Open Access Article
Open Access ArticleThe secondary metabolites of rare actinomycetes: chemistry and bioactivity
Ting Ding
a,
Luo-Jie Yangc,
Wei-Dong Zhang
*ab and
Yun-Heng Shen *b
*b
aState Key Laboratory of New Drug and Pharmaceutical Process, Shanghai Institute of Pharmaceutical Industry, China State Institute of Pharmaceutical Industry, Shanghai 201203, China. E-mail: wdzhanggy@hotmail.com
bSchool of Pharmacy, The Second Military Medical University, Shanghai 200433, China. E-mail: shenyunheng@hotmail.com
cInstitute of Interdisciplinary Integrative Medicine Research, Shanghai University of Traditional Chinese Medicine, Shanghai 201203, China
First published on 16th July 2019
Abstract
Actinomycetes are outstanding and fascinating sources of potent bioactive compounds, particularly antibiotics. In recent years, rare actinomycetes have had an increasingly important position in the discovery of antibacterial compounds, especially Micromonospora, Actinomadura and Amycolatopsis. Focusing on the period from 2008 to 2018, we herein summarize the structures and bioactivities of secondary metabolites from rare actinomycetes, involving 21 genera.
1 Introduction
Actinomycetes can produce novel secondary metabolites, most of which possess significant antibacterial activity. So far, actinomycetes have contributed about two-thirds of available antibiotics, more than 70% of which are produced using Streptomyces.1 In 2013, Chavan et al. introduced marine actinomycetes and their biotechnological applications.2 In recent years, because of the increasing prevalence of bacterial resistance, it is important to discover new antibiotics.3 However, with continuous research on Streptomyces having been carried out, it has become difficult to find novel compounds with potent antibacterial activity from Streptomyces. On the contrary, since many successful antibiotics have been produced using rare actinomycetes, such as rifamycins produced by Amycolatopsis mediterranei, erythromycin produced by Saccharopolyspora erythraea and teicoplanin produced by Actinoplanes teichomyceticus, the significant ability of rare actinomycetes to produce antibiotics has obtained great attention.4 The proportion of antibiotics produced by rare actinomycetes has increased to being up to 25–30% of known antibiotics in the last two decades.5 Therefore, rare actinomycetes have become potential resources for the discovery of compounds with significant antibacterial activity.According to Lechevalie, when using traditional separation methods, the frequency of rare actinomycetes is far below the frequency of Streptomyces.6 Later, Okami corrected the concept of rare actinomycetes; he thought that rare actinomycetes had a slow growth rate and small population compared to Streptomyces. In addition, unusual growth conditions are necessary, and they result in distinctive physiological properties. Different from Streptomyces, it is more difficult to isolate rare actinomycetes from soil or other media. Therefore, research into secondary metabolites of rare actinomycetes has been less frequent than Streptomyces research, which has made rare actinomycetes a significant resource for finding new antibiotics. In this review, we mainly summarize the chemical structures and biological activities of the secondary metabolites of rare actinomycetes discovered from 2008 to 2018, involving 21 genera: Amycolatopsis, Actinomadura, Nonomuraea, Pseudonocardia, Micromonospora, Saccharothrix, Actinosynnema, Nocardia, Actinoplanes, Saccharopolyspora, Streptosporangium, Actinokineospora, Microbispora, Lechevalieria, Planomonospora, Microbacterium, Kribbella, Dietzia timorensis, Catenuloplanes, Catenulispora and Actinoalloteichus. Soil and insects are the main sources of rare actinomycetes in this review. Beyond these, plants are also important sources.
2 Chemistry and biological activity
2.1 Secondary metabolites from Amycolatopsis
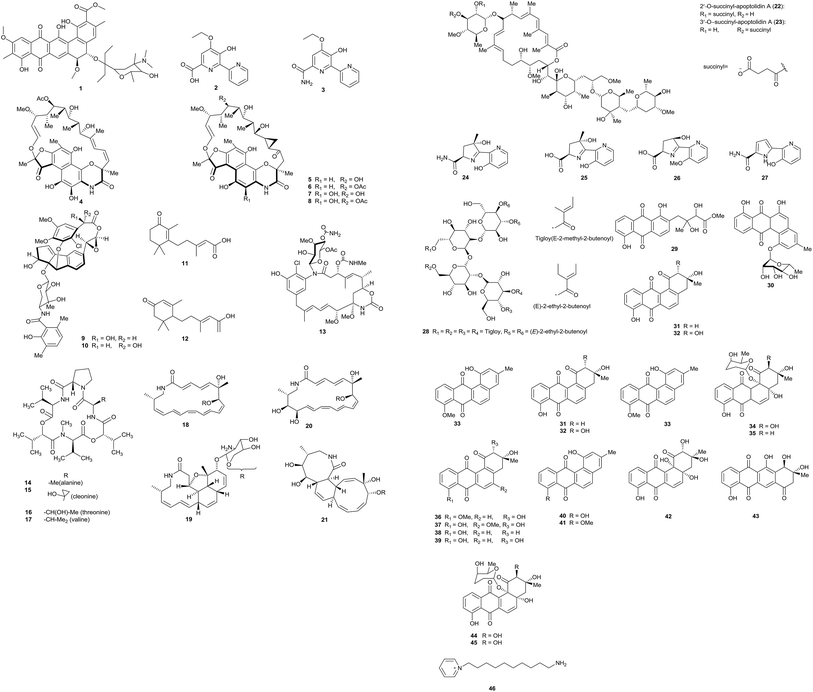
Pradimicin-IRD (1), a new polycyclic antibiotic, was produced by Amycolatopsis sp. IRD-009, which was isolated from the soil of an area of Brazilian rainforest undergoing restoration. Pradimicin-IRD showed antimicrobial activity against many bacteria, such as Streptococcus agalactiae, Pseudomonas aeruginosa and Staphylococcus aureus (all MICs were 3.1 μg mL−1). Beyond that, the compound also showed strong cytotoxicity properties against HCT-116 colon carcinoma cells with an IC50 value of 0.8 μM and against MM 200 melanoma cells with an IC50 value of 2.7 μM (Fig. 1).7From the rare actinomycete Amycolatopsis sp. K16-0194, a new compound named dipyrimicin B (2) was isolated via an interaction with Si-ES resin. In addition, another analog, dipyrimicin A (3), was also found without using Si-ES resin. Dipyrimicin A showed strong antimicrobial activity against Saccharomyces cerevisiae ATCC 9763, Kocuria rhizophila ATCC 9341 and Escherichia coli NIHJ at a dose of 30 μg. It also showed significant cytotoxicity against HeLa 3S, A549, THP-1, Jarkat and HL-60 cells with IC50 ≤ 5 μM. However, dipyrimicin B exhibited distinctly poor cytotoxicity and antimicrobial activity. The authors thought the differences in carboxyl and amide groups between the dipyrimicin species is the first factor involved in determining the cytotoxicity and antimicrobial activity.8
Xiao et al. isolated five unusual macrolactams rifamorpholines A–E (4–8) with an unprecedented 5/6/6/6 ring chromophore structure, representing a new subclass of rifamycin antibiotics. The compounds were produced by Amycolatopsis sp. HCa4, isolated from the guts of locusts (Locusta migratoria). Rifamorpholines B and D showed significant activities towards methicillin-resistant Staphylococcus aureus (MRSA) (MICs = 4.0 and 8.0 μM, respectively).9 Two novel enediyne-derived natural products, amycolamycin A and B (9–10) with a special 2-(cyclopenta[a]-inden-5-yl) oxirane core also were characterized from the same rare actinomycete Amycolatopsis sp. HCa4. Amycolamycin A possessed cytotoxic activity (IC50 = 7.9 μM) against the M231 cell line. With flow cytometry and Western blot analysis, it was demonstrated that amycolamycin A suppressed the proliferation of M231 cells through inducing apoptosis through the activation of caspase-3.10 Li et al. isolated two new abscisic acid-type sesquiterpenes (11–12) and one new ansamycin (13), together with four known ansamycins from Amycolatopsis alba DSM 44262. However, all of these examples showed no evident bacterial inhibition.11
Hashizume et al. discovered a novel cyclic depsipeptide, valgamicin C (14), containing the extremely rare amino acid cleonine and the related valgamicin compounds A, T and V (15–17) from Amycolatopsis sp. ML1-hF4. This is the first report of a cleonine-containing metabolite isolated from a naturally isolated microorganism. Compound 16 showed mild cytotoxicity against human tumor cell lines (MIA Paca 2, HGC-27, NCI-H1650, NB16 and ME-180) with IC50 values from 6.6 to 21.6 μM.12
Amycolatopsis sp. M39 was discovered from a Macrotermes natalensis colony in a termite worker. The strain produced four new 20-membered glycosylated polyketide macrolactams, named macrotermycin A–D (18–21). Macrotermycin A displayed antimicrobial activity against Bacillus subtilis ATCC 6051, Staphylococcus aureus ATCC 25923, Candida albicans ATCC 24433 and Saccharomyces cerevisiae ATCC 9763 (IC50 = 1, 1.5, 10 and 5 μg mL−1, respectively).13
Amycolatopsis sp. ICBB 8242, isolated from a unique black water ecosystem on the island of Borneo in Indonesia, produced two new apoptolidins, 2′-O-succinyl-apoptolidin A (22) and 3′-O-succinyl-apoptolidin A (23). These compounds can remarkably suppress the proliferation and viability of human H292 (IC50s values: 91 and 82 nM, respectively) and HeLa cells (IC50 = 240 and 260 nM, respectively).14
Via a bioassay-guided fractionation and isolation process, three new siderochelins, named siderochelin D (25), E (26), and F (27), along with the known siderochelin A (24), were obtained from Amycolatopsis sp. LZ149 isolated from the rhizosphere of Cynodon dactylon collected at Baicheng beach in Xiamen.15 Chemical analysis of the strain Amycolatopsis sp. HCa1, which came from the guts of grasshoppers (Oxya chinensis), led to the isolation of many chemical constituents. At first, a new tetrasaccharide derivative, actinotetraose L (28), was discovered by Guo et al. But the compound was not shown to display potent immunosuppressive activity and cytotoxicity from T-cell viability and MTT assays.16 One new angucyclinone derivative, amycomycin A (29), and one new angucycline, amycomycin B (30), along with 5 known compounds, tetrangomycin (31), PD116779 (32), X-14881E (33), sakyomicin A (34) and sakyomicin C (35), also were isolated from the rare actinobacterium Amycolatopsis sp. HCa1. Compound 35 displayed moderate cytotoxic activity against the human tumor cell lines BGC 823, HepG2, and A375 (IC50 values: 11.03, 17.36 and 17.5 μM, respectively).17 The same rare actinobacterium also yielded two new angucyclines, named (2R,3R)-2-hydroxy-8-O-methyl-tetrangomycin (36) and (2R,3R)-2-hydroxy-5-O-methyltetrango-mycin (37), together with eight known compounds (38–45). Compounds 38, 39, 43 and 44 displayed significant cytotoxic activities against HeLa cells and the IC50 values were 0.27, 0.11, 0.56, and 0.39 μM, respectively.18
The marine actinomycete Amycolatopsis alba var. nov. DVR D4 produced a novel pyridinium compound, 1-(10-aminodecyl)pyridinium (46). The strain was obtained in marine sediment samples. 1-(10-Aminodecyl)pyridinium showed potent cytotoxicity against HeLa, MCF-7 and U87MG in vitro and it also possessed antibacterial activities against Gram-negative and Gram-positive bacteria.19
