Open Access Article
Open Access ArticleTetranuclear Cu(II)-chiral complexes: synthesis, characterization and biological activity†
Krisana Peewasan*a,
Marcel P. Merkelb,
Kristof Zarschler c,
Holger Stephan
c,
Holger Stephan c,
Christopher E. Anson
c,
Christopher E. Anson a and
Annie K. Powell
a and
Annie K. Powell *ab
*ab
aInstitute of Inorganic Chemistry, Karlsruhe Institute of Technology, Engesserstrasse 15, 76131 Karlsruhe, Germany. E-mail: Krisana.peewasan@kit.edu; christopher.anson@kit.edu
bInstitute of Nanotechnology, Karlsruhe Institute of Technology, Campus North, Hermann-von-Helmholtz-Platz 1, Eggenstein-Leopoldshafen, 76344, Karlsruhe, Germany. E-mail: Marcel.merkel@kit.edu; annie.powell@kit.edu
cInstitute of Radiopharmaceutical Cancer Research, Helmholtz-ZentrumDresden-Rossendorf, D-01328 Dresden, Germany. E-mail: k.zarschler@hzdr.de; h.stephan@hzdr.de
First published on 2nd August 2019
Abstract
Tetranuclear chiral Cu(II)-Schiff-base complexes S-1 and R-1, were synthesised using enantiomerically pure (S)-(H2vanPheol) and (R)-(H2vanPheol) ligands respectively in the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 of Cu(NO3)2 to (S/R)-(H2vanPheol) in MeOH at room temperature. A pair of polynuclear chiral Cu(II)-cluster complexes were characterized using single-crystal X-ray diffraction, elemental analysis, infrared and CD spectroscopy. The results revealed the importance of these chiral ligands encouraging the arrangement of copper metal in non-centrosymmetric polar packing. The potential of the novel [Cu4(S/R-vanPheol)2(S/R-HvanPheol)2(CH3OH)2](NO3)2 complexes as biologically active compounds was assessed in particular regarding their anti-proliferative and anti-microbial properties.
1 of Cu(NO3)2 to (S/R)-(H2vanPheol) in MeOH at room temperature. A pair of polynuclear chiral Cu(II)-cluster complexes were characterized using single-crystal X-ray diffraction, elemental analysis, infrared and CD spectroscopy. The results revealed the importance of these chiral ligands encouraging the arrangement of copper metal in non-centrosymmetric polar packing. The potential of the novel [Cu4(S/R-vanPheol)2(S/R-HvanPheol)2(CH3OH)2](NO3)2 complexes as biologically active compounds was assessed in particular regarding their anti-proliferative and anti-microbial properties.
Introduction
Applications of metal-based drugs are of great interest, in particular to final alternatives to the Pt-based anticancer drug cisplatin.1–4 The metals can have variable coordination number, geometry, and redox states. To date, cisplatin is one of the most widely used anticancer drugs although it shows significant side-effects such as nausea, bone-marrow production suppression, and nephrotoxicity.1 Aiming to diminish the side effects and resistance caused by cisplatin, the development of new therapeutic metal-based treatments are needed. Requirements for the desired high efficiency therapeutic metal-based drugs are (i) high inhibition of tumour angiogenesis with simultaneous tumour cell death, (ii) diminishing the side effects and resistance caused by cisplatin and (iii) biological activity in living organisms.5 To this end, copper is a good candidate in comparison to other transition metals because it acts as a cofactor during the angiogenesis process6–8 where it enhances the production of cytokinase, endothelial cell proliferation and migration. The chelation of copper ions can also suppress angiogenesis and decrease tumour volume in animals.9–12 Recent studies reveal that the use of novel Schiff-base copper(II)-based complexes can remarkably induce apoptosis, inhibit proliferation, suppress migration, metastasis and angiogenesis and thus inhibit the growth of cervical cancer.13–22 Schiff-base ligand derivatives have been widely used in coordination chemistry due to the advantages of their flexible properties and the easy manipulation of their coordination properties via the variation of functional groups such as amine, alcohol, carboxylate and sulfonate.23–26 Out of the small number of Cu(II) complexes formed with pocket ligands only one of these employs a chiral ligand and none contain Cu(II) coordination clusters.Herein we report the development of an ambient condition synthesis with high yields where enantioselectivity provides an atom efficient route to this system. The new enantiomerically pure ligands were selected since they provide a tetradentate coordination pocket which is ideally suited to accommodate Cu(II)-ions to give the enantiopure complexes, [Cu4(S-vanPheol)2(S-HvanPheol)2(CH3OH)2](NO3)2 (S-1) and [Cu4(R-vanPheol)2(R-HvanPheol)2(CH3OH)2](NO3)2 (R-1) via self-assembly in methanol under aerobic conditions. The single-crystal X-ray diffraction, elemental analysis, infrared and circular dichroism (CD) spectroscopy of Cu(II)-complexes and biological activities have been investigated.
