Open Access Article
Open Access ArticleA simple, robust and equipment-free DNA amplification readout in less than 30 seconds†
Michael Glenn Mason * and
José Ramón Botella*
* and
José Ramón Botella*
Plant Genetic Engineering Laboratory, School of Agriculture and Food Sciences, The University of Queensland, St. Lucia, QLD 4072, Australia. E-mail: j.botella@uq.edu.au; Michael.Mason@uq.edu.au; Fax: +61 7 33651699; Tel: +61 7 33651128 Tel: +61 7 33459172
First published on 7th August 2019
Abstract
Molecular based diagnostic methods rely on the amplification of pathogen DNA but naked eye visualization of results is still challenging. We present here a simple and highly reliable DNA amplification readout system for naked eye detection of isothermally or PCR amplified DNA in less than 30 seconds. This system utilizes spermine to precipitate DNA amplicons and initiate bridging flocculation of a mix of charcoal and diatomaceous earth particles in suspension. In the absence of amplification, the charcoal particles remain suspended resulting in a black, non-transparent colloid solution while positive samples in which DNA amplification has occurred can be identified within seconds as the particles flocculate and settle leaving a transparent liquid phase. We have coupled this method with our rapid dipstick DNA purification method and isothermal DNA amplification to create a simple four-step diagnostic system that can be preassembled to reduce unnecessary manipulation in the field. The method's simplicity, low cost, minimal equipment and clear presence/absence readout makes it ideal for rapid diagnostic testing in the laboratory and in situations where users have limited technical training or resources including high school science classes and field-based research.
Introduction
The ability to rapidly screen for diseases in humans or agricultural products is a critical first step in preventing their spread both locally and to other parts of the world. Therefore, the development of user-friendly diagnostics for health carers, farmers, quarantine and protection personnel at the point-of-need (PON) is critically important. To be most effective, diagnostic testing should meet the ASSURED criteria set out by the World Health Organization being: Affordable, Sensitive, Specific, User-friendly, Rapid and robust, Equipment-free and Deliverable to those who need it.1 Diagnostics meeting these criteria will encourage wide-spread adoption by enabling the tests to be performed outside the laboratory environment by individuals that may have limited scientific training.The need for expensive laboratory thermocyclers for molecular-based diagnostics has now been eliminated with the development of isothermal DNA amplification methods such as Loop-Mediated Isothermal Amplification (LAMP), Recombinase Polymerase Amplification (RPA), and Helicase-Dependent Amplification (HDA).2–4 These methods require a single, constant temperature for DNA amplification and thus can be performed in something as simple as a bucket filled with hot water. Recently, a simple, rapid and instrument-free method of nucleic acid extraction was developed,5 which when combined with isothermal DNA amplification simplifies the process to the point where field-based DNA amplification becomes practical. However, visualization of the results in the field is still a problem. In the laboratory, the most common method to analyze amplification products is by agarose gel electrophoresis in the presence of dyes such as ethidium bromide followed by visualization under UV light. Alternate approaches such as real-time PCR or electrochemical detection6 are used however, for field applications and high schools where resources are limited, simpler, cheaper and ideally instrument-free approaches are required.
A number of simplified methods to visualize the success of amplification reactions have been developed, all of which have their own advantages and disadvantages. For example, researchers have measured turbidity changes to monitor the progression of the DNA amplification.7,8 The turbidity is due to an amplification by-product, magnesium-pyrophosphate, which forms an insoluble white precipitate. Although turbidity can be observed with the naked eye by a trained operator, it is unreliable especially in field situations as only strong amplifications will produce obvious turbidity and the apparent level of turbidity is subject to light conditions.9
Numerous color based methods have been developed for equipment-free visualization of DNA amplification; in one approach, gold nanoparticles (AuNP) with covalently attached probes can added to the completed reaction to produce a color change in the presence of the specific amplicon.10–15 In a different approach, the DNA intercalating fluorescent dye SYBR Green I is added to the reaction turning the solution orange in the absence of DNA amplification and yellow/green in the presence of amplified DNA.16–18 However, to achieve a strong difference between the positive and negative DNA amplifications, a high concentration of SYBR needs to be added after the amplification is finished as it would otherwise inhibit the reaction.9 Both SYBR Green and AuNP approaches typically require opening the reaction tubes post-amplification, which creates a risk of cross-contamination however, although a number of approaches have been suggested to address this issue.16,18–20
The metal ion-sensitive indicator hydroxy naphthol blue (HNB) indirectly monitors DNA amplification by changing color in response to the levels of free magnesium in solution.21–26 Unlike SYBR Green I, HNB can be added to the reaction prior to amplification, resulting in a purple solution due to the abundance of free magnesium; as DNA amplification proceeds, free magnesium ions are consumed and the reaction turns sky blue. Despite HNB's advantages over SYBR Green I, naked eye interpretation of results is difficult as the color shift is relatively subtle especially when the amplification reaction does not consume the majority of the free magnesium ions.21,27,28 pH changes due to the production of hydrogen ions during DNA amplification have been exploited by adding pH-color sensitive dyes.28 This method can only be achieved in weakly-buffered or un-buffered solutions, which is likely to lead to inconsistencies in field samples due to the presence of unexpected contaminants.
Recently, three independent research groups developed simple naked-eye DNA detection methods based on the aggregation of paramagnetic particles in the presence of DNA.29–31 In one method, DNA could be quantified by the degree of particle aggregation that occurred after the DNA and paramagnetic particles were combined in a rotating magnetic field.29 The other two methods were relatively similar with the amplified DNA being initially bound to paramagnetic particles, pulled down and compacted by a magnet, washed (and, in the case of Wee et al., resuspended in a low pH buffer), and then gently resuspended. In the absence of DNA, the particles would re-disperse easily, whereas in the presence of DNA the particles would remain aggregated. A significant advantage of this technique is that it provides a binary yes/no readout compared to the spectrum of hues of the colorimetric assays. However, despite this, the published methods are not ideally suited for field-based research due to the large amount of sample manipulation required and the risk misinterpretation of result due to differences in the user's technique. In this study, we sought to take advantage of the flocculation reaction to create a new method that is suited for field-based research. By changing the type of particles and the nature of the chemical flocculent we have created a field-capable, robust, easy to interpret, yes/no readout that can be visualized within 30 seconds post-amplification without any pipetting or wash steps and is unaffected by differences in user technique.