2.2 Secondary metabolites from Actinomadura (Fig. 2)
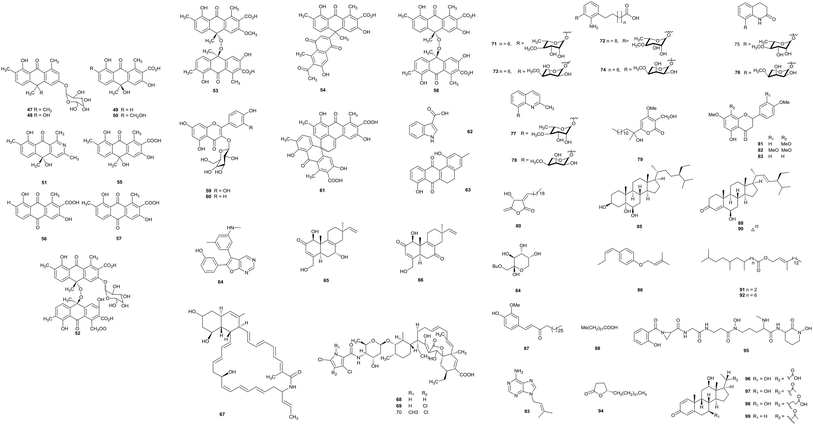
The fermentation of Actinomadura sp. BCC47066 isolated from soil from Thailand produced eight new anthrone derivatives, (+)-oxanthromicin E (47), F (49), and G (50), (±)-hemi-oxanthromicin D (48), azanthromicin A (51), adxanthromicin A2 (52), 3-methoxy oxanthromicin (53), and (±)-oxanthromicin G (54), along with eight known compounds (55–62). Adxanthromicin A2 and compounds 58 and 61 possessed anti-Bacillus cereus activity (IC50 values were in the range of 1.56–12.50 μg mL−1). In addition, compounds 52 and 58 also showed anti-phytopathogenic activity against Colletotrichum capsici and C. gloeosporioides (the IC50 values were all 6.25 μg mL−1).205,6-Dihydro-1,8-dihydroxy-3-methylbenz[a]anthracene-7,12-quinone (63), previously reported as an intermediate in the synthesis of tetrangulol, has been isolated from Actinomadura sp. DS-MS-114. Deep-sea sediment from Sagami Bay was the habitat of the strain. Compound 63 exhibited antibacterial activity against S. aureus NBRC12732T and its inhibition zone was 12.7 mm at 100 μg.21
Upon the screening of antibacterial endolithic actinobacteria from a granite rock sample, Bhattacharjee et al. separated the rare actinomycete Actinomadura sp. AL2. A novel antibacterial compound CCp1 (64) was characterized from Actinomadura sp. AL2 culture. CCp1 showed antimicrobial activity towards Bacillus subtilis (MIC = 2.00 μg mL−1). From in silico studies relating to its ability to bind DNA gyrase and 30S RNA molecules, researchers found that the compound demonstrated more favorable docking with DNA gyrase, 30S RNA molecules and OmpF porins.22
A new secondary metabolite actinomadurol (65), a rare member of the bacterial C-19 norditerpenoid class, and a known compound JBIR-65 (66) were reported from cultures of Actinomadura strain KC 191, isolated from a soil sample. Actinomadurol displayed significant antibacterial activity against Staphylococcus aureus, Proteus hauseri and Kocuria rhizophila (MICs = 0.39–0.78 μg mL−1), but JBIR-65 did not show antimicrobial activity.23 Sagamilactam (67) is a new polyene macrocyclic lactam, which was separated from Actinomadura sp. K13-0306 isolated from a soil sample collected in Kanagawa Prefecture, Japan. With strong antitrypanosomal activity, its IC50 value was 0.25 ± 0.11 μM.24
From a soil sample, the rare actinobacterium Actinomadura sp. 2EPS was isolated. The strain produced three known compounds as spirotetronate antibiotics, BE-45722A (68), BE-45722B (69) and BE-45722C (70). The three compounds all possessed potent antibacterial activity against Gram-positive bacteria, Staphylococcus aureus ATCC 25923, Bacillus cereus ATCC 14579, and B. subtilis ATCC 6633 (MICs = 0.08–5.0 μg mL−1). Moreover, both 68 and 70 also displayed significant inhibitory activities against Clostridium perfringens S107 (all MICs = 0.63 μg mL−1) and C. difficile 630 (all MICs = 0.08 μg mL−1). This is the first report of spirotetronate antibiotics possessing activity against pathogenic Clostridium spp.25
Actinomadura sp. BCC27169 obtained from soil (from Nam Nao National Park, Phetchabun province) was the source of eight new 3-oxyanthranilic acid derivatives (71–78). Compounds 73 and 78 had antitubercular activity (MIC = 50 μg mL−1). Only compound 73 displayed cytotoxicity against KB cells with an IC50 value of 18.63 μg mL−1.26
A previously undescribed γ-pyrone, miaolienone (79), and a new butanolide, miaolinolide (80), along with 13 known compounds (81–93), were obtained from Actinomadura miaoliensis BCRC 16873, which was isolated from a soil sample collected in Miaoli County, Taiwan. Compounds 79 and 80 could inhibit the production of TNF-α in U937 cells in vitro significantly, and the IC50 values were 0.59 and 0.76 μM, respectively. Compounds 81–85 showed mild inhibitory activities towards LPS-induced TNF-α production.27 The same rare actinomycete also yielded one new γ-lactone, actinomiaolone (94), along with seven known compounds, 3,4-methylenedioxy-5-methoxycinnamyl alcohol, myristic acid, casticin, (+)-glaberide I, (+)-syringaresinol, (+)-yangambin, and trans-cinnamaldehyde.28
From chemical research into the rare actinomycete Actinomadura sp. DSMZ 13491, a bioactive metabolite madurastatin C1 (95) was obtained.29 Fermentation of Actinomadura sp. SBMs009 from tissue sections of the marine sponge Suberites japonicas produced three 3-keto sterols, bendigole D–F (96–98), along with the previously described bendigole B (99). Bendigole D displayed cytotoxicity against the L929 cell line with an IC50 value of 30 μM and it displayed significant inhibitory activity towards GR-translocation; at the same time, compound 98 was the most effective inhibitor of NF-κB nuclear translocation (IC50 = 71 μM).30
2.3 Secondary metabolites from Nonomuraea
Nonomuraea belongs to the Streptococcus family of actinomycetes, and soil and sediment are its main habitats. From 2009–2012, the number of new species belonging to this genus has increased with the addition of 12 novel species,31 implying that Nonomuraea species could produce more new metabolites with significant biological activity (Fig. 3).Two new metabolites, nonomuric acid (100) and 3-hydroxy deoxydaunorubicinol aglycone (101), together with two known compounds, ε-rhodomycinone (102) and 7-deoxy-13-dihydrocarminomycinone (103), were separated from Nonomuraea rhodomycinica NR4-ASC07 from soil from the Sirindhorn peat swamp forest. Compounds 100 and 102 displayed antimalarial activity, and their IC50 values were 8.00 and 8.88 μg mL−1, respectively. Meanwhile, compound 103 displayed inhibitory activity against Mycobacterium tuberculosis and Bacillus cereus with MIC values of 50.0 and 12.50 μg mL−1, respectively.32
Madurahydroxylactone (104), as a new inhibitor of Staphylococcus aureus FtsZ, and its derivatives 105–108 have been obtained from Nonomuraea sp. AN100570 from soil collected near Gongju city, Korea. Madurahydroxylactone showed potent anti-multidrug-resistant bacterial activities against S. aureus FtsZ (IC50 value of 53.4 μM), S. aureus and MRSA (all MIC values: 1 μg mL−1). In cell division phenotype assays, compound 104 could induce cell elongation, suggesting that madurahydroxylactone exerted potent antibacterial activity against FtsZ.33
An investigation into secondary metabolites from the culturing of Nonomuraea species from the soil of a hypogeal ecosystem afforded a new S-bridged pyronaphthoquinone dimer, hypogeamicin A (109), along with its monomeric precursors hypogeamicin B–D (110–112). Compound 109 had significant cytotoxic activity against the colon cancer derived cell line TCT-1 with IC50 = 6.4–12.8 μM, but it did not show antibacterial activity. On the contrary, its monomeric precursors did not showed observable cytotoxicities but possessed antimicrobial activities towards B. subtilis (MIC = 7–28 μg mL−1).34
A novel family of thiopeptides, thiomuracin A–H (113–121), which are decorated with thiazoles and defined by a central six-membered heterocyclic ring system, has been obtained from Nonomuraea strain Bp3714-39. The complicated biosynthetic pathway to this class of antibiotics was revealed through mining the genomes of thiopeptide-producing strains.35
A new trehalose-derived metabolite, brartemicin (122), was found from Nonomuraea strain TP-A0870. The strain was separated from a dried leaf of Artemisia vulgaris L. The compound possessed inhibitory activity towards the invasion of murine colon carcinoma 26-L5 cells (IC50 = 0.39 μM) in a concentration-dependent manner and showed no cytotoxic effects.36
Five new cyclic tetrapeptides, 123–127, were separated by Terui et al. from Nonomuraea sp. TA-0426 from a soil sample collected in Henan province, China. Compounds 124–127 exhibited potent inhibitory activity towards GlyT1. A preliminary structure–activity relationship (SAR) study demonstrated that a cyclic tetrapeptide structure is necessary for inhibitory activity towards GlyT.37
Two new macrolactams, 6-desmethyl-N-methylfluvirucin A1 (128) and N-methylfluvirucin A1 (129), have been obtained from Nonomuraea turkmeniaca MA7364, which was isolated from a soil sample collected in Mexico. 6-Desmethyl-N-methylfluvirucin A1 showed activity against Haemonchus contortus larvae in vitro with an EC90 value of 15 ± 5 μg mL−1.38
2.4 Secondary metabolites from Pseudonocardia
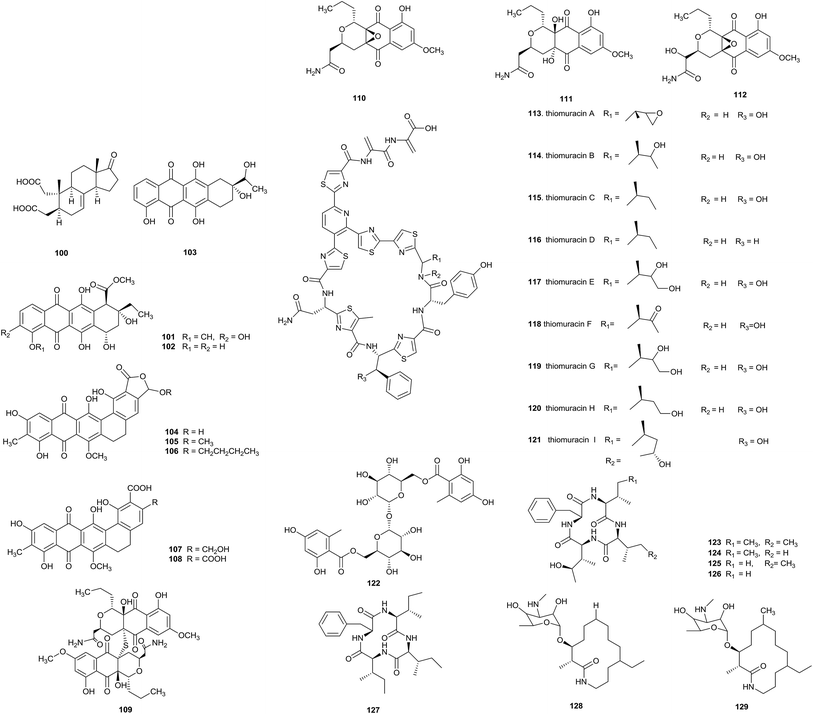
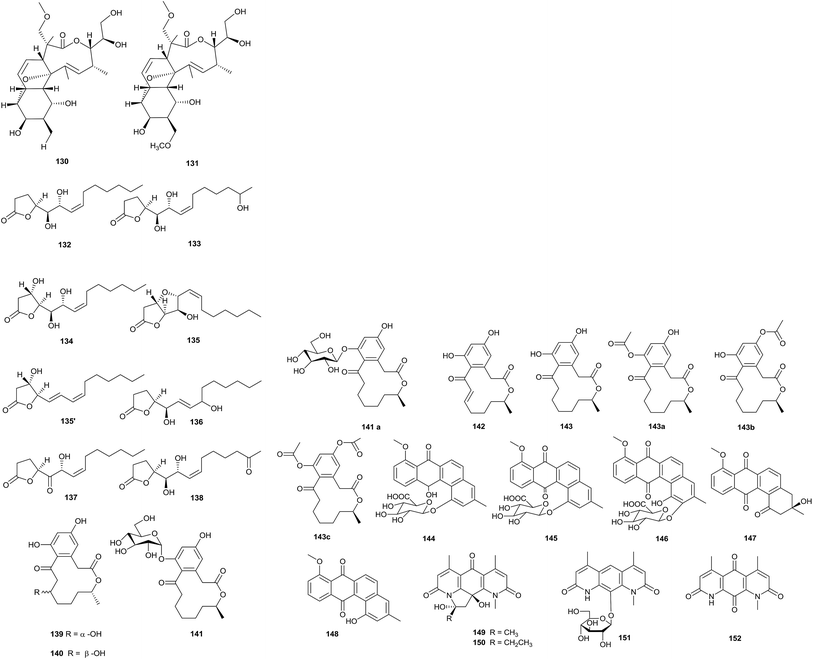
The deep sea is a potential source of new antibiotics, because it is an extreme environment with high pressure, low temperature, darkness, high salinity, and low oxygen concentrations.39 Two new macrolide antibiotics, branimycin B (130) and C (131), were reported from a culture of Pseudonocardia carboxydivorans M-227, which was isolated from a water column at a depth of 3000 m in the Cantabrian Sea. These compounds showed antibacterial activities against Gram-positive bacteria, including Corynebacterium urealyticum (MIC values of 8 and 16 μg mL−1, respectively), Clostridium perfringens (MIC values of 32 and 16 μg mL−1, respectively), and Micrococcus luteus (MIC values of 1 and 16 μg mL−1, respectively), and against the Gram-negative bacterium Neisseria meningitides (MIC values of 32 and 64 μg mL−1, respectively). In addition, branimycin B exhibited mild antibacterial activity against other Gram-negative bacteria, such as Bacteroides fragilis (MIC = 32 μg mL−1), Haemophilus influenza ATCC 492471 (MIC = 32 μg mL−1), and Escherichia coli (MIC = 64 μg mL−1). At the same time, branimycin C displayed antimicrobial activity toward the Gram-positive Enterococcus faecalis 10544 (MIC = 64 μg mL−1) and methicillin-sensitive and methicillin-resistant Staphylococcus aureus (for Staphylococcus aureus ATCC 25923, MIC = 64 μg mL; for Staphylococcus aureus ATCC 6538P, MIC = 32 μg mL−1) (Fig. 4).40Seven new γ-butyrolactones, pseudonocardide A–G (132–138), were isolated from Pseudonocardia sp. YIM M13669. The strain was isolated from a marine sediment sample from the South China Sea (2448 m depth). From paper disc diffusion assays, none of the compounds showed antimicrobial activity against M. smegmatis MC2155 at 30 μg per disc.41