Results and discussion
The synthesis of the Schiff-base ligands S/R-(H2vanPheol) (Scheme 1) was performed by the condensation reaction between 2-hydroxy-3-methoxybenzaldehyde (o-vanillin) and enantiomerically pure S- or R-2-amino-3-phenyl-1-propanol (phenylalaninol) under reflux in absolute ethanol.27 The corresponding chiral Schiff-base ligand S-(H2vanPheol) or R-(H2vanPheol) was reacted with Cu(NO3)2 in methanol. Green block-like crystals of the complex S-1 or R-1 with formula [Cu4(S/R-vanPheol)2(S/R-HvanPheol)2(CH3OH)2](NO3)2 were obtained after 7 days.Single-crystal X-ray analysis shows that S-1 and R-1 are isostructural and crystallize in the polar space group P21 with Z = 2 (Table 1).
| S-1 | R-1 | |
|---|---|---|
| Empirical formula | C71H82Cu4N6O21 | C70.7H80.8Cu4N6O21 |
| Formula weight | 1609.58 | 1604.77 |
| Temperature/K | 180(2) | 180.15 |
| Crystal system | Monoclinic | Monoclinic |
| Space group | P21 | P21 |
| a [Å] | 10.6198(7) | 10.5835(8) |
| b [Å] | 21.4213(14) | 21.529(2) |
| c [Å] | 15.6347(12) | 15.5711(12) |
| α [°] | 90 | 90 |
| β [°] | 94.833(6) | 94.696(6) |
| γ [°] | 90 | 90 |
| Volume [Å3] | 3544.1(4) | 3536.0(5) |
| Z | 2 | 2 |
| ρcalc [g cm−3] | 1.508 | 1.507 |
| μ [mm−1] | 1.263 | 1.266 |
| F(000) | 1668 | 1662 |
| Crystal size [mm3] | 0.34 × 0.18 × 0.16 | 0.58 × 0.23 × 0.18 |
| Radiation | MoKα (λ = 0.71073) | MoKα (λ = 0.71073) |
| 2θ range for data collection [°] | 3.232 to 53.462 | 3.784 to 52.742 |
| Index ranges | −13 ≤ h ≤ 13, −24 ≤ k ≤ 27, −16 ≤ l ≤ 19 | −11 ≤ h ≤ 13, −26 ≤ k ≤ 26, −19 ≤ l ≤ 19 |
| Reflections collected | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 165 165 |
28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 025 025 |
| Independent reflections | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 517 [Rint = 0.0493, Rsigma = 0.0652] 517 [Rint = 0.0493, Rsigma = 0.0652] |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 367 [Rint = 0.0873, Rsigma = 0.0750] 367 [Rint = 0.0873, Rsigma = 0.0750] |
| Data/restraints/parameters | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 517/29/940 517/29/940 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 367/34/935 367/34/935 |
| Goodness-of-fit on F2 | 0.938 | 0.978 |
| Final R indices [I > 2σ(I)] | 0.0380 | 0.0431 |
| Final wR1 indices [all data] | 0.0936 | 0.1200 |
| Largest diff. peak/hole [e Å−3] | 0.53/−0.62 | 0.73/−0.63 |
| Flack parameter | −0.010(7) | −0.003(15) |
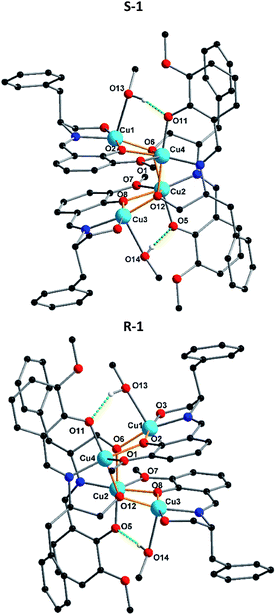