Materials and methods
Safety and hazards
The chemicals and materials used in this study do not pose a significant threat to safety if handled as outlined in their respective safety data sheets.Plants and pathogens
Plant materials used in this study included Arabidopsis thaliana ecotype Columbia infected with Pseudomonas syringae pv. tomato strain DC3000, and tomato (Solanum lycopersicum cv. Moneymaker) leaf tissue infected with Cucumber mosaic virus.Nucleic acid amplification
Nucleic acid amplification was performed by either polymerase chain reaction (PCR), or loop mediated isothermal amplification (LAMP). For PCR amplification, 50 μl reactions were performed using 25 μl of GoTaq Green Master Mix (Promega), 50 pmol of both forward and reverse primers and 10 ng purified Arabidopsis genomic DNA. PCR cycling parameters were as follows: 95 °C for two minutes, 41 cycles of 95 °C for 15 seconds, 50 °C for 15 seconds, 72 °C for 75 seconds, followed by final extension of 72 °C for one minute. Arabidopsis G-protein gamma subunit 2 (AGG2) forward primer used: TGTATCCAACCAGTAACAAATGG, AGG2 reverse primer sequence: CAATTACATCAAATTCACTGCCG.LAMP reactions were performed by in a solution containing 20 mM Tris (pH 8.8), 10 mM (NH4)2SO4, 50 mM KCl, 0.1% (v/v) Tween-20, 0.8 M betaine, 8 mM MgSO4, 1.2 mM dNTPs, 0.32 U μl−1 Bst 2.0 WarmStart (NEB Biolabs, USA), 0.8 μM of FIP and BIP primers and 0.2 μM of F3 and B3 primers. Reactions were incubated at 63 °C for 50 minutes followed by a five minute incubation at 80 °C to denature the enzyme. For colorimetric assays, hydroxy naphthol blue (HNB) was added to the reaction mix post-amplification to a final concentration of 120 μM. Primers used to detect Cucumber mosaic virus: F3: GGATACATGAGTGTCCCTCAAGTG, B3: ACAACAGCAAAACACCGCTT, FIP: CTTGTCGCCTAGATCAGCTAAGTATCGAACAGTTTCTACCGATGCTGAAGG, BIP: AGCAGTGCGTCACATTACATAACCTGTCTCCATGGGACAATCATACG. Primers used to detect Pseudomonas syringae: F3: AAAGCCGCATATCCCCCA, B3: TCAGATACCGTCTCCTCACAC, FIP: GGCATTTAACTCAAAAGCCGGCCGGCCACCCAATAGCAATGTAACCAAT, BIP: CGGCATCCCCGACAAAAACGGGAGGAGGCAAAATGTCTGTG.
Amplicon purification
100 μl of LAMP or PCR amplified amplicons were pooled together and purified using Agencourt AMPure XP PCR Purification kit (Beckman Coulter) following the manufacturer's recommendations. Briefly, 100 μl amplification reaction was mixed with 180 μl of paramagnetic particles. The mixture was incubated at room temperature for five minutes and then placed against a magnet for five minutes to pull down DNA bound paramagnetic particles. After supernatant was removed, paramagnetic particles were washed twice with 75% (v/v) ethanol and then air dried for five minutes. The bound DNA was eluted by resuspending the particles in TE buffer (10 mM Tris (pH 8), 1 mM ETDA) and incubating at room temperature for one minute before pulling the particles down to the bottom of the tube by magnet. The purified amplicons were quantified on a spectrophotometer.Particle size analysis
Charcoal and diatomaceous earth particles were viewed on a Zeiss Axio light microscope using a 20× objective. The images were analysed in ImageJ using the Particle Size Analyser (PSA) version 12 macro (https://code.google.com/archive/p/psa-macro) with the following settings: rolling ball background removal, automatic thresholding mode, particle diameter range 1–999 μm, circularity between 0.1 and 1. A total of six or eight images of activated charcoal and diatomaceous earth images, respectively were analysed and the results averaged.Flocculation assay using paramagnetic beads
The low pH flocculation solution was made by placing a tube with 100 μl paramagnetic beads from the Agencourt AMPure XP PCR Purification kit (Beckman Coulter) against a magnet so that the supernatant could be discarded and replaced with 300 μl of 125 mM glycine (pH 3.6). Similarly, spermine based flocculation solution was made by replacing the supernatant of 30 μl of paramagnetic particles with 100 μl of 50 mM Tris pH 8) containing either 5 or 10 mM spermine. For each assay with the paramagnetic beads, 20 μl of flocculation solution was combined with 10 μl LAMP amplification reaction or dilution of crude bacterial extract and mixed by flicking the tube. The magnetic beads were then placed against a magnet to pellet the paramagnetic beads. The tubes were then tapped by hand and the tubes held vertically to observe the results.Flocculation using colored silica particles
20 μl of flocculation solution containing 50 mM Tris (pH 8), 10 mM spermine and 25 μg μl−1 of either Mason Stains (MS): Deep Sea (6244) or Yellow (6433) (Mason color, Ohio, USA) or Northcote pottery supplies (NPS) Dark red onglaze (11M10) (Northcote pottery supplies, Victoria, Australia) was added to 5 μl of 50 ng μl−1 of salmon sperm DNA or water, mixed briefly and held vertically to view the results.Flocculation solution manufacture and readout assay
The final, optimised flocculation solution is made from 100–400 mesh activated charcoal (Sigma, St. Louis, USA) and powdered diatomaceous earth (Absorbacide, Mt Sylvia Diatomite, ACT, Australia) that had been ground separately in a coffee grinder for 45 seconds to break up any large particles. 400 mg of activated charcoal and 600 mg of diatomaceous earth were combined in a 50 ml solution containing 50 mM Tris (pH 8), 10 mM spermine and 1% (v/v) PEG 8000. The flocculation solution can either be stored at 4 °C or −20 °C for at least a year without loss of activity.During the optimisation of the flocculation solution, the amount of activated charcoal was kept constant and the other reagents were altered as described in the text. Unless otherwise stated, all flocculation assays during solution development and testing were performed by first agitating the flocculation solution to suspend all the particles. 30 μl of flocculation solution was then added to 10 μl of an amplification reaction in a 0.2 ml tube. The tube was gently flicked 20 times to mix the solution and allow the DNA and particles to flocculate together in large clumps. The tubes were then held vertically and the results visualised by naked eye for several minutes. Positive reactions occur in the presence of DNA and is indicated by the particles clumping together and settle to the bottom of the tube. In reactions with little or no DNA, the solution remains black and largely non-transparent for more than a minute and was then considered negative.