Pseudonocardia sp. HS7, isolated from the cloacal aperture of sea cucumber (Holothuria moebii), produced an unknown macrolide curvularin-7-O-α-D-glucopyranoside (141) with a rare α-D-glucopyranose substituent, and four other curvularin macrolides (139–140, 142–143). Compound 142 showed potential cytotoxicity toward six tested cancer cell lines (C6, U87-MG, SHG-44, U251, HCT-15 and SW620), and the IC50 values were from 0.59 to 3.39 μM.42
vFrom a chemical study of Pseudonocardia sp. EC080529-01, which was isolated from the fungus-growing ant Apterostigma dentigerum, pseudonocardone A–C (144–146) were yielded as new members of the angucycline class of antibiotics, together with the known antibiotics 6-deoxy-8-O-methylrabelomycin (147) and X-14881 E (148). Compounds 147 and 148 had antibacterial activity against Bacillus subtilis 3610 (MIC values of 25 and 3.13 μg mL−1, respectively) and liver-stage Plasmodium berghei (IC50 = 18.5 and 3.0 μM, respectively).43
Three new diazaanthraquinone derivatives, pseudonocardian A–C (149–151), along with the previously synthesized compound deoxynyboquinone (152), were discovered from Pseudonocardia sp. SCSIO 01299. The habitat of this strain was a sediment sample from a depth of 3258 m from the South China Sea. Interestingly, most of the diazaanthraquinone species showed strong antitumor or antibacterial activities, which highlighted their potential for drug development. From bioassays, pseudonocardian A–C displayed significant cytotoxicities towards the three tumor cell lines SF-268, MCF-7 and NCI-H460 (IC50 values between 0.01 and 0.21 μM), and they also displayed antibacterial activities towards Staphylococcus aureus ATCC 29213, Enterococcus faecalis ATCC 29212 and Bacillus thuringiensis SCSIO BT01 (MIC = 1–4 μg mL−1).44
2.5 Secondary metabolites from Micromonospora
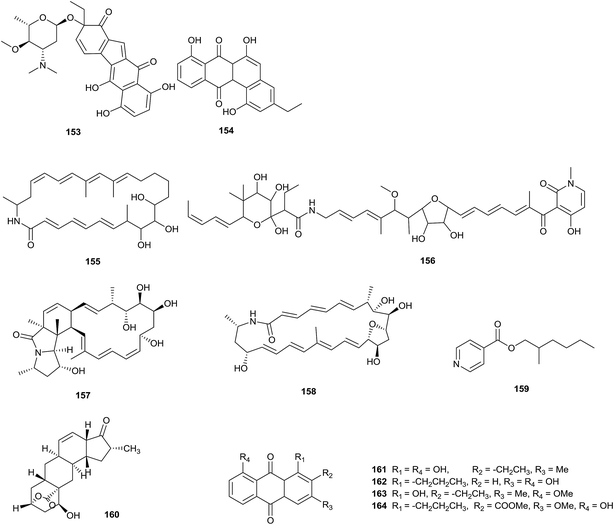
A novel benzo[b]fluorene compound, nenestatin A (153), was isolated from Micromonospora echinospora SCSIO 04089, obtained from the deep sea. Interestingly, the biosynthetic intermediate homo-dehydrorabelomycin E (154) was also afforded via gene disruption experiments in vivo, which verified the biosynthetic gene cluster of nenestatin A (Fig. 5).45FW05328-1 (155) is a new 26-membered polyene macrolactam, which was discovered from Micromonospora sp. FIM05328. The strain was separated from a soil sample collected from the East China Sea. A known compound, aurodox (156), was also identified during the same experiment. Compounds 155 and 156 displayed fantastic antiproliferative activities towards KYSE30, KYSE180 and EC109 human tumor cell lines.46
Two novel macrolactams, dracolactam A (157) and B (158), were obtained from a combined-culture of Micromonospora species and Tsukamurella pulmonis TP-B0596, which is a mycolic-acid containing bacterium (MACB). The combined-culture was proposed by Bode in a one-strain-many-compounds (OSMAC) strategy, which is aimed at increasing the number of secondary metabolites from a single microbial source.47 In this work, the cryptic oxidase genes of Micromonospora species were activated by MACB, which maybe induced downstream polyene macrolactam cyclization.48
An oily bioactive compound, 2-methylheptylisonicotinate (159), was obtained from Micromonospora auratinigra HK-10, isolated from a soil sample from an Indo-Burma biodiversity hotspot in Northeast India. 2-Methylheptylisonicotinate showed antimicrobial activity against Staphylococcus aureus, Bacillus subtilis, Proteus vulgaris, Escherichia coli, Pseudomonas aeruginosa and Mycobacterium abscessus (MIC values of 70, 40, 80, 60, 60, and 50 μg mL−1, respectively).49
The rare actinomycete Micromonospora sp. NPS2077, separated from an unidentified marine sponge (from Uranouchi Bay, Kochi, Japan), was the source of a novel β-hydroxyl-δ-lactone compound, neomacquarimicin (160). The compound did not show significant inhibitory activity against human acidic sphingomyelinase and rat neutral sphingomyelinase at 0.07–700 μM. Meanwhile, it showed no potential activity against Bacillus subtilis ATCC 43223 and Escherichia coli ATCC 10536 at concentrations ranging from 50 to 0.50 μM.50
Two new anthraquinone derivatives, 2-ethyl-1,8-dihydroxy-3-methyl-anthraquinone (161) and 3,8-dihydroxy-1-propylanthraquinone (162), along with two known anthraquinone derivatives (163–164), were produced from Micromonospora rhodorangea isolated from marine soil.51
2.6 Secondary metabolites from Saccharothrix
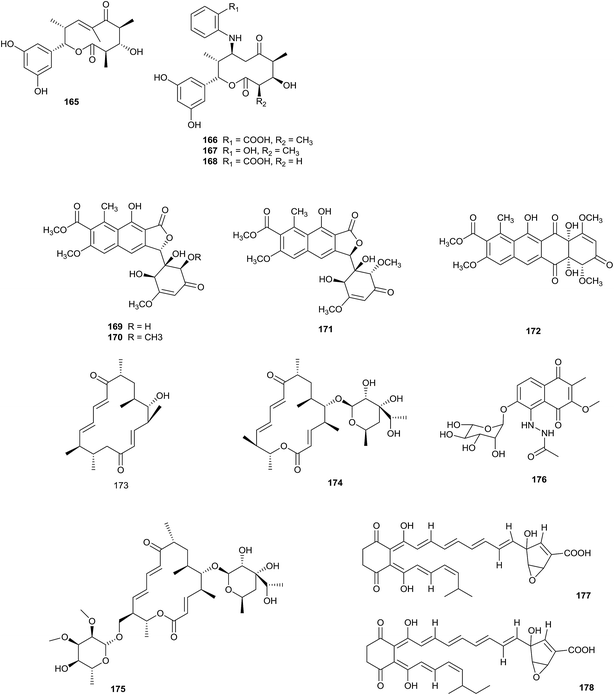
The highly reactive precursor molecule presaccharothriolide X (165) was separated from Saccharothrix sp. A1506. Through Michael addition, saccharothriolide B (166) exhibited unique masking/activating properties towards 2-aminophenol. From a SAR study involving human fibrosarcoma HT1080 cells and fission yeast cells, presaccharothriolide X was proposed to be the real source of the cytotoxicity of compound 166 (Fig. 6).52An investigation into secondary metabolites from a culture of the rare actinomycete Saccharothrix sp. A1506 (from a soil sample) yielded three new 10-membered macrolides, saccharothriolide D–F (166–168). Compounds 167 and 168 displayed cytotoxicity towards human fibrosarcoma HT1080 cells with IC50 values of 29.2 and 66.4 μM, respectively.53
Saccharothrixone A–C, as new aromatic polyketide seco-tetracenomycins (169–171), and a new tetracenomycin analogue, saccharothrixone D (172), have been discovered from Saccharothrix sp. 10–10. The strain was separated from a marine sediment sample. Importantly, this is the first seco-tetracenomycin where the quinone ring B is cleaved and re-formed into a furanone ring. Saccharothrixone D exhibits cytotoxic activity towards the HepG2 cancer cell line (IC50 = 7.5 μM).54
A chemical investigation into the crude extract of Saccharothrix xinjiangensis NRRL B-24321 led to the isolation of two new 16-membered macrolactone-type polyketides, tianchimycin A (173) and B (174), along with a known compound, swalpamycin B (175). From antibacterial screening, compound 174 displayed wonderful selective inhibition towards Enterococcus faecalis ATCC 29212 (MIC value of 6.25 μg mL−1), while tianchimycin A and B displayed weaker antimicrobial activities than swalpamycin B.55
Nakae et al. reported a new naphthoquinone derivative sacchathridine A (176) with an acetylhydrazino moiety, obtained from Saccharothrix sp. MI559-46F5 isolated from a soil sample collected from Higashikagawa, Kagawa Prefecture, Japan. As a prostaglandin release inhibitor, the compound showed inhibitory activity towards prostaglandin E2 release in human synovial sarcoma SW982 cells, without affecting cell growth.56
The new compounds A4 (177) and A5 (178) were reported from a culture of Saccharothrix SA198. The strain was discovered from a Saharan soil sample from Algeria. A4 and A5 showed significant inhibition towards the fungi M. ramannianus (MIC = 5–1 μg mL−1), A. carbonarius (MIC values from 10 to 2 μg mL−1) and P. expansum (all MIC values were 2 μg mL−1).57
2.7 Secondary metabolites from Actinosynnema
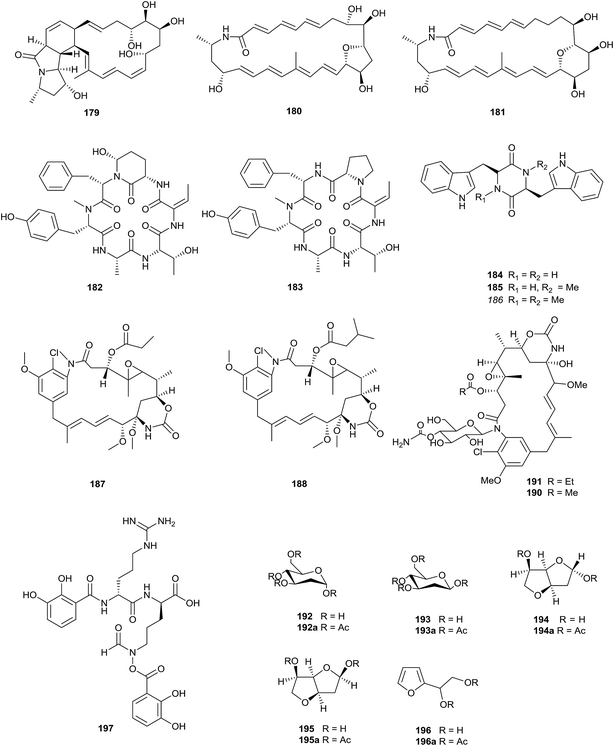
Three novel polycyclic macrolactams, mirilactam C–E (179–181), were obtained from a combined culture of Actinosynnema mirum NBRC 14064 and the mycolic acid-containing bacterium, Tsukamurella pulmonis TP-B0596, which can activate cryptic natural product biosynthesis in co-cultures with actinobacteria. In 2017, Hoshino et al. also found two novel macrolactams, dracolactam A (156) and B (157), under co-culture conditions. None of the compounds displayed antimicrobial activity (towards Candida albicans, Bacillus cereus, and methicillin-sensitive Staphylococcus aureus) or cytotoxicity towards HeLa, MCF-7 and A549 at concentrations of up to 100 μM (Fig. 7).58The chemical analysis of Actinosynnema pretiosum ATCC31565 led to the isolation of two novel cyclic hexapeptides, actinosynneptide A (182) and B (183), along with three tryptophan-containing diketopiperazines (184–186). Importantly, actinosynneptide A was the first cyclic peptide to be reported that contained 3-amino-6-hydroxy-2-piperidone. Actinosynneptide A and B displayed mild cytotoxicities towards HeLa (IC50 = 34.4 and 13.0 μM), PC3 (IC50 = 31.0 and 19.8 μM) and HCT116 (IC50 = 50 and 32.0 μM).59 Two new ansamitocin analogs, maytansinol-9-methyl ether-3-propionate (187) and maytansinol-9-methyl ether-3-3′-methy-butyrate (188), were isolated from Actinosynnema pretiosum FIM06-0063. However their bioactivities are unknown.60
Actinosynnema pretiosum ssp. auranticum ATCC 31565 could produce two new ansamitocins, N-(6-O-carbamoyl-b-D-glucopyranosyl)-N-demethylansamitocin P2 (189) and N-(4-O-carbamoyl-b-D-glucopyranosyl)-N-demethylansamitocin P1 (190), together with the known N-(O-carbamoylglucopyranosyl)ansamitocin (191).61 The same rare actinomycete also yielded five special hexose derivatives, 2-deoxy-α-D-arabino-hexopyranose (192), 2-deoxy-β-D-arabino-hexopyranose (193), 3,6-anhydro-2-deoxy-α-D-arabino-hexofuranose (194), 3,6-anhydro-2-deoxy-β-D-arabino-hexofuranose (195), and 2-(D-glycerol-1,2-dihydroxyethyl)furan (196)62.
