Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nutritional characteristics of marine fish Sardinella zunasi Bleeker and immunostimulatory activities of its glycoprotein
Yu
Han
a,
Huili
Hao
a,
Lihong
Yang
ab,
Guolian
Chen
a,
Yucong
Wen
a and
Riming
Huang
 *a
*a
aGuangdong Provincial Key Laboratory of Food Quality and Safety, College of Food Science, South China Agricultural University, Guangzhou 510642, China. E-mail: hanyu@stu.scau.edu.cn; hhlhaohuili@stu.scau.edu.cn; guolianchen@scau.edu.cn; vtchong@hotmail.com; huangriming@scau.edu.cn; Tel: +86 20 8528 3448
bShenzhen Shajing People's Hospital, Guangzhou University of Chinese Medicine, Shenzhen, China. E-mail: 13570253038@163.com
First published on 24th September 2019
Abstract
Sardinella zunasi Bleeker, an edible and medicinal marine fish, is largely distributed in tropical oceans. However, the chemical composition and nutritional properties of this species have not yet been investigated. In the present study, proximate composition, fatty acids, amino acids, taurine, and minerals of S. zunasi Bleeker were characterized, and the immunostimulatory properties of its glycoprotein were evaluated. The results indicated the presence of crude protein (19.66%), crude lipid (6.29%) and carbohydrate (0.74%) in S. zunasi Bleeker; monounsaturated fatty acids and polyunsaturated fatty acids in the fatty acid composition of S. zunasi Bleeker were 25.00% and 31.01%, respectively; S. zunasi Bleeker was rich in taurine (219 mg/100 g) and essential amino acids (5.57 g/100 g). In addition, the glycoprotein of S. zunasi consisted of protein and sugars, with a total content of 34.25% and 16.27%, respectively. The glycoprotein showed significant effects on promoting NO, TNF-α and IL-6 in a dose-dependent manner in RAW264.7 macrophage cells. Thus, these findings provide a scientific basis for the further utilization of glycoprotein from S. zunasi Bleeker.
1. Introduction
Fish is a healthy food with numerous benefits to human health. It has been consumed in large quantities not only because it is a source of high nutritional quality protein, but also as a significant reserve of polyunsaturated fatty acids.1 Fish muscle is more digestible than other animal protein due to its lower level of connective tissue. Fish fat is one of the few natural food sources of vitamin D and contains important amounts of vitamins A and E.2 Fish is a perfect supplement to a high cereal diet because of its high lysine content.3 There is plenty of evidence that the consumption of fish reduces the risk of coronary heart disease.4–7 Marine fish is a rich source of high-quality protein, lipids, as well as all kinds of vitamins and minerals. It has been reported that the long-chain polyunsaturated (omega-3) fatty acids rich in marine fish, and these polyunsaturated fatty acids can reduce various pathophysiologic abnormalities including anoxic ventricular arrhythmia, atherogenesis, hypertension, blood clotting and cardiovascular disease.8,9S. zunasi Bleeker, an edible and medicinal marine fish, belonging to genus Sardinella, family Clupeidae, is widely distributed in the coastal areas of tropical oceans, such as the Philippines, Japan, Korea and China. The species of genus Sardinella are taken as a good source of bioactive components. Previous investigation of genus Sardinella species has exhibited that the chemical or nutritional composition is rich in proteins, lipids (especially polyunsaturated fatty acids), and minerals10–13 and significant biological properties such as antioxidant,14,15 antibacterial,16 anticancer,17 hepatoprotective and nephroprotective,18 hypolipidemic, antiobesity and cardioprotective effects.19 However, there is still a lack of more data about the chemical or nutritional composition of genus Sardinella, and there is no report about the chemical or nutritional composition of S. zunasi Bleeker. Thus, it's necessary to investigate the chemical and nutritional composition of S. zunasi Bleeker before any in-depth study.
In order to make more effective utilization of S. zunasi Bleeker, the present work aims to determine the chemical and nutritional components including crude protein, crude fat, total carbohydrates, amino acids, taurine, fatty acids and minerals. In addition, the immunomodulatory effect of its glycoprotein on the murine RAW264.7 macrophage cell lines, including evaluating the effects on the proliferation of RAW264.7 cells, phagocytic uptake, the production of nitric oxide (NO), TNF-α, and IL-6 on RAW264.7 cells, was investigated. The results of this work might provide a useful information for further investigation of biological components from S. zunasi Bleeker and their utilization.