Nucleic acid purification dipstick manufacture
Dipsticks were created as described previously.5 Briefly, Whatman No. 1 filter paper was partially dipped into molten wax (Paraplast Plus, Fluka). After the wax had set, filter paper was cut into strips that were approximately 2 mm wide and 42 mm long so that each dipstick contained a 2 × 4 mm area of uncoated filter paper (nucleic acid binding area) and a 2 × 40 mm wax coated handle.3D-printed tube lid insert manufacture
Tube lid inserts were designed using 3D computer aided design (CAD) software which was then fabricated using a 3D printer (Fabrikator Mini (V1.5), http://HobbyKing.com) with Acrylonitrile–Butadiene–Styrene (ABS) filament at 0.1 mm print resolution and 10% infill. After printing, 50 μl of acetone was added to the reaction well of the insert and then immediately removed to fuse the layers of ABS together to create a smooth water impenetrable surface. The inserts were then placed under vacuum for 20 minutes to remove residual traces of acetone.Single tube LAMP DNA amplification and readout system
For each assay, a reaction tube was first setup in which 225 μl flocculation solution was added to the bottom of a 1.5 ml tube and a 3D printed cap insert was placed into the lid. 25 μl of LAMP reaction mix was added into the reaction well of the lid insert. Subsequently, leaf tissue (approximately 200 mm2) from healthy or infected plants was added to a 2 ml screw capped tube containing 500 μl cell lysis buffer (20 mM Tris, 25 mM NaCl, 2.5 mM EDTA, 0.05% (v/v) SDS) and two ball bearings. The plant tissue was macerated by shaking tube for approximately eight seconds. The dipstick was dipped (3×) into extract to bind nucleic acids then dipped (3×) into 500 μl of wash buffer (10 mM Tris (pH 8)) and then finally the bound nucleic acids were eluted by dipping (15×) directly into 25 μl amplification reaction mix located inside the well of the 3D-printed lid insert. After elution, the dipstick was discarded, and reaction tube closed and then transferred to a water bath set at 63 °C. After 50 minutes, 5 μl of the reaction was removed to run on an agarose gel and then the tubes immediately closed and inverted 20 times to allow the flocculation solution to mix with the amplification reaction. The tubes were then held vertically to view the results within 30 seconds. A positive reaction in which amplification has occurred is indicated by the particles clumping together and settle to the bottom of the tube. If the solution remains black and largely non-transparent for more than a minute it is considered negative.Results
Simplifying the flocculation readout
A distinct advantage of the previously developed bridging flocculation methods for DNA amplification detection29–31 is their ability to be interpreted as a simple binary readout, that is, it is easy to determine whether significant amplification has or has not occurred. The method developed by Wee et al., in which the flocculation reaction is performed in tubes appears to provide the clearest readout, however, the requirement for approximately 10 minutes of post-amplification processing and significant care not flick the tube with too much force is not practical for field-based point-of-need applications. We hypothesized that the need for multiple buffers and washing steps could be avoided if the paramagnetic beads were suspended in low pH flocculation buffer and added directly to the amplification mix. As predicted, once the amplified DNA was mixed with the paramagnetic beads in flocculation buffer and compacted together by magnet, the particles stayed clumped together due to bridging flocculation after gentle flicking of the tube (Fig. S1A†).Next, we assessed whether the modified flocculation solution could tolerate the elevated temperatures (63 °C) used for many isothermal amplification methods such as LAMP. For this purpose, the flocculation solution with paramagnetic beads was carefully placed into the lids of 0.2 ml tubes containing a DNA solution at the base of the tube. After incubation of the tubes at 63 °C for different amounts of time, the paramagnetic beads were combined with the DNA by flicking the tube. A slight deterioration in flocculation was observed after 10 minutes incubation at 63 °C, which became significantly more obvious after 20 minutes incubation, before completely failing after 30 minutes (Fig. S1A†). These results indicate that a single tube amplification and flocculation reaction based on paramagnetic particles and low pH buffer will not be possible for LAMP amplification, which typically requires a 50–60 minutes incubation at 63 °C.
DNA compaction agents, such as spermine, were examined as a potential replacement for low pH due to their ability to destabilize DNA in solution,32,33 which we hypothesized would promote interaction with the paramagnetic particles and result in flocculation. The paramagnetic beads were suspended in a buffer containing spermine and placed in the lids of 0.2 ml tubes with previously amplified LAMP products at the base of the tube. After incubation of the tubes for 60 minutes at 63 °C the paramagnetic beads were combined with the DNA by flicking the tube. The spermine-containing solution successfully induced flocculation of the paramagnetic particles despite extended incubation at 63 °C while no flocculation was observed in the controls samples (Fig. S1B†).