Isolation of the rare actinomycete Actinosynnema mirum yielded a siderophore, named mirubactin (197), containing an O-acyl hydroxamic acid ester, which is a rare type of chemical functionality in secondary metabolites. Meanwhile, mirubactin is the first siderophore isolated from the genus Actinosynnema, and a complete bioinformatic overview of the mrb-gene cluster has been demonstrated.63
2.8 Secondary metabolites from Nocardia
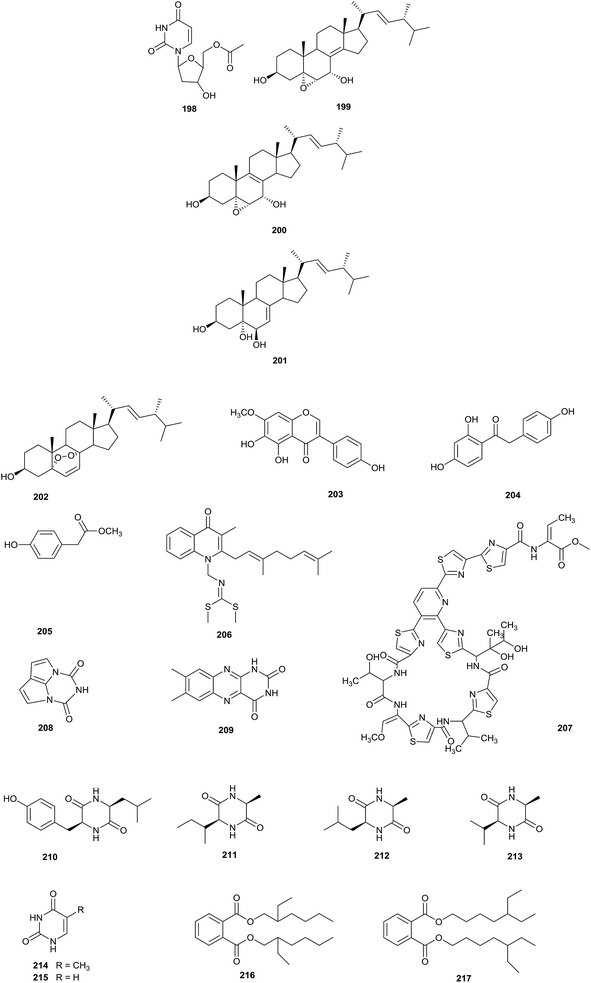
Eight different types of compound (198–205) have been discovered from Nocardia sp. YIM 64630, of which compound 198, 5′-O-acetyl-2′-deoxyuridine, was a new natural nucleotide, seen for the first time. The rare actinomycete was isolated from healthy root tissue of Tripterygium wilfordii, which was collected in Kunming, in Yunnan province, China (Fig. 8).64Chemical research into Nocardia sp. ML96-86F2, which was separated from a soil sample collected in Tokyo, led to the isolation of a new antitumor compound, intervenolin (206). Intervenolin displayed excellent antitumor activity through modulating the interactions between stromal cells and tumor cells, which can inhibit the progression of tumors. Towards HCT-15 cells and CCD-18Co stromal cells, the compound showed significant inhibitory activities at 25 mg kg−1 in female nude mice. In addition, intervenolin displayed potent antimicrobial activities against H. pylori, H. pylori JCM12093 and H. pylori JCM12095 strains, with MIC values ranging from 0.016 to 0.008 μg mL−1.65
Nocardithiocin (207), a novel thiopeptide antibiotic with thiazole, and natural and unnatural amino acid moieties, was isolated from pathogenic Nocardia pseudobrasiliensis IFM 0757 collected from a patient at the Health Center, University of Texas. It is difficult to isolate pathogenic actinomycetes, which makes pathogenic actinomycetes a relatively unknown resource for the discovery of novel bioactive compounds. Compound 207 displayed significant antimicrobial activities towards rifampicin-resistant and -sensitive Mycobacterium tuberculosis strains, most of which were inhibited at concentrations of 0.025–1.56 μg mL−1.66
Chemical research into the rare actinomycete Nocardia alba sp. nov, which was separated from a soil sample collected from Yunnan province, yielded a novel compound nocarsin A (208), along with seven known compounds: lumichrome (209), cyclo(L-Leu-L-Tyr) (210), cyclo(L-Ala-L-Ile) (211), cyclo(L-Ala-L-Leu) (212), cyclo(L-Val-L-Ala) (213), 5-methyluracil (214) and uracil (215).67
The fermentation of Nocardia levis MK-VL_113 obtained from laterite soils from the Guntur region produced two new phthalate esters, bis-(2-ethyl-hexyl)phthalate (216) and bis-(5-ethylheptyl)phthalate (217). Compounds 216 and 217 showed antimicrobial activities against the tested bacteria (Staph. aureus, Bacillus cereus, Bacillus megaterium, Bacillus subtilis, Corynebacterium diphtheriae, Escherichia coli, Proteus vulgaris, and Pseudomonas aeruginosa) with MIC values ranging from 30 to 175 μg mL−1 and 50 to 225 μg mL−1, respectively. Meanwhile, the compounds were found to display inhibitory effects towards Aspergillus flavus, Aspergillus niger, Alternaria alternate, Curvularia maculans, Curvularia lunata, Fusarium oxysporum and Penicillium citrinum, with MIC values varying from 50 to 300 μg mL−1 and 75 to 400 μg mL−1, respectively. Overall, bis-(2-ethylhexyl)phthalate displayed more significant antimicrobial activities than bis-(5-ethylheptyl)phthalate. In addition, this is the first report of bis-(5-ethylheptyl)phthalate and bis-(2-ethylhexyl)phthalate from the genus Nocardia.68
2.9 Secondary metabolites from Actinoplanes
A novel prenylated indole derivative, 3-hydroxy-6-dimethylallylindolin (DMAIN)-2-one (218), and its biosynthetic precursors, 6-dimethylallyltryptophan (DMAT, 219) and 6-DMAI (220), were isolated from Actinoplanes missouriensis NBRC 102363. The complete genome of A. missouriensis was searched to discover the biosynthesis genes of these compounds, and the authors found that the gene cluster was composed of an iptA homolog (AMIS_22580, named iptA-Am) and a putative tryptophanase gene (AMIS_22590, named tnaA-Am) (Fig. 9).69From a bioactivity-guided approach, an antifungal metabolite, 5-hydroxyl-5-methyl-2-hexenoic acid (221), was found from Actinoplanes sp. HBDN08 obtained from a soil sample collected in Harbin, China. Compound 221 displayed significant antifungal activities in vitro towards Botrytis cinerea, Cladosporium cucumerinum and Corynespora cassiicola (IC50 values of 32.45, 27.17, and 30.66 mg L−1, respectively), and could effectively control in vivo diseases caused by B. cinerea, C. cucumerinum and C. cassiicola, with inhibitory rates of 71.42%, 78.63% and 65.13%, respectively, at 350 mg L−1. In summary, 5-hydroxyl-5-methyl-2-hexenoic acid could be a potential candidate for new antifungal drugs.70 The strain also produced a new chlorinated isoflavone, 3′,8-dichlorogenistein (222), along with 8-chlorogenistein (223). 3′,8-Dichlorogenistein and 8-chlorogenistein displayed significant antioxidant activities (IC50 = 5.2 and 7.5 μM, respectively), which were stronger than the positive control genistein (IC50 value: 13.6 μM).71
A new isofuranonaphthoquinone, 7,8-dihydroxy-1-methylnaphtho[2,3-c]furan-4,9-dione (224), was obtained from Actinoplanes sp. ISO06811. The strain was found in a soil sample collected in the state of Colorado using in situ diffusion technology. The compound showed no antimicrobial activities against Bacillus subtilis 1A1 or Escherichia coli K12, but it is able to complex with Fe(III) at circumneutral pH values.72
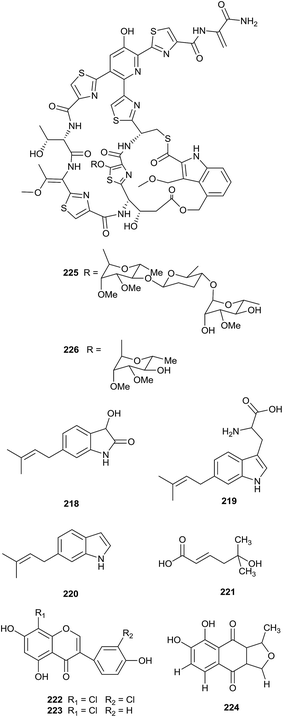
From bioassay-guided isolation, philipimycin (225), a new thiazolyl peptide glycoside, and an acid-catalyzed degradation product (226) were identified from Actinoplanes philippinensis MA7347. All compounds possessed excellent antimicrobial activities against Gram-positive bacteria, including MRSA (MIC = 0.015–1 μg mL−1). Importantly, philipimycin displayed excellent antimicrobial activity in an in vivo mouse model of S. aureus infection (ED50 = 8.4 mg kg−1).73
2.10 Secondary metabolites from Saccharopolyspora
An investigation into Saccharopolyspora cebuensis MCCC 1A09850, which was isolated from deep-sea sediment from the Atlantic Ocean, afforded one new cyclic tetrapeptide, saccharopolytide A (227), and two known polyketides (228, 229), along with six miscellaneous compounds (230–235). Compound 230 displayed a weak anti-allergic effect (IC50 = 55.75 μg mL−1). In addition, compound 228 could weakly inhibit the proliferation of HeLa and H1299 tumor cell lines (Fig. 10).744′′-O-glucosyl tenvermectin A (236) and 4′′-O-glucosyl tenvermectin B (237), two new 16-membered macrocyclic lactone antibiotics, have been obtained from Saccharopolyspora erythraea ATCC 11635 via the microbial conversion of tenvermectin A and B. Compounds 236 and 237 showed weak nematocidal activities (LC50 = 6.7984 mg L−1 and 5.7980 mg L−1, respectively), which were stronger than those of the precursors tenvermectin A and B. At the same time, the compounds showed inhibitory abilities towards T. cinnabarinus adult mites with LC50 values of 0.0156 and 0.0113 mg L−1, respectively.75
Research into Saccharopolyspora sp. BCC 21906 obtained from soil collected in the Evergreen forest, Thailand, yielded three new angucyclinones (238–240), saccharosporone A, B and C, and the known compounds (+)-ochromycinone, (+)-rubiginone B2, tetrangulol methyl ether and fujianmycin A. Saccharosporone A and B showed antimicrobial activities towards Plasmodium falciparum K1. The IC50 values were 4.1 and 3.9 μM, respectively. In addition, compounds 238 and 239 possessed cytotoxicities towards KB (IC50 values of 9.1 and 4.9 μM), MCF-7 (IC50 = 3.4 and 3.6 μM), NCI-H187 (IC50 = 7.7–4.5 μM) and nonmalignant Vero cells (IC50 = 9.1–12.3 μM).76