Crystallization in a polar space group with Flack χ parameters of zero highlights the efficient transfer of chirality from the chiral Schiff-base ligand to the coordination compound. Due to the fact that S-1 and R-1 are a pair of enantiomers S-1 is selected as a representative to describe the specific structure here. According to the X-ray crystallographic analyses, S-1 is composed of an independent tetranuclear Cu(II) cluster combining four Schiff-base ligands, which are oriented around the cluster (Fig. 1). In each cluster, two ligands are doubly-deprotonated, with their alkoxo oxygens (O6 or O12) forming μ3-bridges between copper centres, while the other two are singly-deprotonated, with their alkoxy O–H groups (O3 or O9) forming hydrogen bonds to nitrate counterions. In each tetranuclear Cu(II) cluster compound, the core structure can be regarding being based on a Cu4O4 cubane unit, in which the Cu2+ centres are bridged by four deprotonated oxygens from ligands. Although the two alkoxo oxygen (O6 and O12) form μ3-bridges, the two phenoxo oxygens O2 and O8 only form μ2-bridges. This results in a partial opening of the tetracubane core, with the Cu3⋯O2 and Cu1⋯O8 edges now both over 3.2 Å. The edges Cu1⋯Cu4 and Cu2⋯Cu3 are each further supported by a hydrogen bond between the O–H of a methanol ligand and a deprotonated but non-bridging phenoxo oxygen.
The charge balance is provided by two nitrate anions, which form intermolecular hydrogen bonds with propanol groups of the ligand. The Cu–O distances range from 1.904(4) to 2.423(4) Å, whereas the Cu–N distances are between 1.930(5) and 1.948(5) Å. In addition, the O–Cu–O angles lie between 106.9(19)° to 127.1(2)° and the N–Cu–O angles between 82.31(19)° and 172.8(2)° (Table S1†). In R-1, the unidentate ligand coordinating to Cu1 is not pure methanol, but a disordered superposition of methanol (70%) and water (30%). This disordered mixture influences the position of the neighbouring nitrate since through the presence of coordinated H2O, the nitrate is pulled closer to the core structure than in methanol case. The FTIR spectra in the region 4000–400 cm−1 for complexes S-1 and R-1 have been evaluated (Fig. S5†). The comparison of these complexes with the free ligand revealed that the strong and sharp bands in the region 1629–1630 cm−1 can be assigned to the azomethine ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) stretching frequency of the coordinated ligands in S-1 and R-1. The shift of this band towards a lower frequency compared to that of the free ligands (1642–1605 cm−1) indicates the coordination of the imine nitrogen atom to the metal centre. The broad band around 3551–3167 cm−1 is due to the ν(OH) of the alcohol coordinated to the metal centre. The other ν(C–H), ν(C
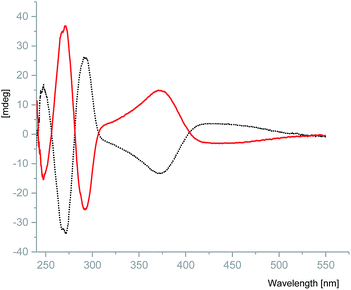