Spermine destabilizes DNA by binding to the major and minor grooves of the DNA helix and neutralizing the repelling negative charges between adjacent phosphate groups.33 The low pH used in the one-step flocculation method described above also neutralize the negative charges on the DNA molecules however, low pH will also affect the charges of other molecules in solution including proteins. Therefore, we hypothesized that the spermine buffer would make the flocculation less sensitive to contaminants commonly introduced from sample lysates. To test this hypothesis, we boiled Salmonella enterica cultures as an example of a complex mixture of cellular components including nucleic acids, proteins and lipids. Several dilutions of the original lysate were prepared and the two flocculation buffers tested. The low pH buffer induced flocculation in all dilutions tested, whereas the spermine buffer only induced flocculation in the presence of undiluted culture indicating that it is significantly less sensitive to cellular contaminants (Fig. S1C†).
Fine tuning the flocculation assay
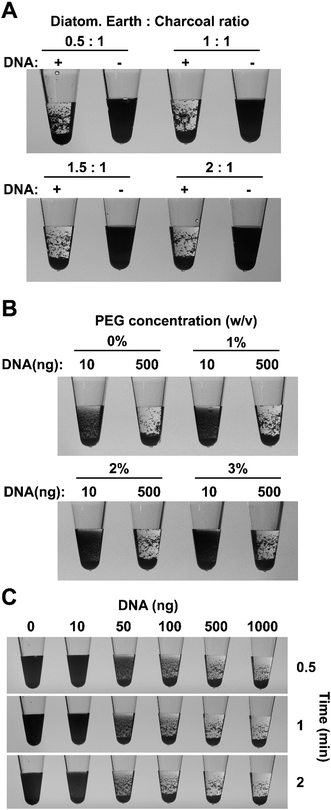
To reduce costs and further improve the flocculation system we sought to replace the relatively expensive paramagnetic beads and the requirement for magnetic pull down. We tested the suitability of a number of materials as a source of particles including colored silica particles used by ceramic artists (Fig. S2†) and found that the flocculation-based readout for DNA amplification is not dependent on a specific particle type. However, we found that a combination of activated charcoal and diatomaceous earth were ideal for such an assay as they are cheap sources of small, low-density particles, which can remain suspended in solution for extended periods and their irregular and highly porous structures provide a large surface area to aid interaction with DNA. The charcoal particles remain suspended for several minutes creating a black, non-transparent solution while the diatomaceous earth particles help to aggregate the fine charcoal particles and reduce the time for the flocculated particles to settle to the bottom of the tube. A flocculation solution containing charcoal and diatomaceous earth particles produced clearly visible aggregation when added to a tube containing a LAMP amplification product with clumping apparent even during mixing process (Movie S1†). After mixing, the bulk of the particles in the positive amplification sample settled to the bottom of the tube within 10 seconds while negative controls remained opaque black for approximately five minutes. The success of these results warranted further assessment of the charcoal–diatomaceous earth flocculation system.To further develop the new flocculation system, a large quantity of LAMP amplified DNA was purified using the Agencourt AMPure PCR purification system (Beckman Coulter, USA) to remove unincorporated dNTPs and oligonucleotides and quantified for use in subsequent assays (Fig. S3A†). As charcoal and diatomaceous earth particles provide different characteristics to the flocculation reaction, experiments were conducted to determine the ideal ratio providing the greatest difference between positive and negative amplifications. In these experiments, the amount of activated charcoal was kept constant while the amount of diatomaceous earth was varied. When a ratio of 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w) diatomaceous earth
1 (w/w) diatomaceous earth![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) charcoal was used, the charcoal particles flocculated in the presence of 500 ng of DNA and rapidly settled on the bottom of the tube however, the finer charcoal particles tended to stick to the walls of the tube instead of dropping to the bottom (Fig. 1A). Increasing the proportion of diatomaceous earth helped to alleviate this problem and a ratio of 1.5
charcoal was used, the charcoal particles flocculated in the presence of 500 ng of DNA and rapidly settled on the bottom of the tube however, the finer charcoal particles tended to stick to the walls of the tube instead of dropping to the bottom (Fig. 1A). Increasing the proportion of diatomaceous earth helped to alleviate this problem and a ratio of 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w) diatomaceous earth
1 (w/w) diatomaceous earth![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) charcoal was selected for further experiments.
charcoal was selected for further experiments.
In practical situations, it is not uncommon to obtain weak, non-specific amplification in some samples however, in a binary (presence/absence) system such as ours it is important to avoid flocculation in these circumstances. The speed of particle sedimentation in a solution can be modified by adjusting the viscosity of the solution, therefore we used this general physical principle to maximize the visual differences between positive and negative reactions. The effect different concentrations of polyethylene glycol (PEG) 8000 was examined on the flocculation of positive control samples, containing 500 ng of purified LAMP products, and synthetic negative controls, which contained 10 ng of LAMP amplified DNA in order to mimic the presence of non-template DNA in a negative reaction. The absence of PEG in the flocculation solution, resulted in obvious particle settling within five minutes in negative (10 ng) samples, potentially leading to misinterpretation of the results (Fig. 1B and S3B†). The addition of PEG 8000 reduced the flocculation speed and a final concentration of 1% (v/v) PEG was chosen for all future experiments since it significantly delayed particle sedimentation speed in the negative samples while having only a small effect in the positive samples (Fig. S3B†). Sedimentation speed was further decreased by higher concentrations of PEG providing additional flexibility for specific experimental situations.