2. Materials and methods
2.1. Specimens of fish
S. zunasi Bleeker was purchased from Zhanjiang, Guangdong Province, China in August 2017. A voucher specimen (no. 20170803) was deposited in Guangdong Provincial Key Laboratory of Food and Safety, South China Agricultural University, China. After collection, fresh fish was packed in plastic bags, preserved in ice and taken immediately to the laboratory, it was preserved in a −20 °C refrigerator for about half a month before further treatments. The whole fish were thoroughly washed with running water, cut into pieces and then homogenised in a mincer before the analysis of nutritional characteristics and preparation of crude glycoprotein (H1) and pure glycoprotein (H2). Approximately 20 kilograms of fish was used for the entire study.2.2. Standards and reagents
Standard mixtures of fatty acid methyl ester were purchased from Nu-Chek Prep (Inc., Elysian, MN, USA). All sugars (fructose, rhamnose, arabinose, xylose, mannose, glucose and galactose) were purchased from Aladdin Industrial Corporation (Shanghai, China). Murine RAW264.7 macrophage cells were obtained from Jinan University (Guangzhou, China). Lipopolysaccharides (LPS) and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) were purchased from Sigma Co. (Mo, U.S.A.). Dulbecco Modified Eagle Medium (DMEM), fetal bovine serum (FBS), and penicillin–streptomycin were purchased from Gibco Life Technologies (Grand Island, NY.). Nitric oxide (NO) kit was purchased from Beyotime Biotechnology Co. Ltd. Mouse TNF-α, and IL-6 ELISA kits were purchased from NeoBioscience Biotechnology Co. Ltd. DEAE-Cellulose 52 and Sephadex G-200 were purchased from Shanghai Yuanye Bio-Technology Co. Ltd. The other chemicals used in this research were of analytical grade.2.3. Proximate composition analysis
The fish was washed and removed the surface water on the skin before composition analysis. The moisture, crude protein, crude lipid, and ash contents were analysed according to AOAC official methods.20 Moisture content was determined using a hot-air oven at 105 °C for 6 h until constant weight was reached. Nitrogen content was measured by the Kjeldahl method adapted from where protein content is estimated by multiplying the nitrogen content by 6.25. Crude lipids were determined by Soxhlet extraction with petroleum ether as the solvent. Ash content was measured by heating the fish in a muffle furnace at 550 °C until the resultant ash was light grey in color. Carbohydrate content was determined using the phenol-sulfuric acid method.212.4. Nutritional composition analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mixed solution and the flushing fluid also transferred to separating funnel. After shaking for 5 min, the ether layer was collected, and then evaporated to dryness. 8 mL of 2% sodium hydroxide methanol solution and 7 mL of 15% boron trifluoride methanol solution were added to the extraction during heating in an 80 °C water bath for 5 min, and then cooled to room temperature quickly. 20 mL of n-heptane was added, after 2 min shaking, saturated sodium chloride solution was added. After stratification, 5 mL of n-heptane was taken from the top to a tube and shook with 4 g of anhydrous sodium sulfate for 1 min, the upper solution was taken for further analysis.
1) mixed solution and the flushing fluid also transferred to separating funnel. After shaking for 5 min, the ether layer was collected, and then evaporated to dryness. 8 mL of 2% sodium hydroxide methanol solution and 7 mL of 15% boron trifluoride methanol solution were added to the extraction during heating in an 80 °C water bath for 5 min, and then cooled to room temperature quickly. 20 mL of n-heptane was added, after 2 min shaking, saturated sodium chloride solution was added. After stratification, 5 mL of n-heptane was taken from the top to a tube and shook with 4 g of anhydrous sodium sulfate for 1 min, the upper solution was taken for further analysis.