N) stretching frequency of the coordinated ligands in S-1 and R-1. The shift of this band towards a lower frequency compared to that of the free ligands (1642–1605 cm−1) indicates the coordination of the imine nitrogen atom to the metal centre. The broad band around 3551–3167 cm−1 is due to the ν(OH) of the alcohol coordinated to the metal centre. The other ν(C–H), ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) and ν(C–O) vibrations are found in the normal ranges for these types of linkages.26 The maxima in the UV/vis absorption spectra of complexes S-1 and R-1 at 376 nm (π–π*) and 660 nm (n–π*), Cu(II) charge transfer transition, were found to be blue shifted from 419 nm in comparison to the free ligand. (Fig. S6†). No fluorescence emission was observed for either complex possibly as a result of the quenching effect of unoccupied d orbitals of the copper(II) through the nonradiative dissipation process.28 Crystalline samples of S-1 and R-1 were prepared as 50 μM solution in MeOH and their optical properties were evaluated using circular dichroism (CD). S-1 and R-1 display opposite Cotton effects over the measured range to give the expected mirror image spectra. S-1 exhibits an intermediate positive peak at 247 nm and strong negative peak at 271 nm plus a broad negative peak at 371 nm. The mirror image complex R-1, displays absorption peaks of the opposite sign in the same wavelength range (Fig. 2).
C) and ν(C–O) vibrations are found in the normal ranges for these types of linkages.26 The maxima in the UV/vis absorption spectra of complexes S-1 and R-1 at 376 nm (π–π*) and 660 nm (n–π*), Cu(II) charge transfer transition, were found to be blue shifted from 419 nm in comparison to the free ligand. (Fig. S6†). No fluorescence emission was observed for either complex possibly as a result of the quenching effect of unoccupied d orbitals of the copper(II) through the nonradiative dissipation process.28 Crystalline samples of S-1 and R-1 were prepared as 50 μM solution in MeOH and their optical properties were evaluated using circular dichroism (CD). S-1 and R-1 display opposite Cotton effects over the measured range to give the expected mirror image spectra. S-1 exhibits an intermediate positive peak at 247 nm and strong negative peak at 271 nm plus a broad negative peak at 371 nm. The mirror image complex R-1, displays absorption peaks of the opposite sign in the same wavelength range (Fig. 2).
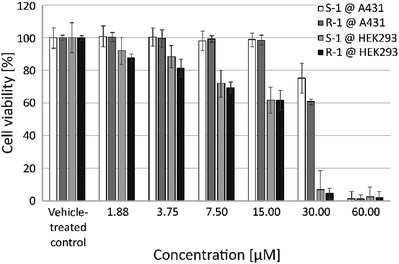
The effect of S-1 and R-1 on the viability of living human cells was investigated using the two cell lines A431 and HEK293 (Fig. 3). These in vitro studies revealed a comparable concentration-dependent decrease in the number of metabolically active cells for both enantiomers. This suggests that the observed effects of the complexes are independent of their chirality. Interestingly, the human embryonic kidney cell line HEK293 seems to be more sensitive to exposure to S-1 and R-1 compared to the more robust epidermoid human cancer cell line A431 (Fig. 3). Consequently, the respective IC50 values for HEK293 are less than half the values of A431 and are in the same range as cisplatin (Table 2).