The sensitivity of the new flocculation system was tested using a range of purified LAMP amplified DNA concentrations and a positive correlation between DNA content and the rate of particle settling was evident. The charcoal and diatomaceous earth particles in samples with ≥50 ng of DNA showed obvious settling within 30 seconds. Samples containing ≥500 ng of DNA flocculated in large clumps and rapidly settled to the bottom of the tube within 30 seconds showing two clearly observable phases (Fig. 1C). Samples containing 100 ng of DNA resulted in smaller clumps that took longer to settle while in the 50 ng DNA samples, flocculation was obvious however some fine charcoal particles remained suspended in solution giving the liquid a slightly dark appearance that remained even after several minutes. Samples containing 0 or 10 ng DNA did not display any obvious flocculation with a large proportion of the particles remaining suspended for the duration of the experiment. Similar results were observed when spermine was replaced with hexaminecobalt(III) chloride, another DNA compaction agent, indicating that the sensitivity is not specific to spermine but rather a result of the physical–chemical properties of bridging flocculation (Fig. S3C†). LAMP amplicons range from 300 bp up to 10 kb in size2 raising the possibility that the new flocculation system might not work as well for other amplification systems. We therefore repeated the sensitivity experiments described above using a 1.2 kb PCR amplification fragment from the Arabidopsis thaliana AGG2 gene (At3g22942).34 The results mirrored those for LAMP amplicons with all samples that contained ≥50 ng DNA displaying obvious flocculation (Fig. S3D†). The particles in the samples with ≥500 ng DNA had either settled to the bottom or had form clumps on the sides of the tubes within 30 seconds. In contrast, samples with 10 ng of DNA or less did not show display obvious flocculation and remained black for the duration of the experiment.
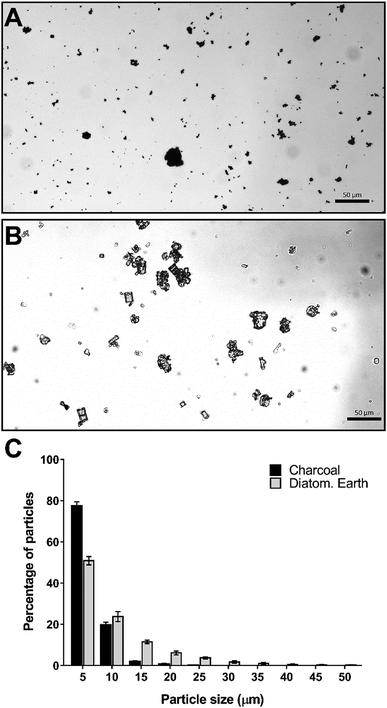
To further characterize the flocculation solution we examined the particle sizes of the charcoal and diatomaceous earth under a light microscope. The differences observed in the particle size distribution between charcoal and diatomaceous earth were consistent with our observations of the particles in solution. The charcoal component, which settles relatively slowly in water (no DNA present) is composed almost entirely of particles below 5 μm in diameter with less than 3% of particles larger than 10 μm (Fig. 2A and C). In contrast, 25% of the diatomaceous earth particles were larger than 10 μm (Fig. 2B and C). These larger particles can be seen rapidly settling in the flocculation mix regardless of whether DNA is present or not; however, in the presence of DNA, these large particles aggregate together with the DNA and smaller particles and contribute to the rapid clearing of the liquid phase.
Testing the system's robustness
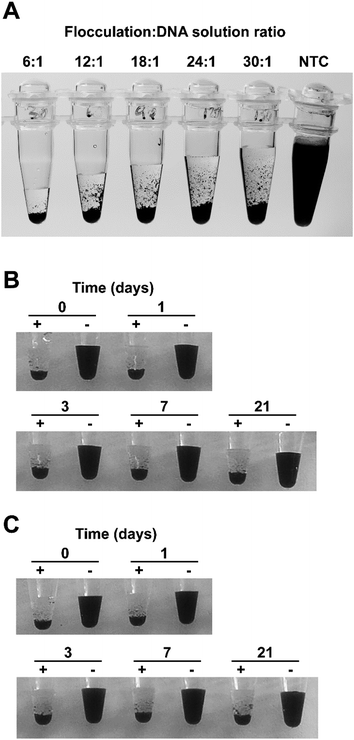
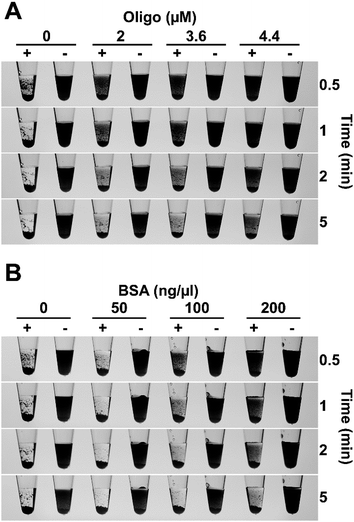
For a diagnostic test to be useful in field conditions, it must be robust and tolerate less than ideal conditions, for example, non-specialist operators cannot be relied upon to accurately measure small volumes or be familiar with the use of micropipettes. To assess the flocculation method's robustness, we added different amounts of flocculation solution to 500 ng of purified LAMP amplicon and observed rapid and obvious flocculation occurred when six to 24 volumes of flocculation mix were added to the DNA solution (Fig. 3A). Flocculation also occurred using higher ratios of flocculation mix but it became apparent that some fine charcoal particles remained in suspension. Field assays may result in exposure to elevated temperatures for long periods especially in tropical/sub-tropical regions therefore, we incubated our flocculation solution at 37 °C for up to 3 weeks and found that this did not affect its ability to distinguish between positive and negative LAMP samples (Fig. 3B). Lyophilisation (freeze-drying) is commonly used to extend the shelf life of solutions especially when there is limited access to refrigeration. Freeze drying the flocculation solution and subsequently re-hydrating it did not affect its ability to flocculate in the presence of DNA or its thermal stability (Fig. 3C). Finally, we explored the effect of long term storage with the flocculation solution remaining functional after 11 months of storage at 4 °C (Fig. 6D and E, and S3D†).We also examined the method's capability to discriminate between double-stranded DNA amplicons and single stranded oligonucleotides. Amplification reactions contain oligonucleotides at concentrations of 2 μM for PCR and 3.6–4.4 μM for LAMP. As spermine specifically binds to double stranded DNA,35 oligonucleotides were not expected to induce flocculation and our results confirmed that none of the tested oligonucleotide concentrations induced flocculation in the negative control reactions (Fig. 4A). However, the presence of excessive amounts of oligonucleotides slightly delayed flocculation in positive samples. Similarly, the presence of protein contamination in the final amplification mix can be an issue as DNA purity, especially in field-processed samples, is not always optimal. We found that the presence of protein (bovine serum albumin (BSA)) does not induce flocculation in the negative control samples (Fig. 4B), but did delay flocculation of positive samples at BSA concentrations ≥ 100 ng μl−1. However, the positive samples were still clearly distinguishable from the negative controls within 2–5 minutes (Fig. 4B).