Fatty acids were determined by a gas chromatography 7820A (Agilent Technologies, USA), equipped with a flame ionization detector (FID) and a HP88 capillary column (100 m length × 0.25 mm ID × 0.2 μm film thickness). Purified nitrogen was the carrier gas at a flow rate of 1 mL min−1. Sample of 1.0 μL was injected with a split ratio of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The injector and detector temperature were 270 °C and 280 °C, respectively. The column temperature was set at 100 °C for 13 min, followed by a 10 °C min−1 heating ramp to 180 °C, which was held for 6 min. Then the temperature was increased to 200 °C at a rate of 1 °C min−1 and held for 20 min. Finally, it was increased to 230 °C at a rate of 4 °C min−1 and held for 10.5 min. Fatty acids were identified by comparison of retention time with standard mixtures of fatty acid methyl ester, the composition of fatty acids was expressed in relative percentage of total fatty acids according to their peak areas.22
1. The injector and detector temperature were 270 °C and 280 °C, respectively. The column temperature was set at 100 °C for 13 min, followed by a 10 °C min−1 heating ramp to 180 °C, which was held for 6 min. Then the temperature was increased to 200 °C at a rate of 1 °C min−1 and held for 20 min. Finally, it was increased to 230 °C at a rate of 4 °C min−1 and held for 10.5 min. Fatty acids were identified by comparison of retention time with standard mixtures of fatty acid methyl ester, the composition of fatty acids was expressed in relative percentage of total fatty acids according to their peak areas.22
2.5. Glycoproteins extraction and purification
The extraction and purification of glycoprotein from S. zunasi Bleeker were carried out using the methods described previously.23 In brief, the fish was cut into pieces and ground up in a blender, then extracted twice by using a hot water method with the conditions of liquid/solid ratio of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, temperature of 95 °C and extraction time of 2 h. The combined aqueous extracts were concentrated in a rotary evaporator under reduced pressure at 50 °C and then filtered. The free proteins in the extracts were removed by using Sevag reagent (chloroform/n-butanol, v/v = 4
1, temperature of 95 °C and extraction time of 2 h. The combined aqueous extracts were concentrated in a rotary evaporator under reduced pressure at 50 °C and then filtered. The free proteins in the extracts were removed by using Sevag reagent (chloroform/n-butanol, v/v = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The deproteinized solution was precipitated with addition of ethanol to reach a final ethanol concentration of 75% at 4 °C for 24 h. Following centrifugation at 12
1). The deproteinized solution was precipitated with addition of ethanol to reach a final ethanol concentration of 75% at 4 °C for 24 h. Following centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min, the precipitates were washed sequentially with anhydrous ethanol and acetone, and then dialyzed against deionized water for 2 days and lyophilized as crude glycoprotein (H1).
000 rpm for 15 min, the precipitates were washed sequentially with anhydrous ethanol and acetone, and then dialyzed against deionized water for 2 days and lyophilized as crude glycoprotein (H1).
The crude glycoprotein (H1) was dissolved in distilled water and separated by a DEAE-Cellulose 52 column (2.0 cm × 40 cm). The column was eluted with distilled water and a stepwise NaCl gradient (0–1 M). One independent elution peak of the glycoprotein was obtained by phenol-sulfuric acid method at 490 nm. The glycoprotein fraction was collected, dialyzed, concentrated, and further loaded into Sephadex G-200 column (1.8 cm × 50 cm) with distilled water at a flow rate of 0.5 mL min−1. One independent elution peak of the glycoprotein was obtained by phenol-sulfuric acid method at 490 nm. One main fraction was finally obtained, dialyzed, lyophilized and denoted as H2.