A new compound, JBIR-66 (241), was discovered by Takagi et al. from Saccharopolyspora sp. SS081219JE-28, which was isolated from an unidentified tunicate collected from the sea shore of Tateyama city, Japan.77 PM070747 (242) is a new benz[a]anthraquinone, which was obtained from Saccharopolyspora taberi PEM-06-F23-019B collected from a marine sponge near the coast of Tanzania. The compound showed cytotoxicity activities against MDA-MB-231 cells, HT-29 cells and A-549 cells (GI50 values of 0.71, 1.42 and 3.28 μM, respectively; TGI values of 1.06, 1.97 and 4.64 μM, respectively; and LC50 values of 1.72, >2.73 and 6.82 μM, respectively).78
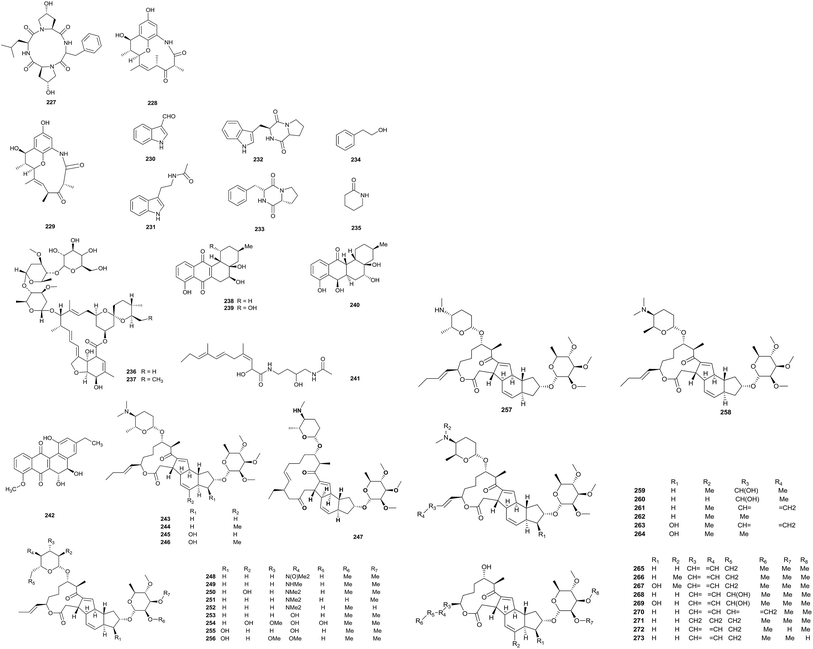
An investigation into the metabolites from a 400 L culture of Saccharopolyspora pogona (NRRL30141) afforded 31 new macrolides (243–273). Most of the metabolites were different from the ‘classical’ spinosyn series because of the presence of a but-1-enyl group at C-21 instead of an ethyl group. In addition to this, there were lots of other new functional groups, such as rare forosamine sugar replacements, hydroxylations and new 14-membered macrolide ring derivatives.79
2.11 Secondary metabolites from Streptosporangium
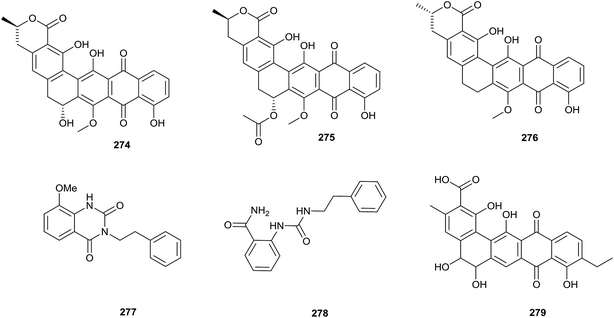
To research the secondary metabolites of Streptosporangium sp. CGMCC 4.7309, which was isolated from sediment from the Huanghai Sea, genome mining was applied. Draft genome sequencing showed that there were more than 20 silent biosynthetic clusters, including a type-II polyketide synthase-containing cluster. Genome mining could allow for the choosing of suitable detection methods and avoid isolating known secondary metabolites through predicting the skeletons of compounds.80 By this method, three pentangular polyphenols, hexaricin A (274), B (275), and C (276), were found from Streptosporangium sp. CGMCC 4.7309 (Fig. 11).81Two new compounds, amethysione (277) and amethysamide (278), along with six known related derivatives, were isolated from Streptosporangium amethystogenes BCC 27081, which was isolated from soil under a banana tree collected from an evergreen forest. Amethysione displayed weak cytotoxicities towards KB and NCI-H187 (IC50 = 16.94 μg mL−1 and 36.99 μg mL−1, respectively).82
A new angucyclinone, compound R2 (279), was discovered from Streptosporangium sp. Sg3 through chemical analysis. Compound R2 displayed excellent antimicrobial activity towards Micrococcus luteus ATCC 9314 and Bacillus subtilis ATCC 6633 with MIC values of 0.5 and 1 μg mL−1, respectively, and mild activity towards Staphylococcus aureus CIP 7625 with an MIC value of 10 μg mL−1. Meanwhile, the compound also exhibited weak activities against Listeria monocytogenes CIP 82110 and Mycobacterium smegmatis ATCC 607 with MIC values of 40 and 50 μg mL−1, respectively.83
2.12 Secondary metabolites from Actinokineospora
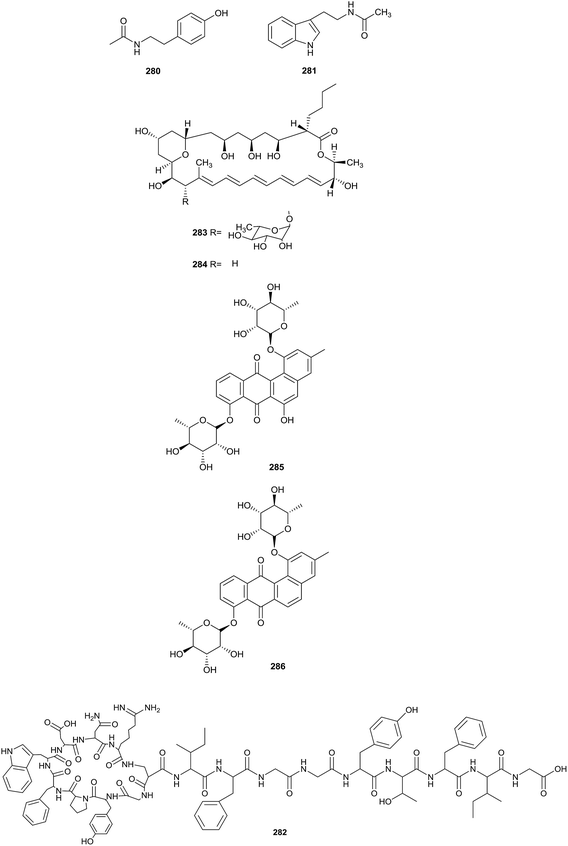
Heidari et al. found two alkaloid compounds, N-acetyltyramine (280) and N-acetyltryptamine (281), which came from Actinokineospora sp. UTMC 968. The strain was collected from a soil sample (Isfahan province, Iran). Compounds 280 and 281 exhibited excellent antiradical activities, as shown by antiradical activity bioassays. The IC50 values showing the DPPH radical scavenging capabilities of N-acetyltyramine and N-acetyltryptamine were 64.7 ± 0.5 and 131.3 ± 1.8 μg mL−1, respectively (Fig. 12).84Actinokineosin (282), a new antibacterial peptide, was reported from a culture broth of Actinokineospora spheciospongiae using genome mining, which is a helpful tool to enable the discovery of more secondary metabolites. Actinokineosin displayed antibacterial activity towards M. luteus with a diameter of 8 mm at a concentration of 50 μg per disk.85
Chemical research into Actinokineospora bangkokensis strain 44EHW led to the isolation of thailandin A (283) and B (284), two new polyene macrolactone antibiotics. Compounds 283 and 284 possessed broad-spectrum antifungal activity against Colletotrichum gloeosporioides DoA d0762, Colletotrichum gloeosporioides DoA c1060 and Colletotrichum capsici DoA c1511, which could lead to anthracnose disease. Additionally, the pathogenic yeasts Candida albicans MT 2013/1, Candida parasilopsis DKMU 434 and Cryptococcus neoformans MT 2013/2 also were inhibited by the compounds (MIC values = 16–32 μg mL−1).86
Applying an OSMAC strategy, trying different fermentation approaches, led to the isolation of two new antioxidant actinosporin analogues, actinosporin C (285) and D (286). Differently from a general culture, a culture of Actinokineospora sp. strain EG49 included calcium alginate. The marine sponge Spheciospongia vagabunda was the habitat of this strain. At a concentration of 1.25 μM, both compounds displayed excellent antioxidant and protective activities towards genomic damage in the HL-60 cell line induced by hydrogen peroxide.87
2.13 Secondary metabolites from Microbispora
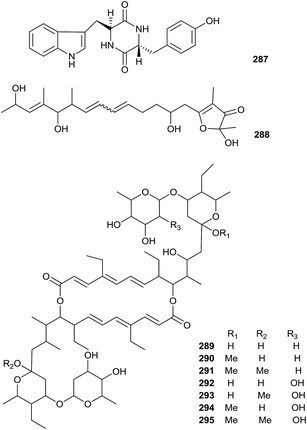
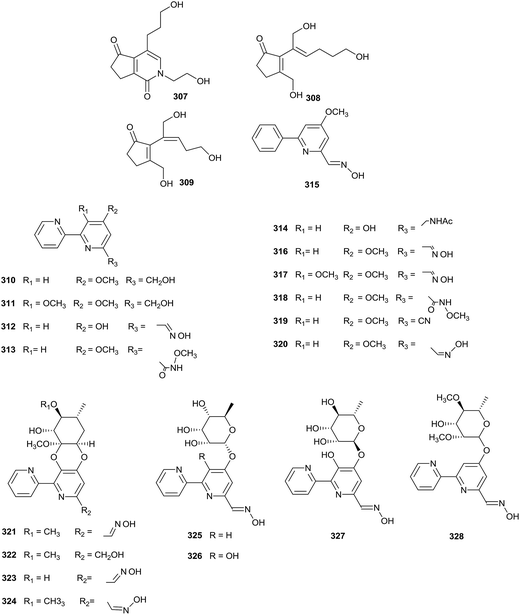
A new natural diketopiperazine, trans-cyclo-(D-tryptophanyl-L-tyrosyl) (287), was obtained from Microbispora aerata strain imbas-11A isolated from penguin excrement collected on Livingston Island in the Antarctic. Compound 287 displayed weak anti-proliferative and cytotoxic effects towards L-929 mouse fibroblast cells, K-562 human leukemia cells (all GI50 values = 50.0 μg mL−1) and HeLa human cervix carcinoma (CC50 = 47.0 μg mL−1). However, trans-cyclo-(D-tryptophanyl-L-tyrosyl) did not show obvious antibacterial activity at a dose of 50 mg per disc (Fig. 13).88Linfuranone A (288) is a new furanone-containing polyketide, which was discovered in a fermentation culture of Microbispora sp. GMKU 363 isolated from the root of a Thai medicinal plant (Clinacanthus siamensis Bremek.). However, the compound did not show significant activity in antimicrobial and cytotoxic research.89
Seven new 20-membered ring macrodiolide antibiotics (289–295), bispolide A1, A2, A3, B1, B2a, B2b and B3, were isolated from Microbispora sp. A34030 collected from a soil sample from a forest station. All compounds showed antimicrobial activities against Gram-positive bacteria, especially methicillin-resistant Staphylococcus aureus (MRSA) at concentrations ranging from 1 to 4 μg mL−1.90
2.14 Secondary metabolites from Lechevalieria
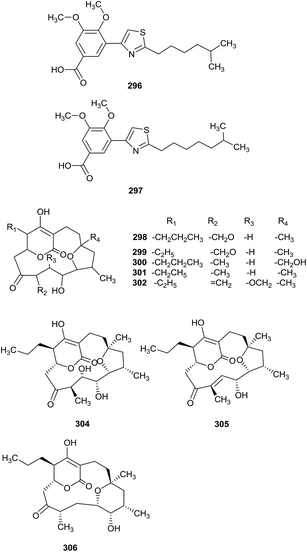
Two new thiazolyl pyridine compounds, pyrizomicin A (296) and B (297), have been obtained from Lechevalieria aerocolonigenes K10-0216, which was separated from mangroves (Iriomote Island). Pyrizomicin A and B displayed antimicrobial activities towards Bacillus subtilis ATCC 6633, Kocuria rhizophila ATCC 9341, Escherichia coli NIHJ, Xanthomonas campestris pv. oryzae KB 88, and Candida albicans ATCC 64548, with a diameter of 6 mm at 10 μg using a paper disk method. In addition, compound 296 also showed inhibition against Staphylococcus aureus ATCC 6538P, K. rhizophila ATCC 9341, X. campestris pv. oryzae KB 88 and E. coli NIHJ (MICs = 32, 32, 32 and 64 μg mL−1, respectively).91 The same rare actinomycetes also produced six rare cyclopentadecane skeletons, mangromicin D, E, F, G, H and I (298–303), with a tetrahydrofuran unit and 5,6-dihydro-4-hydroxy-2-pyrone moiety. From bioassay results, all compounds displayed radical scavenging activities inhibiting DPPH free radicals and NO generation from LPS-stimulated RAW264.7 cells.92 Mangromicin A (304) and B (305), two new cyclopentadecane antibiotics with a tetrahydrofuran unit and 5,6-dihydro-4-hydroxy-2-pyrone moiety, were also discovered from the same strain. The secondary metabolites had excellent inhibitory activities towards Trypanosoma brucei brucei GUT (IC50 = 2.4 μg mL−1). Additionally, they showed more potent cytotoxicities against MRC-5 cells than control drugs (suramin and eflornithine). However, mangromicin B was weaker than mangromicin A in terms of antitrypanosomal and cytotoxic activities.93 Finally, the same research group obtained a new analog, mangromicin C (306), which possessed a tetrahydropyran ring, from the same rare actinomycete Lechevalieria aerocolonigenes K10-0216. Mangromicin C displayed more significant scavenging activity against DPPH free radicals with an IC50 value of 3.8 μM (Fig. 14).942.15 Secondary metabolites from Actinoalloteichus