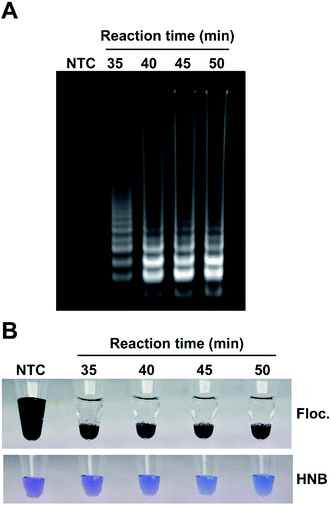
A number of factors can affect amplification, especially in field conditions, therefore it is important to establish the effectiveness of the visualization method in sub-optimal amplification conditions when there is a reduced amount of amplicon produced. To simulate this scenario, LAMP reactions were incubated for 35, 40, 45 or 50 minutes, an aliquot of each sample was run on an agarose gel (Fig. 5A) and flocculation and the popular hydroxyl naphthol blue (HNB)21,36 colorimetric assay was performed on aliquots from the same reaction (Fig. 5B). Flocculation assays provided clear visual confirmation of all the amplifications even for incomplete reactions. The HNB assay is a naked eye readout that relies on color change from violet to sky blue as a result of the reduction in magnesium concentration during DNA amplification. However, unlike flocculation, the HNB colorimetric test was difficult to interpret due to the subtle changes in color. While the difference between the HNB-treated control and complete amplification (50 minutes) samples was relatively obvious when compared side-by-side with sufficient illumination, samples with incomplete amplification could not be reliably interpreted as positive or negative despite amplification being clearly visible on the agarose gel.
Complete flocculation-based diagnostic for low-resource environments
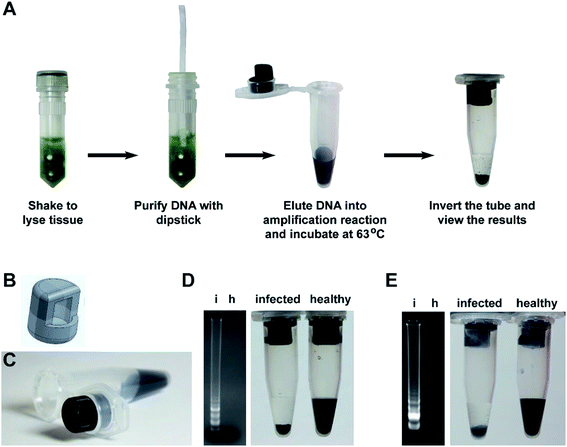
Building on our research, we developed a complete and easy-to-perform diagnostic assay that combines our flocculation readout with our previously developed rapid nucleic acid extraction dipstick method5 (Fig. 6A). To make the flocculation reaction method more suited for field-based research, we designed a tube insert (Fig. 6B) using 3D computer aided design (CAD) software, which was then produced using a 3D printer and Acrylonitrile–Butadiene–Styrene (ABS) filament. The tube insert fits tightly into the lid of a standard 1.5 ml tube and creates a well to hold the amplification reaction (Fig. 6C). The amplification mix is therefore added to the 3D tube insert while the flocculation solution is added to the bottom of the 1.5 ml tube to keep it separate from the LAMP reaction until the reaction is complete. To test the practicality of this system, we used the cellulose dipsticks to extract genomic DNA from leaves of either Arabidopsis plants infected with the bacterial pathogen Pseudomonas syringae pv. tomato strain DC3000 or tomato plants infected with the RNA virus Cucumber Mosaic Virus (CMV). The DNA was then directly eluted the DNA into the LAMP reaction mix contained within the tube insert. After incubation at 63 °C for 60 minutes, 5 μl of the reaction was analyzed by gel electrophoresis while the remaining reaction was left in the tube insert, the lid closed, and the tube inverted several times to mix the LAMP reaction with the flocculation solution. The flocculation-based assay correctly identified the P. syringae and CMV infected plant samples within 30 seconds and accurately reflected the results observed by agarose gel electrophoresis (Fig. 6D and E).Discussion
Molecular diagnostic tests involve three key steps: nucleic acid extraction, amplification and detection of the amplified product. For some applications, agarose gel electrophoresis analysis of the amplification products is needed however, in many instances users only want to know whether an amplicon was generated, for example when using PCR to screen samples for the presence of a disease. The aim of our research was to develop a low-cost, simple to interpret DNA amplification readout method that could rapidly identify the reactions that produce an amplicon without any additional equipment. Such a system would not only benefit researchers in modern laboratory settings but would be amenable for low resource environments and for users with limited scientific training (e.g. high school science classes and field-based research). The method described here utilizes the amplified DNA as a trigger for flocculation of suspended particles. The use of DNA-induced flocculation as a readout method has been previously reported29–31 however, our work uses a radically different approach that makes the system considerably faster, easier to perform and is ideal for low resource and field applications. The detection limit of our flocculation readout is approximately 50 ng of DNA (Fig. 1C and S3D†), high enough to avoid interference from the DNA present in the sample and low enough to trigger flocculation upon amplification.In our assay, the DNA amplification and flocculation reactions can be performed in a single-tube without opening the tube in between the amplification and flocculation reactions. This is a critically important feature as opening tubes post-amplification creates a serious cross-sample contamination risk,16,18,19 especially if the test is performed by operators with limited technical training. The single tube system takes advantage of surface tension to hold the amplification solution into the 3D printed insert (Fig. 6A and B). The rough layered surface of 3D-printed part creates a large surface area for the amplification solution to adhere to and prevents the liquid from falling out even when the tube is bumped or held horizontally. However, the 3D-printed insert used here is shown only as an example of a system that can provide the necessary physical separation between the amplification and flocculation solutions. Alternative designs could be developed to achieve the same goal.
