma, J. M. Robinson and A. R. Wheeler, Anal. Chem., 2012, 84, 8805–8812 CrossRef CAS PubMed.
- M. H. Shamsi, K. Choi, A. H. C. Ng and A. R. Wheeler, Lab Chip, 2014, 14, 547–554 RSC.
- S. K. Cho, H. Moon and C.-J. Kim, J. Microelectromech. Syst., 2003, 12, 70–80 CrossRef.
- S. K. Cho and Y. Zhao, Lab Chip, 2007, 7, 490–498 RSC.
- R. B. Fair, Microfluid. Nanofluid., 2007, 3, 245–281 CrossRef CAS.
- X. Ding, P. Li, S. C. S. Lin, Z. S. Stratton, N. Nama, F. Guo, D. Slotcavage, X. Mao, J. Shi and F. Costanzo, Lab Chip, 2013, 13, 3626–3649 RSC.
- Z. Guttenberg, H. Müller, H. Habermüller, A. Geisbauer, J. Pipper, J. Felbel, M. Kielpinski, J. Scriba and A. Wixforth, Lab Chip, 2005, 5, 308–317 RSC.
- Z. Wang and J. Zhe, Lab Chip, 2011, 11, 1280–1285 RSC.
- L. Y. Yeo and J. R. Friend, Annu. Rev. Fluid Mech., 2014, 46, 379–406 CrossRef.
- Y. Ding, J. Li, W. Xiao, K. Xiao, J. Lee, U. Bhardwaj, Z. Zhu, P. Digiglio, G. Yang and K. S. Lam, Anal. Chem., 2015, 87, 10166–10171 CrossRef CAS.
- G.-S. Du, J.-Z. Pan, S.-P. Zhao, Y. Zhu, J. M. den Toonder and Q. Fang, Anal. Chem., 2013, 85, 6740–6747 CrossRef CAS.
- Y. Lin, Z. Wu, Y. Gao, J. Wu and W. Wen, Appl. Surf. Sci., 2018, 442, 189–194 CrossRef CAS.
- H. Wu, X. Chen, X. Gao, M. Zhang, J. Wu and W. Wen, Anal. Chem., 2018, 90, 4303–4309 CrossRef CAS.
- A. Kulesa, J. Kehe, J. E. Hurtado, P. Tawde and P. C. Blainey, Proc. Natl. Acad. Sci. U. S. A., 2018, 201802233 Search PubMed.
- Y. Zhang and T.-H. Wang, Theranostics, 2012, 2, 631 CrossRef CAS.
- V. J. Bailey, H. Easwaran, Y. Zhang, E. Griffiths, S. A. Belinsky, J. G. Herman, S. B. Baylin, H. E. Carraway and T.-H. Wang, Genome Res., 2009, 19(8), 1455–1461 CrossRef CAS.
- C.-Y. Zhang, H.-C. Yeh, M. T. Kuroki and T.-H. Wang, Nat. Mater., 2005, 4, 826 CrossRef CAS.
- T. R. Pisanic, Y. Zhang and T. Wang, Analyst, 2014, 139, 2968–2981 RSC.
- Y. Ho, H. H. Chen, K. W. Leong and T. Wang, J. Controlled Release, 2006, 116, 83–89 CrossRef CAS.
- Y. Zhang, D. J. Shin and T.-H. Wang, Lab Chip, 2013, 13, 4827–4831 RSC.
- N. Hesari, A. Alum, M. Elzein and M. Abbaszadegan, Enzyme Microb. Technol., 2016, 83, 22–28 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra03469b |
| This journal is © The Royal Society of Chemistry 2019 |