| IC50 [μM] | ||
|---|---|---|
| A431 | HEK293 | |
| S-1 | 45.28 ± 10.21 | 13.47 ± 1.97 |
| R-1 | 39.16 ± 7.95 | 12.10 ± 1.57 |
| Cisplatin | 7.8 ± 7.95 | 8.1 ± 7.95 |
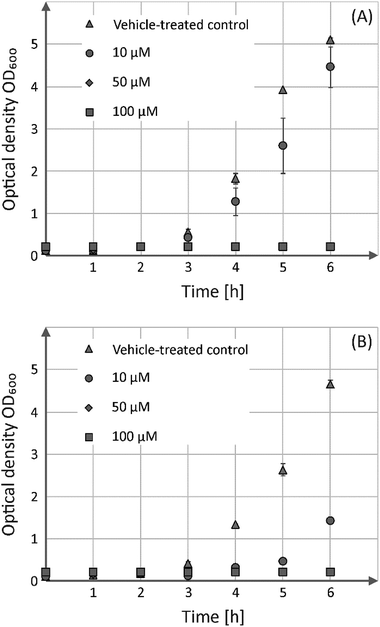
In addition to the viability assessment with regard to human cells, the antibacterial activity of both compounds was evaluated in the batch cultures, Gram-positive B. subtilis (Fig. 4) as well as Gram-negative E. coli (Fig. S7†).
These analyses showed that both enantiomers possess comparable growth inhibitory effects towards the Gram-positive strain, but have no bactericidal activity against its Gram-negative counterpart. Incubation of B. subtilis in the presence of 50 or 100 μM of either S-1 or R-1 prevents their growth entirely, whereas addition of 10 μM of both substances led only to growth delay.
Conclusions
We have developed a high yield synthetic route to chiral Schiff-base pocket ligands. A pair of chiral Schiff-base Cu(II)-clusters have been successfully prepared under aerobic conditions and these are the first examples of Cu(II) coordination clusters captured in chiral Schiff-base pocket ligands. Single-crystal X-ray analysis shows that S-1 and R-1 are isostructural and crystallize in the polar space group P21 with Z = 2. The crystallization in a polar space group highlights the efficient transfer of chirality from the chiral Schiff-base ligand to the coordination compound which exhibit the expected mirror image spectra for the Cotton effect using CD spectroscopy. Both enantiomerically pure Cu(II)-complexes exhibit anti-proliferative properties against human cell lines and anti-microbial activity against Gram-positive bacteria, although the observed effects of the complexes are independent of their chirality. Additionally, these Cu(II) chiral complexes will be further investigated as catalysts for asymmetric reactions. In addition we plan to study complexes formed with other M(II) ions.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We thank the DFG for funding through SFB/TRR 88 “3MET”, Helmholtz POF STN, and Helmholtz Association (VH-VI-422) and Dr Stephan Sinn for the CD measurement. We are furthermore grateful to Utta Herzog for excellent technical assistance.References
- S. Dasari and P. B. Tchounwou, Eur. J. Pharmacol., 2014, 740, 364–378 CrossRef CAS PubMed.
- Y. Ho, S. C. F. Au-Yeung and K. K. W. To, Med. Res. Rev., 2003, 23, 633–655 CrossRef CAS PubMed.
- N. Graf and S. J. Lippard, Adv. Drug Delivery Rev., 2012, 64, 993–1004 CrossRef CAS PubMed.
- K. Yoh, K. Kubota, H. Ohmatsu, K. Goto, S. Niho and Y. Ohe, Anticancer Res., 2012, 32, 4131–4135 CAS.
- N. P. E. Barry and P. J. Sadler, Chem. Commun., 2013, 49, 5106–5131 RSC.
- G. F. Hu, J. Cell. Biochem., 1998, 69, 326–335 CrossRef CAS PubMed.
- C. K. Sen, S. Khanna, M. Venojarvi, P. Trikha, E. C. Ellison, T. K. Hunt and S. Roy, Am. J. Physiol.: Heart Circ. Physiol., 2002, 282, H1821–H1827 CrossRef CAS PubMed.
- C. J. M. D. Griffith and J. P. Parker, Anti-Cancer Agents Med. Chem., 2010, 10, 354–370 CrossRef.
- C. Cox, T. N. Teknos, M. Barrios, G. J. Brewer, R. D. Dick and S. D. Merajver, Laryngoscope, 2001, 111, 696–701 CrossRef CAS PubMed.