An added benefit of our single tube method is that the large volume of flocculation solution raises the humidity inside the tube and prevents evaporation of the amplification reaction solution and thereby negates the need for heated lids or layering mineral oil over the DNA amplification reaction. This is important as it allows LAMP reactions to be performed in the field using any device that can maintain a constant temperature such as electrical or chemical heaters. We have shown the suitability of our method as a field ready pathogen detection system, having demonstrated the successful identification of plants infected with microbial (P. syringae) and viral (Cucumber mosaic virus) pathogens (Fig. 6C and D).
The simplicity and reliability of this method is the result of two key components: a DNA-specific compaction agent and a mixture of small, low density particles that create an opaque colloidal suspension. We used spermine as the DNA compaction agent although we demonstrate that hexaminecobalt(III) chloride, another compaction agent with a similar charge distribution can also be used (Fig. S3C†). The alignment of the positive charges from the compaction agents with the negatively charged phosphate groups on the DNA molecule destabilize the DNA structure32,33,37 and, as a result, enhance DNA binding to the suspended particles. The mechanism behind the aggregation of the charcoal and diatomaceous earth particles is likely to occur via a process known as bridging flocculation, as has been previously suggested for DNA-induced flocculation of paramagnetic particles.30,31 In bridging flocculation, DNA molecules need to have a minimal length to allow binding to multiple particles, causing aggregation and eventually fall out of suspension. The addition of spermine to the flocculation buffer increases specificity for double stranded DNA,35 helping to minimize interference by contaminants such cell debris, proteins or oligonucleotides (Fig. 3 and S1†).
The particles used in the flocculation solution are a combination of activated charcoal and diatomaceous earth that provide a large surface area for interaction with DNA molecules due to their high degree of porosity and irregular shape (Fig. 4A and B). Additionally, their relatively low densities ensure that they will remain suspended for extended periods unless they are induced to flocculate. Although we found that flocculation can be achieved using either individual component, the combination takes advantage of their different properties to provide an optimal visual effect. The charcoal particles create a black, non-transparent solution that enables positive and negative samples to be easily distinguished in any light environment while the diatomaceous earth particles help to aggregate the fine charcoal particles and reduce the time to settle at the bottom of the tube. The size of most charcoal and diatomaceous earth particles used in our study range between 1 to 5 μm, consistent with previous results that found the use of particles ≥1 μm in size were ideal for flocculation in the presence of DNA.30 While charcoal and diatomaceous earth was our preferred choice, our DNA amplification readout is not dependent specifically on these particles, or their exact size distribution, as we successfully detected DNA amplification using other particles including colored silica particles used by ceramic artists (Fig. S2†).
Conclusions
The combination of our previously developed rapid DNA extraction method,5 isothermal amplification and the simple DNA amplification readout system described here is closely aligned with the World Health Organisation's ASSURED criteria for diagnostics (Affordable, Sensitive, Specific, User-friendly, Rapid, Equipment-free, and Deliverable to those who need it).1 The method has been demonstrated to work with PCR and LAMP and can theoretically be applied to any amplification technology provided the amplicon is large enough to induce the bridging flocculation mechanism. The flocculation reaction is very robust and can tolerate variations in operating procedures and environmental conditions making it ideal for field based research. Moreover, this method is amenable to non-specialized users with limited technical training or resources and can be easily incorporated into existing diagnostic platforms aimed at detecting human, plant and animal diseases. Aside from the obvious diagnostics applications, our method is especially suited for learning environments in high schools were students can perform nucleic acid amplification experiments and easily observe the results of their efforts without the use of expensive or dangerous chemicals, thus facilitating the teaching of these technologies in pre-university environments.Funding
This work was supported by the Australian Centre for International Agricultural Research (ACIAR) HORT/2014/027.Conflicts of interest
The research generated in this manuscript has been used to file for a provisional patent application in Australia to protect the intellectual property that has arisen from this work (Applicant: The University of Queensland; Filing date: May 21, 2019). Prior to the submission of this manuscript, the technology has not been licensed or sold to any company and it is not currently being commercialized.Acknowledgements
The authors would like to thank Stone Cat Ceramics (http://www.stonecat.com.au) for supplying the colored silica particles used in this study.References
- H. Kettler, K. White and S. Hawkes, Mapping the landscape of diagnostics for sexually transmitted infections, WHO, 2004 Search PubMed.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS.
- O. Piepenburg, C. H. Williams, D. L. Stemple and N. A. Armes, PLoS Biol., 2006, 4, e204 CrossRef PubMed.
- M. Vincent, Y. Xu and H. Kong, EMBO Rep., 2004, 5, 795–800 CrossRef CAS PubMed.
- Y. Zou, M. G. Mason, Y. L. Wang, E. Wee, C. Turni, P. J. Blackall, M. Trau and J. R. Botella, PLoS Biol., 2017, 15, e2003916 CrossRef PubMed.
- F. Zhang, J. Wu, R. Wang, L. Wang and Y. B. Ying, Chem. Commun., 2014, 50, 8416–8419 RSC.
- Y. Mori, K. Nagamine, N. Tomita and T. Notomi, Biochem. Biophys. Res. Commun., 2001, 289, 150–154 CrossRef CAS PubMed.
- M. Enosawa, S. Kageyama, K. Sawai, K. Watanabe, T. Notomi, S. Onoe, Y. Mori and Y. Yokomizo, J. Clin. Microbiol., 2003, 41, 4359–4365 CrossRef CAS.
- J. Fischbach, N. C. Xander, M. Frohme and J. F. Glokler, BioTechniques, 2015, 58, 189–194 CrossRef CAS PubMed.