Chemical research into Actinoalloteichus nanshanensis NEAU 119, isolated from the rhizosphere of a fig tree, afforded three new cyclopentenone derivatives (307–309). Towards A549, K562 and ACHN cell lines, compound 307 displayed cytotoxic properties (IC50 = 14.67, 11.87, and 23.36 μg mL−1, respectively). However other compounds did not display inhibition towards the tested cell lines at 100 μg mL−1 (Fig. 15).95Caerulomycin F–J, five new bipyridine alkaloids (310–314), a new phenylpyridine alkaloid, caerulomycin K (315), and five known analogues (316–320), caerulomycin A, caerulomycin C, caerulomycinamide, caerulomycinonitrile, and (Z)-4-methoxy-2,20-bipyridine-6-carbaldehyde oxime, have been obtained from Actinoalloteichus cyanogriseus WH1-2216-6. The strain came from marine sediment from the seashore in Weihai, China. Compounds 310–319 displayed inhibitory activities towards HL-60, K562, KB, and A549 cell lines (IC50 values range from 0.26 to 15.7 μM). In addition, caerulomycin A and C exhibited antimicrobial activities towards Escherichia coli, Aerobacter aerogenes, Pseudomonas aeruginosa, and Candida albicans (MIC values range from 9.7 to 38.6 μM).96 Further chemical studies of Actinoalloteichus cyanogriseus WH1-2216-6 yielded four new glycosidic bipyridine derivatives, cyanogriside A–D (321–324), containing a rare cyclic glycoside, which was formed from the vicinal hydroxyl groups of an aglycone with both the anomeric center and adjacent carbonyl of a keto sugar. Cyanogriside A displayed potent inhibition against K562, KB, and MCF-7 cells (IC50 = 1.2, 4.7, and 9.8 μM, respectively). At the same time, the IC50 values of cyanogriside C against K562 and KB cells were 0.73 and 4.7 μM, respectively. Additionally, cyanogriside B could reverse the multiple drug resistances of K562/A02, MCF-7/Adr and KB/VCR cells at 10 μM.97 Cyanogriside E–H (325–328), as new acyclic bipyridine glycosides, also have been discovered from Actinoalloteichus cyanogriseus WH1-2216-6 via the inactivation of the O-methyltransferase gene crmM. Cyanogriside E and H showed moderate cytotoxicities towards K562 (IC50 values = 6.0 and 0.8 μM). In addition, cyanogriside F and G could inhibit HCT116 and HL-60 cells with IC50 values of 0.8/3.6, and 3.1/2.0 μM, respectively.98
2.16 Secondary metabolites from other genera
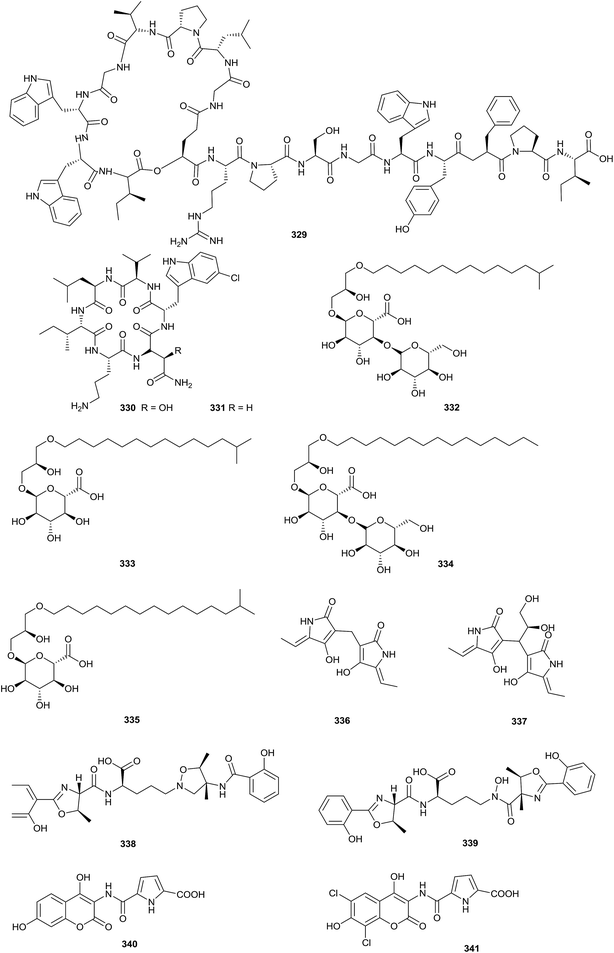
Sphaericin (329), a new lasso peptide, was obtained from Planomonospora sphaerica. At a concentration of 50 μg per disk, sphaericin exhibited potential antimicrobial activity against Micrococcus luteus (Fig. 16).99Nicrophorusamide A and B (330–331), two new chlorinated cyclic hexapeptides, were isolated from the gut of the carrion beetle Nicrophorus concolor. Compound 330 possessed antimicrobial activities towards Staphylococcus aureus ATCC 25923, Enterococcus faecalis ATCC 19433, Enterococcus faecium ATCC 19434, and Salmonella enterica ATCC 14028 (MIC values = 8–16 μg mL−1).100
An investigation of the fermentation broth of Kribbella sp. MI481-42F6, which was isolated from a soil sample collected in Japan, yielded kribelloside A–D (332–335), four new alkyl glyceryl ethers. Kribelloside A–D showed inhibitory activities towards Cet1p (RNA 5′-triphosphatase) from Saccharomyces cerevisiae in vitro (IC50 = 5–8 μM). Beyond that, all compounds possessed antifungal activities towards S. cerevisiae (MIC values = 3.12–100 μg mL−1).101
Two novel tetramic acid dimers (336–337), dietziamide A and B, were obtained from Dietzia timorensis MZ-3 from a soil sample. Dietziamide A and B exhibited mild DPPH radical scavenging activities, with IC50 values of 34 and 37 μM, respectively.102
Catenulobactin A (338) and B (339) are two new heterocyclic peptide isomers obtained from Catenuloplanes sp. RD067331. It's remarkable that a co-culture of Catenuloplanes sp. RD067331 and the mycolic acid-containing bacterium T. pulmonis TP-B0596 could increase the output of compounds. Catenulobactin B could form a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Fe(III) complex, which displayed characteristic 56Fe- and 54Fe-containing positive ion species. In addition, it could inhibit P388 murine leukemia cells (IC50 = 22.4 μM). However, catenulobactin A showed no cytotoxicity, even at 100 μM.103
1 Fe(III) complex, which displayed characteristic 56Fe- and 54Fe-containing positive ion species. In addition, it could inhibit P388 murine leukemia cells (IC50 = 22.4 μM). However, catenulobactin A showed no cytotoxicity, even at 100 μM.103
Using genome mining, a new aminocoumarin gene cluster was identified from Catenulispora acidiphila DSM 44928. Then, two new aminocoumarins, cacibiocin A and B (340–341), were discovered from the strain. Interestingly, heterologous expression with Streptomyces coelicolor M1152 could increase the production of cacibiocin from 4.9 mg L−1 to 60 mg L−1.104
3 Conclusions
As this review mentions, among the 341 compounds reported, most metabolites belong to the classes of cyclic peptides and quinones. Over the period from 2008 to 2018, the rare actinomycetes with the most potential were Amycolatopsis, Actinomadura, Nonomuraea and Micromonospora. The most preferable habitat for rare actinomycetes is soil. In addition, marine habitats, plants and insects are important habitats for unknown rare actinomycetes (Fig. 17).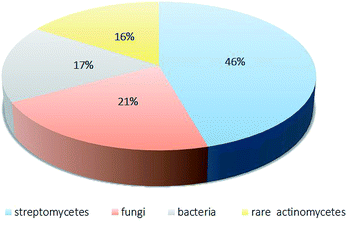 | ||
| Fig. 17 The distribution of the sources of antibiotics from microorganisms.4 | ||
In recent years, rare actinomycetes have produced numerous secondary metabolites with diverse structures and significant bioactivities, such as cytotoxicity and antimicrobial activity. Because of their excellent bioactivities, those secondary metabolites show great commercial prospects. According to Lazzarini,4 there have been more than 8000 antimicrobial products, of which 45.6% came from streptomycetes and 16% came from to rare streptomycetes. More importantly, the proportion of rare actinomycetes has been increasing.4 According to our review, 2′-O-succinyl-apoptolidin A (22), 3′-O-succinyl-apoptolidin A (23), miaolienone (79) and brartemicin (122) showed significant antitumor activities. More importantly, using brartemicin as a lead compound, Jiang et al. synthesized 19 brartemicin analogs and studied their structure–activity relationships as anti-invasive agents. One analog was as potent as brartemicin against the invasion of murine colon 26-L5 carcinoma cells in vitro.105 As potent antibacterial agents, actinomadurol (65) (for Staphylococcus aureus, Proteus hauseri and Kocuria rhizophila; MIC values = 0.39–0.78 μg mL−1)23 and nocardithiocin (207) (towards rifampicin-resistant and -sensitive Mycobacterium tuberculosis strains, most of which were inhibited at concentrations of 0.025–1.56 μg mL−1)66 have shown excellent antimicrobial activities. As for potent antifungal agents, the new compounds A4 (177) and A5 (178) have displayed significant inhibition towards the fungi M. ramannianus, A. carbonarius and P. expansum. Based on their excellent bioactivities and diverse structures, secondary metabolites of rare actinomycetes can draw more attention from the scientific community.