2.6. Physicochemical characterization of H2
2.7. Immunomodulatory activity of H2
| Cell viability (%) = (A2 − A0)/(A1 − A0) × 100 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were added, and then the cells were lysis for 30 min at room temperature. The absorbance was determined at 540 nm by microplate reader (Molecular Devices, Austria). The phagocytic rate was calculated by the following formula:
1) were added, and then the cells were lysis for 30 min at room temperature. The absorbance was determined at 540 nm by microplate reader (Molecular Devices, Austria). The phagocytic rate was calculated by the following formula:| Phagocytosis rate (%) = (A2 − A0)/(A1 − A0) × 100 |
2.8. Statistical analysis
Data were expressed as mean ± SD, at least 3 independent experiments for each sample except fatty acids analysis of S. zunasi Bleeker and composition analysis and amino acid composition of H2. Statistical significance was calculated by one-way analysis of variance ANOVA followed by Turkey's test to determine the difference between groups (GraphPad Prism 5.0). Values of P < 0.05 were considered as statistically significant.3. Results and discussion
3.1. Proximate composition
The proximate composition of S. zunasi Bleeker is presented in Table 1. Moisture (68.75% ± 0.79%) was the most abundant component in S. zunasi Bleeker, similar to Pomatomus saltatrix (70.87% ± 0.59%) and Engraulis encrasicolus (66.95% ± 0.64%).28 The protein content29 ranged from 17 g/100 g to 20 g/100 g for freshwater and 18 g/100 g to 22 g/100 g for marine fish. Crude protein content of S. zunasi Bleeker was 19.66% ± 0.15%, higher than that of Mullus barbatus (14.54% ± 0.00%), Pomatomus saltatrix (15.24% ± 0.55%) and Scorpaena porcus (16.91% ± 0.04%).28 Fishes were often classified as lean fish (lipid content < 5%), medium fat fish (5–10%), and fatty fish (>10%) on the basis of their fat content.30 Based on this classification, S. zunasi Bleeker was medium fat fish with 6.29% ± 0.74% lipid content, was similar to values reported in muscle of other fish species, such as sardines and mackerel.1 The ash content (6.16% ± 0.28%) of S. zunasi Bleeker was much higher than those of Pampus argenteus (2.25% ± 0.62%) and Harpadon nehereus (0.93% ± 0.11%).31 The major carbohydrate composition in fish muscle is glycogen that is a polymer of glucose. A typical muscle in a live fish may contain between 0.1 and 1% glycogen.32 In our study, we found carbohydrate content of S. zunasi Bleeker was 0.74% ± 0.01%, which agreed with the value reported in literature.323.2. Nutritional composition analysis
| Fatty acids | Contenta (%) |
|---|---|
| a Values expressed as wet weight. b SFA: saturated fatty acids. c MUFA: monounsaturated fatty acids. d PUFA: polyunsaturated fatty acids. | |
| C12:0 | 0.14 |
| C13:0 | 0.16 |
| C14:0 | 8.60 |
| C14:1 | 0.11 |
| C15:0 | 0.91 |
| C16:0 | 21.8 |
| C16:1 | 7.60 |
| C17:0 | 1.40 |
| C17:1 | 1.40 |
| C18:0 | 8.50 |
| C18:1 | 12.40 |
| C18:2 | 3.10 |
| C18:3 | 1.30 |
| C18:4 | 0.75 |
| C20:0 | 1.10 |
| C20:1 | 1.50 |
| C20:2 | 0.15 |
| C20:3 | 0.21 |
| C20:4 (ARA) | 2.70 |
| C20:5 (EPA) | 9.60 |
| C21:0 | 0.32 |
| C22:0 | 0.50 |
| C22:1 | 0.89 |
| C22:5 (EPA) | 2.50 |
| C22:6 (DHA) | 10.70 |