- J. Yoshii, H. Yoshiji, S. Kuriyama, Y. Ikenaka, R. Noguchi, H. Okuda, H. Tsujinoue, T. Nakatani, H. Kishida, D. Nakae, D. E. Gomez, M. S. De Lorenzo, A. M. Tejera and H. Fukui, Int. J. Cancer, 2001, 94, 768–773 CrossRef CAS PubMed.
- Q. Pan, C. G. Kleer, K. L. van Golen, J. Irani, K. M. Bottema, C. Bias, M. de Carvalho, E. A. Mesri, D. M. Robins, R. D. Dick, G. J. Brewer and S. D. Merajver, Cancer Res., 2002, 62, 4854–4859 CAS.
- L. Finney, S. Vogt, T. Fukai and D. Glesne, Clin. Exp. Pharmacol. Physiol., 2009, 36, 88–94 CrossRef CAS PubMed.
- X. Y. Qin, L. C. Yang, F. L. Le, Q. Q. Yu, D. D. Sun, Y. N. Liu and J. Liu, Dalton Trans., 2013, 42, 14681–14684 RSC.
- A. E. Stacy, D. Palanimuthu, P. V. Bernhardt, D. S. Kalinowski, P. J. Jansson and D. R. Richardson, J. Med. Chem., 2016, 59, 4965–4984 CrossRef CAS PubMed.
- X.-Y. Qin, Y.-N. Wang, X.-P. Yang, J.-J. Liang, J.-L. Liu and Z.-H. Luo, Dalton Trans., 2017, 46, 16446–16454 RSC.
- S. Zehra, T. Roisnel and F. Arjmand, ACS Omega, 2019, 4, 7691–7705 CrossRef CAS.
- P. Daneshmand, J. L. Jime, M. Aragon--alberti and F. Schaper, Organometallics, 2018, 37, 1751–1759 CrossRef CAS.
- C. M. Rajesh and M. Ray, Dalton Trans., 2014, 43, 12952–12960 RSC.
- J. Pejic, D. Vušak, G. Szalontai, B. Prugovečki, D. Mrvoš-Sermek, D. Matković-Čalogović and J. Sabolovic, Cryst. Growth Des., 2018, 18, 5138–5154 CrossRef CAS.
- X. Zhou, Q. Sun, L. Jiang, S. Li, W. Gu, J. Tian, X. Liu and S. Yan, Dalton Trans., 2015, 44, 9516–9527 RSC.
- M. Shibata, K. Nakajima and Y. Nishibayashi, Chem. Commun., 2014, 50, 7874–7877 RSC.
- I. Castillo, V. Pérez, I. Monsalvo, P. Demare and I. Regla, Inorg. Chem. Commun., 2013, 38, 1–4 CrossRef CAS.
- M. J. MacLachlan, M. K. Park and L. K. Thompson, Inorg. Chem., 1996, 35, 5492–5499 CrossRef CAS PubMed.
- M. Andruh, Dalton Trans., 2015, 44, 16633–16653 RSC.
- H. Yu, M. Liu, X. Gao and Z. Liu, Polyhedron, 2017, 137, 217–221 CrossRef CAS.
- S. Karmakar and S. Khanra, CrystEngComm, 2014, 16, 2371 RSC.
- P. Adão, M. L. Kuznetsov, S. Barroso, A. M. Martins, F. Avecilla and J. C. Pessoa, Inorg. Chem., 2012, 51, 11430–11449 CrossRef PubMed.
- M. Hazra, T. Dolai, A. Pandey, S. K. Dey and A. Patra, J. Saudi Chem. Soc., 2017, 21, S240–S247 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section, X-ray structure determinations, elemental analysis (CHN), NMR spectra, IR spectra, UV/vis spectra, cell culture and CIFs of complexes S-1 and R-1. CCDC 1912163 and 1912164. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ra03586a |
| This journal is © The Royal Society of Chemistry 2019 |