Great progress has been made in the research of secondary metabolites from rare actinomycetes recently. Although many new species and rare actinomycetes have been identified, it is estimated that only 0.1–1% of actinomycete species have been isolated and researched. Therefore, actinomycetes are still a rich resource waiting to be investigated. On one hand, unexplored habitats have become a wonderful option as sources for the isolation of new genera that may produce novel secondary metabolites. On the other hand, lots of novel technologies should be tried to discover more novel compounds with excellent bioactivities. The OSMAC (one-strain-many-compounds) strategy combined with gene scanning will increase the chances of discovering novel compounds. Gene scanning technology could be used to find new biosynthetic gene clusters of microbial secondary metabolites. At the same time, the structures and some physical properties of the compounds can be predicted from the analysis of gene clusters, which is conducive to separation and identification.106 Because of advances in microbiological and biotechnological technology, we do believe that the ability of rare actinomycetes to produce novel secondary metabolites will be better exploited. As a result, more and more antibiotics will be produced using rare actinomycetes.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work was supported by the Key Research and Development Program of China (2017YFC1702002), the NSFC (31870327, 81573318, 81230090, 81520108030, 1302658), Professor of Chang Jiang Scholars Program, the Shanghai Engineering Research Center for the Preparation of Bioactive Natural Products (10DZ2251300), the Scientific Foundation of Shanghai China (16401901300, 17431902800), the National Major Project of China (2011ZX09307-002-03) and the National Key Technology R&D Program of China (2012BAI29B06).References
- Z. H. Liu, Modern biology and biotechnology of actinomycetes, Institute of Microbiology, Chinese Academy of Sciences, 2004, p. 646 Search PubMed.
- S. S. Mulaje, D. V. Chavan and R. Y. Mohalkar, Int. J. Pharm. Sci. Res., 2013, 4, 1730–1742 Search PubMed.
- V. Agarwal and S. K. Nair, Antibiotics for Emerging Pathogens, Springer, New York, 2013, pp. 497–511 Search PubMed.
- A. Lazzarini, L. Cavaletti, G. Toppo and F. Marinelli, Antonie van Leeuwenhoek, 2001, 78, 399–405 CrossRef.
- K. Tiwari and R. K. Gupta, Crit. Rev. Biotechnol., 2012, 32, 108–132 CrossRef CAS.
- H. A. L. And and M. P. Lechevalier, Annu. Rev. Microbiol., 1967, 21, 71–100 CrossRef.
- A. Bauermeister, F. A. Calil, F. C. L. P. Das, T. Medeiros, L. C. Almeida, L. J. Silva, I. M. De, T. D. Zucchi, L. V. Costalotufo and L. Moraes, Nat. Prod. Res., 2019, 33, 1713–1720 CrossRef CAS.
- S. Izuta, S. Kosaka, M. Kawai, R. Miyano, H. Matsuo, A. Matsumoto, K. Nonaka, Y. Takahashi, S. Ōmura and T. Nakashima, J. Antibiot., 2018, 535–537, DOI:10.1038/s41429-018-0028-0.
- Y. S. Xiao, B. Zhang, M. Zhang, Z. K. Guo, X. Z. Deng, J. Shi, W. Li, R. H. Jiao, R. X. Tan and H. M. Ge, Org. Biomol. Chem., 2017, 15, 3909–3916 RSC.
- S. Y. Ma, Y. S. Xiao, B. Zhang, F. L. Shao, Z. K. Guo, J. J. Zhang, R. H. Jiao, Y. Sun, Q. Xu, R. X. Tan and H. M. Ge, Org. Lett., 2017, 19, 6208–6211 CrossRef CAS.
- X. M. Li, X. M. Li and C. H. Lu, J. Asian Nat. Prod. Res., 2017, 19, 946–953 CrossRef CAS.
- H. Hashizume, K. Iijima, K. Yamashita, T. Kimura, S. I. Wada, R. Sawa and M. Igarashi, J. Antibiot., 2018, 71, 129–134 CrossRef CAS.
- C. Beemelmanns, T. R. Ramadhar, K. H. Kim, J. L. Klassen, S. Cao, T. P. Wyche, Y. Hou, M. Poulsen, T. S. Bugni, C. R. Currie and J. Clardy, Org. Lett., 2017, 19, 1000–1003 CrossRef CAS.
- Y. Sheng, S. Fotso, J. D. Serrill, S. Shahab, D. A. Santosa, J. E. Ishmael, P. J. Proteau, T. M. Zabriskie and T. Mahmud, Org. Lett., 2015, 17, 2526–2529 CrossRef CAS.
- C. H. Lu, F. W. Ye and Y. M. Shen, Chin. J. Nat. Med., 2015, 13, 69–72 CrossRef CAS PubMed.
- G. Zhi-Kai, Y. Wei, T. Ren-Xiang and G. Hui-Ming, J. Asian Nat. Prod. Res., 2015, 17, 1109–1113 CrossRef.
- Z. K. Guo, S. B. Liu, R. H. Jiao, T. Wang, R. X. Tan and H. M. Ge, Bioorg. Med. Chem. Lett., 2012, 22, 7490–7493 CrossRef CAS.
- Z. K. Guo, T. Wang, Y. Guo, Y. C. Song, R. X. Tan and H. M. Ge, Planta Med., 2011, 77, 2057–2060 CrossRef CAS.
- V. R. Dasari, M. K. Muthyala, M. Y. Nikku and S. R. Donthireddy, Microbiol. Res., 2012, 167, 346–351 CrossRef CAS.
- N. Bunbamrung, K. Supong, C. Intaraudom, A. Dramae, P. Auncharoen and P. Pittayakhajonwut, Phytochem. Lett., 2018, 25, 109–117 CrossRef CAS.
- A. Kurata, M. Sugiura, K. Kokoda, H. Tsujimoto, T. Numata, C. Kato, K. Nakasone and N. Kishimoto, Biotechnol. Biotechnol. Equip., 2017, 31, 1000–1006 CrossRef CAS.
- K. Bhattacharjee, S. Kumar, N. R. Palepu, P. K. Patra, K. M. Rao and S. R. Joshi, World J. Microbiol. Biotechnol., 2017, 33, 178 CrossRef.
- B. Shin, B. Y. Kim, E. Cho, K. B. Oh, J. Shin, M. Goodfellow and D. C. Oh, J. Nat. Prod., 2016, 79, 1886–1890 CrossRef CAS PubMed.
- T. Kimura, M. Iwatsuki, Y. Asami, A. Ishiyama, R. Hokari, K. Otoguro, A. Matsumoto, N. Sato, K. Shiomi and Y. Takahashi, J. Antibiot., 2016, 69, 818–824 CrossRef CAS.
- E. Jirayut, et al., World J. Microbiol. Biotechnol., 2015, 31, 391–398 CrossRef.
- C. Intaraudom, A. Dramae, S. Supothina, S. Komwijit and P. Pittayakhajonwut, Tetrahedron, 2014, 70, 2711–2716 CrossRef CAS.
- M. Tseng, Y. S. Su, M. J. Cheng, T. W. Liu, I. S. Chen, M. D. Wu, H. S. Chang and G. F. Yuan, Chem. Biodiversity, 2013, 10, 303–312 CrossRef CAS.
- M.-J. Cheng, M. Tseng, G.-F. Yuan and I.-S. Chen, Chem. Nat. Compd., 2013, 49, 226–228 CrossRef CAS.
- E. Mazzei, M. Iorio, S. I. Maffioli, M. Sosio and S. Donadio, J. Antibiot., 2012, 65, 267–269 CrossRef CAS.
- L. Simmons, K. Kaufmann, R. Garcia, G. Schwär, V. Huch and R. Müller, Bioorg. Med. Chem., 2011, 19, 6570–6575 CrossRef CAS.
- R. Sungthong and N. Nakaew, J. Basic Microbiol., 2015, 55, 554–565 CrossRef.
- K. Supong, P. Sripreechasak, W. Phongsopitanun, S. Tanasupawat, K. Danwisetkanjana, N. Bunbamrung and P. Pittayakhajonwut, Nat. Prod. Res., 2019, 33, 2285–2291 CrossRef CAS.
- B. M. Kim, H. Y. Choi, G. W. Kim, C. J. Zheng, Y. H. Kim and W. G. Kim, J. Microbiol. Biotechnol., 2017, 27, 1994–1998 CrossRef CAS.
- D. Derewacz, C. R. Mcnees, G. Scalmani, C. L. Covington, G. Shanmugam, L. J. Marnett, P. L. Polavarapu and B. O. Bachmann, J. Nat. Prod., 2014, 77, 1759–1763 CrossRef CAS.
- R. P. Morris, et al., J. Am. Chem. Soc., 2009, 131(16), 5946–5955 CrossRef CAS.
- Y. Igarashi, T. Mogi, S. Yanase, S. Miyanaga, T. Fujita, H. Sakurai, I. Saiki and A. Ohsaki, J. Nat. Prod., 2009, 72, 980–982 CrossRef CAS.
- Y. Terui, Y. W. Chu, J. Y. Li, T. Ando, T. Fukunaga, T. Aoki and Y. Toda, ChemInform, 2010, 40, 6321–6323 Search PubMed.
- S. Ayers, D. L. Zink, J. S. Powell, C. M. Brown, A. Grund, O. Genilloud, O. Salazar, D. Thompson and S. B. Singh, J. Nat. Prod., 2007, 70, 1371–1373 CrossRef CAS PubMed.
- A. T. Bull, A. C. Ward and M. Goodfellow, Microbiol. Mol. Biol. Rev., 2000, 64, 573–606 CrossRef CAS.
- A. F. Brana, A. Sarmiento-Vizcaino, I. Perez-Victoria, L. Otero, J. Fernandez, J. J. Palacios, J. Martin, M. de la Cruz, C. Diaz, F. Vicente, F. Reyes, L. A. Garcia and G. Blanco, J. Nat. Prod., 2017, 80, 569–573 CrossRef CAS.
- X.-M. Zhang, D.-F. Zhang, W.-J. Li and C.-H. Lu, Helv. Chim. Acta, 2016, 99, 191–196 CrossRef CAS.
- X. Ye, K. Anjum, T. Song, W. Wang, S. Yu, H. Huang, X. Y. Lian and Z. Zhang, Nat. Prod. Res., 2016, 30, 1156–1161 CrossRef CAS.
- C. Gavin, E. R. Derbyshire, C. Eric, C. R. Currie and C. Jon, J. Nat. Prod., 2012, 75, 1806–1809 CrossRef.
- S. Li, X. Tian, S. Niu, W. Zhang, Y. Chen, H. Zhang, X. Yang, W. Zhang, W. Li and S. Zhang, Mar. Drugs, 2011, 9, 1428–1439 CrossRef CAS.
- X. Jiang, Q. Zhang, Y. Zhu, F. Nie, Z. Wu, C. Yang, L. Zhang, X. Tian and C. Zhang, Tetrahedron, 2017, 73, 3585–3590 CrossRef CAS.
- Y. L. Nie, Y. D. Wu, C. X. Wang, R. Lin, Y. Xie, D. S. Fang, H. Jiang and Y. Y. Lian, Nat. Prod. Res., 2017, 32, 2133–2138 CrossRef.
- H. B. Bode, B. Bethe, R. Höfs and A. Zeeck, ChemBioChem, 2002, 3, 619–627 CrossRef CAS.
- S. Hoshino, M. Okada, T. Awakawa, S. Asamizu, H. Onaka and I. Abe, Org. Lett., 2017, 19, 4992–4995 CrossRef CAS.