- P. Valentini, R. Fiammengo, S. Sabella, M. Gariboldi, G. Maiorano, R. Cingolani and P. P. Pompa, ACS Nano, 2013, 7, 5530–5538 CrossRef CAS PubMed.
- Y. Zhang, J. Hu and C. Y. Zhang, Anal. Chem., 2012, 84, 9544–9549 CrossRef CAS.
- S. Chomean, N. Wangmaung, P. Sritongkham, C. Promptmas and W. Ittarat, Clin. Chim. Acta, 2014, 437, 197–202 CrossRef CAS.
- S. Chomean, N. Wangmaung, P. Sritongkham, C. Promptmas, S. Mas-Oodi, D. Tanyong and W. Ittarat, Analyst, 2014, 139, 813–822 RSC.
- C. P. Ma, W. S. Wang, A. Mulchandani and C. Shi, Anal. Biochem., 2014, 457, 19–23 CrossRef CAS PubMed.
- A. Muangchuen, P. Chaumpluk, A. Suriyasomboon and S. Ekgasit, Sensors, 2014, 14, 14472–14487 CrossRef PubMed.
- M. Hong, L. Zha, W. L. Fu, M. J. Zou, W. J. Li and D. G. Xu, World J. Microbiol. Biotechnol., 2012, 28, 523–531 CrossRef CAS PubMed.
- Z. K. Njiru, A. S. J. Mikosza, T. Armstrong, J. C. Enyaru, J. M. Ndung'u and A. R. C. Thompson, PLoS Neglected Trop. Dis., 2008, 2, e147 CrossRef PubMed.
- X. Zhang, H. Zhang, J. J. Pu, Y. X. Qi, Q. F. Yu, Y. X. Xie and J. Peng, PLoS One, 2013, 8, e82841 CrossRef PubMed.
- H. H. Xu, L. Zhang, G. Q. Shen, C. Feng, X. Y. Wang, J. Yan and Y. J. Zhang, J. Virol. Methods, 2013, 194, 21–25 CrossRef CAS PubMed.
- L. Wang, Z. C. Huang, R. Wang, Y. B. Liu, C. Qian, J. Wu and J. W. Liu, ACS Appl. Mater. Interfaces, 2018, 10, 4409–4418 CrossRef CAS.
- M. Goto, E. Honda, A. Ogura, A. Nomoto and K. I. Hanaki, BioTechniques, 2009, 46, 167–172 CrossRef CAS.
- J. A. Tomlinson, N. Boonham and M. Dickinson, Plant Pathol., 2010, 59, 465–471 CrossRef CAS.
- C. Gosch, R. A. Gottsberger, K. Stich and T. C. Fischer, Eur. J. Plant Pathol., 2012, 134, 835–845 CrossRef CAS.
- M. Goto, K. Shimada, A. Sato, E. Takahashi, T. Fukasawa, T. Takahashi, S. Ohka, T. Taniguchi, E. Honda, A. Nomoto, A. Ogura, T. Kirikae and K. I. Hanaki, J. Microbiol. Methods, 2010, 81, 247–252 CrossRef CAS PubMed.
- S. J. Harper, L. I. Ward and G. R. G. Clover, Phytopathology, 2010, 100, 1282–1288 CrossRef CAS PubMed.
- X. J. Ma, Y. L. Shu, K. Nie, M. Qin, D. Y. Wang, R. B. Gao, M. A. Wang, L. Y. Wen, F. Han, S. M. Zhou, X. A. Zhao, Y. H. Cheng, D. X. Li and X. P. Dong, J. Virol. Methods, 2010, 167, 214–217 CrossRef CAS PubMed.
- M. A. Almasi, M. Aghapour-Ojaghkandi, K. Bagheri, M. Ghazvini and S. M. Hosseyni-Dehabadi, Appl. Biochem. Biotechnol., 2015, 175, 3599–3616 CrossRef CAS PubMed.
- N. A. Tanner, Y. H. Zhang and T. C. Evans, BioTechniques, 2015, 58, 59–68 CrossRef CAS PubMed.
- D. C. Leslie, J. Y. Li, B. C. Strachan, M. R. Begley, D. Finkler, L. A. L. Bazydlo, N. S. Barker, D. M. Haverstick, M. Utz and J. P. Landers, J. Am. Chem. Soc., 2012, 134, 5689–5696 CrossRef CAS PubMed.
- C. Q. Lin, Y. X. Zhang, X. Zhou, B. Yao and Q. Fang, Biosens. Bioelectron., 2013, 47, 515–519 CrossRef CAS PubMed.
- E. Wee, H. Lau, J. Botella and M. Trau, Chem. Commun., 2015, 51, 5828–5831 RSC.
- L. C. Gosule and J. A. Schellman, J. Mol. Biol., 1978, 121, 311–326 CrossRef CAS PubMed.
- J. C. Murphy, J. A. Wibbenmeyer, G. E. Fox and R. C. Willson, Nat. Biotechnol., 1999, 17, 822–823 CrossRef CAS PubMed.
- M. G. Mason and J. R. Botella, Biochim. Biophys. Acta, Gene Struct. Expression, 2001, 1520, 147–153 CrossRef CAS.
- B. C. Hoopes and W. R. McClure, Nucleic Acids Res., 1981, 9, 5493–5504 CrossRef CAS PubMed.
- M. Safavieh, M. U. Ahmed, E. Sokullu, A. Ng, L. Braescuac and M. Zourob, Analyst, 2014, 139, 482–487 RSC.
- S.-Y. Wang, A. Y.-L. Lee, Y.-L. Lee, Y.-H. Lai, J. J. W. Chen, W.-L. Wu, J.-M. P. Yuann, W.-L. Su, S.-M. Chuang and M.-H. Hou, PLoS One, 2012, 7, e47101 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra04725e |
| This journal is © The Royal Society of Chemistry 2019 |