| C24:0 | 0.24 |
| C24:1 | 1.10 |
| SFAb | 43.67 |
| MUFAc | 25.00 |
| PUFAd | 31.01 |
| Amino acids | Contenta (g/100 g) | Percentage of TAAc (%) |
|---|---|---|
| a Values expressed as wet weight. b Essential amino acids. c TAA: total amino acids. | ||
| Asx (Asp + Asn) | 1.45 ± 0.02 | 9.48 |
| Thrb | 0.68 ± 0.01 | 4.44 |
| Ser | 0.64 ± 0.01 | 4.18 |
| Glx (Glu + Gln) | 2.41 ± 0.02 | 15.75 |
| Gly | 1.31 ± 0.03 | 8.56 |
| Ala | 1.19 ± 0.02 | 7.77 |
| Valb | 0.70 ± 0.01 | 4.58 |
| Metb | 0.49 ± 0.01 | 3.20 |
| Trpb | 0.11 ± 0.00 | 0.72 |
| Ileb | 0.56 ± 0.01 | 3.66 |
| Leub | 1.13 ± 0.01 | 7.39 |
| Tyr | 0.46 ± 0.01 | 3.00 |
| Pheb | 0.60 ± 0.00 | 3.92 |
| His | 0.37 ± 0.01 | 2.42 |
| Lysb | 1.30 ± 0.02 | 8.50 |
| Arg | 0.98 ± 0.01 | 6.40 |
| Pro | 0.92 ± 0.01 | 6.01 |
| EAA | 5.57 | 36.41 |
| NEAA | 9.73 | 63.59 |
3.3. Extraction, purification, and physicochemical properties of H2
In this study, crude glycoprotein (H1) was obtained from S. zunasi Bleeker by conventional hot water extraction, alcohol precipitation and deproteinization. Crude glycoprotein was further purified through anion-exchange chromatography according to differences in the existence of ionic groups. One fraction eluted with the 0.2 M sodium chloride solution was collected (Fig. 1a). After dialysis and concentration, it was further purified by Sephadex G-200 column, and then H2 was obtained (Fig. 1b). The physicochemical properties of H2 were shown in Table 5. The content of total sugar and protein were 16.27% and 34.25%, respectively. The main monosaccharide existed in H2 were mannose and galactose. The homogeneity and average molecular weight of H2 were measured by HPGPC shown in Fig. 1c. The GPC curves revealed that the fraction was represented by a single, sharp peak, indicated the glycoprotein was relatively homogeneous in molecular weight distribution. The average molecular weight of H2 was calculated as 7145 Da according to the equation of calibration curve.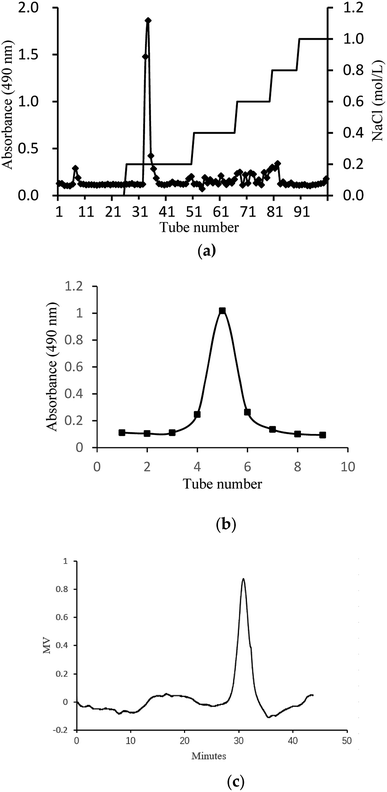 | ||
| Fig. 1 Preparation and physicochemical properties of H2. (a) Elution curve on Cellulose DEAE-52 column. (b) Elution curve on G-200 column and (c) HPGPC profile of H2. | ||
| Total sugar content (%) | Protein content (%) | Sugar composition (molar ratio) | |
|---|---|---|---|
| Mannose | Galactose | ||
| 16.27 | 34.25 | 1.00 | 2.19 |
The FTIR spectra of H2 is shown in Fig. 2, most of the absorption bands could be assigned according to data obtained in previous studies.50–53 The broad stretching intense peak at 3356 cm−1 indicated the presence of hydroxyl and amino groups,54 the 2927 cm−1 region corresponded to the C–H stretching vibration. The band at 1649 cm−1 was the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of amide group. The absorption peak at 1399 cm−1 represented the C–O–H carboxylic acid, and the strong peak observed at 1069 cm−1 was characteristic of all sugar derivatives and hydroxyl group.55 The spectra indicated H2 was a glycoprotein.
O stretching vibration of amide group. The absorption peak at 1399 cm−1 represented the C–O–H carboxylic acid, and the strong peak observed at 1069 cm−1 was characteristic of all sugar derivatives and hydroxyl group.55 The spectra indicated H2 was a glycoprotein.
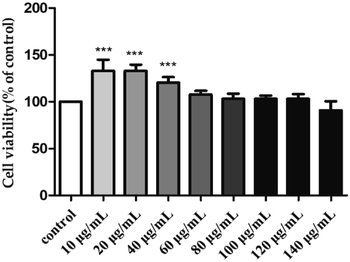
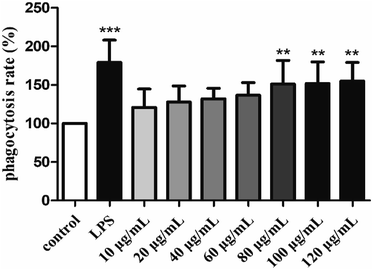
3.4. Immunomodulatory activity of H2
4. Conclusions
To sum up, the study revealed that S. zunasi Bleeker was a valuable marine fish for containing high level of protein and taurine. The amino acids and mineral composition indicated S. zunasi Bleeker was a good source of essential amino acids and micronutrients. Furthermore, S. zunasi Bleeker can be considered as a potential dietary source of DHA and EPA. The glycoprotein from S. zunasi Bleeker showed a significant immunostimulatory activity on RAW264.7 cells. It could not only promote cell proliferation and phagocytosis, but also promote inflammatory cytokines secretion. Thus, S. zunasi Bleeker can be considered as a nutritional supplement with a potential immunostimulatory property.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by Science and Technology Planning Project of Guangdong Province, Guangzhou Planned Program in Science and Technology, Program of Department of Ocean and Fisheries of Guangdong Province, Natural Science Foundation of Guangdong, Guangdong Provincial Key Laboratory of Applied Botany, South China Botanical Garden, Chinese Academy of Sciences, Finance Special Project of Zhanjiang City, Natural Science Foundation of Guangdong, grant number 2017A020217002, 201803020003, GDME2018C014, 2016A030313151, AB2018004, 2018A01044 and 2018A0303070018.Notes and references
- C. E. Fernandes, M. A. D. Vasconcelos, M. D. Ribeiro, L. A. Sarubbo, S. A. C. Andrade and A. B. de Melo, Food Chem., 2014, 160, 67–71 CrossRef CAS PubMed.
- A. K. M. Bhuiyan, W. M. N. Ratnayake and R. G. Ackman, J. Food Compos. Anal., 1993, 6, 172–184 CrossRef.
- S. A. Otitologbon, E. B. Agbaji, O. A. Peters and S. J. Oniye, J. Sci. Food Agric., 1997, 75, 312–314 CrossRef CAS.
- D. Kromhout, E. B. Bosschieter and C. de Lezenne Coulander, N. Engl. J. Med., 1985, 312, 1205–1209 CrossRef CAS.
- D. Kromhout, E. J. Feskens and C. H. Bowles, Int. J. Epidemiol., 1995, 24, 340–345 CrossRef CAS.
- S. E. Norell, A. Ahlbom, M. Feychting and N. L. Pedersen, Br. Med. J., 1986, 293, 426 CrossRef CAS.
- T. A. Dolecek and G. Granditis, World Rev. Nutr. Diet., 1991, 66, 205–216 CAS.
- A. J. McMichael and C. D. Butler, Am. J. Prev. Med., 2005, 29, 322–323 CrossRef.
- R. M. Krauss, R. H. Eckel, B. Howard, L. J. Appel, S. R. Daniels, R. J. Deckelbaum, J. W. J. Erdman, P. Kris-Etherton, I. J. Goldberg, T. A. Kotchen, A. H. Lichtenstein, W. E. Mitch, R. Mullis, K. Robinson, J. Wylie-Rosett, S. St. Jeor, J. Suttie, D. L. Tribble and T. L. Bazzarre, Circulation, 2000, 102, 2284–2299 CrossRef CAS.
- T. M. Tengku-Rozaina, W. S. Jeng and M. A. Amiza, J. Aquat. Food Prod. Technol., 2018, 27, 667–679 CrossRef.
- F. S. Hamdan and N. M. Sarbon, Int. Food Res. J., 2019, 26, 133–140 CAS.
- A. Taheri, N. Sarhaddi, G. A. Bakhshizadeh and S. Sharifian, Iran. J. Fish. Sci., 2017, 16, 1312–1324 Search PubMed.
- O. M. Bahurmiz, F. Adzitey and W. K. Ng, Int. Food Res. J., 2017, 24, 2387–2393 CAS.
- N. Souissi, A. Bougatef, Y. Triki-Ellouz and M. Nasri, Food Technol. Biotechnol., 2007, 45, 187–194 CAS.
- A. Barkia, A. Bougatef, H. Ben Khaled and M. Nasri, J. Food Biochem., 2010, 34, 303–320 CrossRef.
- R. S. C. Som and C. K. Radhakrishnan, Indian J. Geo-Mar. Sci., 2011, 40, 710–716 Search PubMed.
- R. S. C. Som, P. Pillai, S. Lekshmi and C. K. Radhakrishnan, Indian J. Geo-Mar. Sci., 2017, 46, 290–294 Search PubMed.
- Z. Kamoun, A. S. Kamoun, A. Bougatef, R. M. Kharrat, H. Youssfi, T. Boudawara, M. Chakroun, M. Nasri and N. Zeghal, Environ. Sci. Pollut. Res., 2017, 24, 1432–1441 CrossRef CAS.
- I. Jemil, O. Abdelhedi, R. Nasri, L. Mora, R. Marrekchi, K. Jamoussi, A. ElFeki, M. Hajji, F. Toldra and M. Nasri, Life Sci., 2017, 176, 54–66 CrossRef CAS.
- B. E. C. Ziani, W. Rached, K. Bachari, M. J. Alves, R. C. Calhelha, L. Barros and I. C. F. R. Ferreira, J. Funct. Foods, 2019, 53, 237–247 CrossRef CAS.
- M. E. Orqueda, M. Rivas, I. C. Zampini, M. R. Alberto, S. Torres, S. Cuello, J. Sayago, S. Thomas-Valdes, F. Jimenez-Aspee, G. Schmeda-Hirschmann and M. I. Isla, Food Chem., 2017, 216, 70–79 CrossRef CAS.
- G. P. Li, A. J. Sinclair and D. Li, J. Agric. Food Chem., 2011, 59, 1871–1881 CrossRef CAS.
- L. B. Wang, F. C. Liu, A. X. Wang, Z. Y. Yu, Y. Q. Xu and Y. Yang, Food Hydrocolloids, 2017, 66, 357–364 CrossRef CAS.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS.
- R. M. Martinez, D. D. B. Guimaraes, C. R. Berniz, J. P. de Abreu, A. P. M. da Rocha, R. S. de Moura, A. C. Resende and A. J. Teodoro, Foods, 2018, 7, 178 CrossRef CAS.
- X. Liu, J. H. Xie, S. Jia, L. X. Huang, Z. J. Wang, C. Li and M. Y. Xie, Int. J. Biol. Macromol., 2017, 98, 576–581 CrossRef CAS.
- Q. Yu, S. P. Nie, W. J. Li, W. Y. Zheng, P. F. Yin, D. M. Gong and M. Y. Xie, Phytother. Res., 2017, 27, 186–191 CrossRef.
- D. Kocatepe and H. Turan, Lipids, 2012, 47, 635–641 CrossRef CAS PubMed.
- P. Puwastien, K. Judprasong, E. Kettwan, K. Vasanachitt, Y. Nakngamanong and L. Bhattacharjee, J. Food Compos. Anal., 1999, 12, 9–16 CrossRef.
- S. J. Iverson, K. J. Frost and S. L. C. Lang, Mar. Ecol.: Prog. Ser., 2002, 241, 161–181 CrossRef CAS.
- M. Zaman, M. N. Naser, A. T. M. Abdullah and N. Khan, Bangladesh J. Zool., 2014, 42, 251–259 CrossRef.
- A. D. Diwan, H. G. Hingorani and N. Chandrasekhram Naidu, Bull. Environm. Contain. Toxicol., 1979, 21, 269–272 CrossRef CAS.
- R. D. Santos, A. C. M. Gagliardi and H. T. Xavier, Arq. Bras. Cardiol., 2013, 100, 1–40 CAS.
- K. Imura and A. Okada, Nat. Conserv., 1998, 14, 143–148 CAS.
- A. M. M. Jais, R. McCulloch and K. Croft, Gen. Pharmacol., 1994, 25, 947–950 CrossRef CAS.
- G. Y. Wu, Adv. Nutr., 2010, 1, 31–37 CrossRef CAS.
- O. O. Oluwaniyi, O. O. Dosumu and G. V. Awolola, Food Chem., 2010, 123, 1000–1006 CrossRef CAS.
- T. R. Gormley, T. Neumann, J. D. Fagan and N. P. Brunton, Eur. Food Res. Technol., 2007, 225, 837–842 CrossRef CAS.
- M. Imae, T. Asano and S. Murakami, Amino Acids, 2014, 46, 81–88 CrossRef CAS PubMed.
- S. Murakami, Amino Acids, 2014, 46, 73–80 CrossRef CAS.
- Y. Yamori, T. Taguchi, A. Hamada, K. Kunimasa, H. Mori and M. Mori, J. Biomed. Sci., 2010, 17, S6 CrossRef.
- Y. Yamori, T. Taguchi, H. Mori and M. Mori, J. Biomed. Sci., 2010, 17, S21 CrossRef PubMed.
- M. Zhang, L. F. Bi, J. H. Fang, X. L. Su, G. L. Da, T. Kuwamori and S. Kagamimori, Amino Acids, 2004, 26, 267–271 CAS.
- E. O. Elvevoll, K. E. Eilertsen, J. Brox, B. T. Dragnes, P. Falkenberg, J. O. Olsen, B. Kirkhus, A. Lamglait and B. Osterud, Atherosclerosis, 2008, 200, 396–402 CrossRef CAS.
- S. Murakami, Life Sci., 2017, 186, 80–86 CrossRef CAS.
- F. T. Rosa, E. C. Freitas, R. Deminice, A. A. Jordao and J. S. Marchini, Eur. J. Nutr., 2014, 53, 823–830 CrossRef CAS.
- B. P. Mohanty, T. V. Sankar, S. Ganguly, A. Mahanty, R. Anandan, K. Chakraborty, B. N. Paul, D. Sarma, J. S. Dayal, S. Mathew, K. K. Asha, T. Mitra, D. Karunakaran, S. Chanda, N. Shahi, P. Das, P. Das, M. S. Akhtar, P. Vijayagopal and N. Sridhar, Biol. Trace Elem. Res., 2016, 174, 448–458 CrossRef CAS.
- A. Oksuz, A. Ozyilmaz and S. Kuver, Turk. J. Fish. Aquat. Sci., 2011, 11, 69–75 Search PubMed.
- F. Camara, M. A. Amaro, R. Barbera and G. Clemente, Food Chem., 2005, 92, 481–489 CrossRef CAS.
- L. Gong, H. Zhang, Y. G. Niu, L. Chen, J. Liu, S. Alaxi, P. P. Shang, W. J. Yu and L. L. Yu, J. Agric. Food Chem., 2015, 63, 569–577 CrossRef CAS.
- W. Li, X. D. Xia, W. Z. Tang, J. Ji, X. Rui, X. H. Chen, M. Jiang, J. Z. Zhou, Q. Q. Zhang and M. S. Dong, J. Agric. Food Chem., 2015, 63, 3454–3463 CrossRef CAS.
- A. Ebrahiminezhad, F. Moeeni, S. M. Taghizadeh, M. Seifan, C. Bautista, D. Novin, Y. Ghasemi and A. Berenjian, Foods, 2019, 8, 88 CrossRef.
- G. T. Chen, Y. X. Fu, W. J. Yang, Q. H. Hu and L. Y. Zhao, Int. J. Biol. Macromol., 2018, 107, 2150–2156 CrossRef CAS.
- A. E. Abd El-Salam, D. Abd-El-Haleem, A. S. Youssef, S. Zaki, G. Abu-Elreesh and S. A. El-Assar, Braz. J. Biol., 2017, 15, 335–344 Search PubMed.
- S. Busi, S. Karuganti, J. Rajkumari, P. Paramanandham and S. Pattnaik, Water Environ. J., 2017, 31, 97–104 CrossRef CAS.
- Y. Yu, M. Y. Shen, Z. J. Wang, Y. X. Wang, M. Y. Xie and J. H. Xie, Carbohydr. Polym., 2017, 174, 669–676 CrossRef CAS.
- W. X. Chen, W. Y. Zhang, W. B. Shen and K. C. Wang, Cell. Immunol., 2010, 262, 69–74 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2019 |