- M. Talukdar, M. Bordoloi, P. P. Dutta, S. Saikia, B. Kolita, S. Talukdar, S. Nath, A. Yadav, R. Saikia and D. K. Jha, J. Appl. Microbiol., 2016, 121, 973–987 CrossRef CAS.
- S. Sato, F. Iwata, T. Fukae and M. Katayama, J. Antibiot., 2014, 67, 479–482 CrossRef CAS.
- C. M. Xue, L. Tian, W. H. Lin and Z. W. Deng, Nat. Prod. Res., 2009, 23, 533–538 CrossRef CAS.
- S. Lu, S. Nishimura, K. Takenaka, M. Ito, T. Kato and H. Kakeya, Org. Lett., 2018, 20, 4406–4410 CrossRef CAS.
- S. Lu, S. Nishimura, M. Ito, T. Tsuchida and H. Kakeya, J. Nat. Prod., 2016, 79, 1891–1895 CrossRef CAS.
- M. Gan, B. Liu, Y. Tan, Q. Wang, H. Zhou, H. He, Y. Ping, Z. Yang, Y. Wang and C. Xiao, J. Nat. Prod., 2015, 78, 2260–2265 CrossRef CAS.
- X. Wang, J. Tabudravu, M. Jaspars and H. Deng, Tetrahedron, 2013, 69, 6060–6064 CrossRef CAS.
- S. Lu, S. Nishimura, G. Hirai, M. Ito, T. Kawahara, M. Izumikawa, M. Sodeoka, K. Shin-ya, T. Tsuchida and H. Kakeya, Chem. Commun., 2015, 51, 8074–8077 RSC.
- D. Boubetra, N. Sabaou, A. Zitouni, C. Bijani, A. Lebrihi and F. Mathieu, Microbiol. Res., 2013, 168, 223–230 CrossRef CAS.
- S. Hoshino, M. Ozeki, C. P. Wong, H. Zhang, F. Hayashi, T. Awakawa, H. Morita, H. Onaka and I. Abe, Chem. Pharm. Bull., 2018, 66, 660–667 CrossRef CAS.
- C. Lu, F. Xie, C. Shan and Y. Shen, Appl. Microbiol. Biotechnol., 2017, 101, 2273–2279 CrossRef CAS.
- S. Y. Mao, H. Chen, L. Chen, C. X. Wang, W. Jia, X. M. Chen, H. J. Yang, W. Huang and W. Zheng, Nat. Prod. Res., 2013, 27, 1532–1536 CrossRef CAS.
- Y. M. Shen, J. Ma, Y. Zeng and P.-J. Zhao, Helv. Chim. Acta, 2012, 95, 1630–1636 CrossRef.
- C. Lu, L. Bai and Y. Shen, Chem. Nat. Compd., 2008, 44, 594–597 CrossRef CAS.
- T. W. Giessen, K. B. Franke, T. A. Knappe, F. I. Kraas, M. Bosello, X. Xie, U. Linne and M. A. Marahiel, J. Nat. Prod., 2012, 75, 905–914 CrossRef CAS.
- W. Li, X. Yang, Y. Yang, S. Qin, Q. Li, L. Zhao and Z. Ding, Nat. Prod. Res., 2015, 29, 132–136 CrossRef CAS.
- M. Kawada, H. Inoue, S. Ohba, M. Hatano, M. Amemiya, C. Hayashi, I. Usami, H. Abe, T. Watanabe and N. Kinoshita, J. Antibiot., 2013, 66, 543–548 CrossRef CAS.
- A. Mukai, T. Fukai, Y. Hoshino, K. Yazawa, K.-i. Harada and Y. Mikami, ChemInform, 2010, 41, 613–619 Search PubMed.
- Z. G. Ding, J. Y. Zhao, P. W. Yang, M. G. Li, R. Huang, X. L. Cui and M. L. Wen, Magn. Reson. Chem., 2011, 47, 366–370 CrossRef.
- A. Kavitha, P. Prabhakar, M. Vijayalakshmi and Y. Venkateswarlu, Letters in Applied Microbiology, 2010, 49, 484–490 CrossRef.
- R. Satou, M. Izumikawa, Y. Katsuyama, M. Matsui, M. Takagi, K. Shin-Ya and Y. Ohnishi, J. Antibiot., 2014, 67, 231–236 CrossRef CAS.
- Z. Ji, X. J. Wang, Y. J. Yan, J. Ling, J. D. Wang, B. J. Li and W. S. Xiang, Bioresour. Technol., 2010, 101, 8383–8388 CrossRef.
- W. S. Xiang, J. Zhang, J. D. Wang, L. Jiang, B. Jiang, Z. D. Xiang and X. J. Wang, J. Agric. Food Chem., 2010, 58, 1933–1938 CrossRef CAS.
- Q. Zhang, A. J. Peoples, M. T. Rothfeder, W. P. Millett, B. C. Pescatore, L. L. Ling and C. M. Moore, J. Nat. Prod., 2009, 72, 1213–1215 CrossRef CAS PubMed.
- C. Zhang, J. Occi, P. Masurekar, J. F. Barrett, D. L. Zink, S. Smith, R. Onishi, S. Ha, O. Salazar and O. Genilloud, J. Am. Chem. Soc., 2008, 130, 12102–12110 CrossRef CAS PubMed.
- C. L. Xie, S. Niu, J. M. Xia, K. Peng, G. Y. Zhang and X. W. Yang, Nat. Prod. Res., 2017, 32, 1627–1631 CrossRef.
- X. Wan, S. Y. Zhang, H. Zhang, J. Zhai, J. Huang, A. L. Chen and J. D. Wang, J. Antibiot., 2016, 70, 190–192 CrossRef.
- C. Boonlarppradab, C. Suriyachadkun, P. Rachtawee and W. Choowong, J. Antibiot., 2013, 66, 305–309 CrossRef CAS.
- M. Takagi, K. Motohashi, M. Izumikawa, S. T. Khan, J. H. Hwang and K. Shin-Ya, J. Agric. Chem. Soc. Jpn., 2010, 74, 2355–2357 CAS.
- M. Perez, C. Schleissner, P. Rodriguez, P. Zuniga, G. Benedit, F. Sanchez-Sancho and F. de la Calle, J. Antibiot., 2009, 62, 167–169 CrossRef CAS.
- P. Lewer, D. R. Hahn, L. L. Karr, D. O. Duebelbeis, J. R. Gilbert, G. D. Crouse, T. Worden, T. C. Sparks, P. M. Edwards and P. R. Graupner, Bioorg. Med. Chem., 2009, 17, 4185–4196 CrossRef CAS PubMed.
- C. N. Boddy, J. Ind. Microbiol. Biotechnol., 2014, 41, 443–450 CrossRef CAS.
- J. Tian, H. Chen, Z. Y. Guo, N. Liu, J. Li, Y. Huang, W. S. Xiang and Y. H. Chen, Appl. Microbiol. Biotechnol., 2016, 100, 4189–4199 CrossRef CAS PubMed.
- C. Boonlarppradab, C. Suriyachadkun, S. Supothina and P. Laksanacharoen, J. Antibiot., 2016, 69, 459–463 CrossRef CAS PubMed.
- H. Boudjella, A. Zitouni, Y. Coppel, F. Mathieu, M. C. Monje, N. Sabaou and A. Lebrihi, J. Antibiot., 2010, 63, 709–711 CrossRef CAS.
- B. Heidari and F. Mohammadipanah, Mol. Biol. Rep., 2018, 45, 2325–2332 CrossRef CAS.
- N. Takasaka, I. Kaweewan, M. Ohnishi-Kameyama and S. Kodani, Lett. Appl. Microbiol., 2017, 64, 150–157 CrossRef CAS PubMed.
- B. Intra, A. Greule, A. Bechthold, J. Euanorasetr, T. Paululat and W. Panbangred, J. Agric. Food Chem., 2016, 64, 5171–5179 CrossRef CAS.
- T. Grkovic, U. R. Abdelmohsen, E. M. Othman, H. Stopper, R. Edradaebel, U. Hentschel and R. J. Quinn, Bioorg. Med. Chem. Lett., 2014, 24, 5089–5092 CrossRef CAS.
- V. Ivanova, H. Laatsch, M. Kolarova and K. Aleksieva, Nat. Prod. Res., 2013, 27, 164–170 CrossRef CAS PubMed.
- C. Indananda, Y. Igarashi, M. Ikeda, T. Oikawa and A. Thamchaipenet, J. Antibiot., 2013, 66, 675–677 CrossRef CAS PubMed.
- N. Okujo, H. Iinuma and A. Georgea, J. Antibiot., 2007, 60(3), 216–219 CrossRef CAS PubMed.
- T. Kimura, Y. Inahashi, H. Matsuo, T. Suga, M. Iwatsuki, K. Shiomi, Y. Takahashi, S. Ōmura and T. Nakashima, J. Antibiot., 2018, 71, 606–608 CrossRef CAS.
- T. Nakashima, Y. Kamiya, M. Iwatsuki, Y. Takahashi and S. Omura, J. Antibiot., 2014, 67, 533–539 CrossRef CAS PubMed.
- T. Nakashima, M. Iwatsuki, J. Ochiai, Y. Kamiya, K. Nagai, A. Matsumoto, A. Ishiyama, K. Otoguro, K. Shiomi and Y. Takahashi, J. Antibiot., 2014, 67, 253–260 CrossRef CAS PubMed.
- T. Nakashima, Y. Kamiya, M. Iwatsuki, N. Sato, Y. Takahashi and S. Omura, J. Antibiot., 2014, 68, 220–222 CrossRef PubMed.
- X. J. Wang, J. Zhang, P. T. Qian, J. D. Wang, C. X. Liu and W. S. Xiang, J. Asian Nat. Prod. Res., 2014, 16, 587–592 CrossRef CAS PubMed.
- P. Fu, S. X. Wang, K. Hong, X. Li, P. P. Liu, Y. Wang and W. M. Zhu, J. Nat. Prod., 2011, 74, 1751–1756 CrossRef CAS PubMed.
- F. Peng, L. Peipei, L. Xia, W. Yi, W. Shuxia, H. Kui and Z. Weiming, Org. Lett., 2011, 13(22), 5948–5951 CrossRef PubMed.
- F. Peng, Z. Yiguang, M. Xiangui, W. Yi, J. Haijian, Z. Changsheng and Z. Weiming, Org. Lett., 2015, 46, 4264–4267 Search PubMed.
- S. Kodani, Y. Inoue, M. Suzuki, H. Dohra, T. Suzuki, H. Hemmi and M. Ohnishi-Kameyama, Eur. J. Org. Chem., 2017, 2017, 1177–1183 CrossRef CAS.
- Y. H. Shin, S. Bae, J. Sim, J. Hur, S. I. Jo, J. Shin, Y. G. Suh, K. B. Oh and D. C. Oh, J. Nat. Prod., 2017, 80, 2962–2968 CrossRef CAS PubMed.
- M. Igarashi, R. Sawa, M. Yamasaki, C. Hayashi, M. Umekita, M. Hatano, T. Fujiwara, K. Mizumoto and A. Nomoto, J. Antibiot., 2017, 70, 582–589 CrossRef CAS PubMed.
- S. Hoshino, T. Wakimoto, H. Zhang, F. Hayashi, M. Okada and I. Abe, Bioorg. Med. Chem. Lett., 2015, 25, 3953–3955 CrossRef CAS PubMed.
- S. Hoshino, M. Ozeki, T. Awakawa, H. Morita, H. Onaka and I. Abe, J. Nat. Prod., 2018, 81, 2106–2110 CrossRef CAS.
- J. Zettler, H. Y. Xia, N. Burkard, A. Kulik, S. Grond, L. Heide and A. K. Apel, ChemBioChem, 2014, 15, 612–621 CrossRef CAS PubMed.
- Y. L. Jiang, S. Miyanaga, X. Z. Han, L. Q. Tang, Y. Igarashi, I. Saiki and Z. P. Liu, J. Antibiot., 2013, 66, 531–537 CrossRef CAS.
- E. Zazopoulos, K. Huang, A. Staffa, W. Liu, B. O. Bachmann, K. Nonaka, J. Ahlert, J. S. Thorson, B. Shen and C. M. Farnet, Nat. Biotechnol., 2003, 21, 187–190 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |