Open Access Article
Open Access ArticleGlycine betaine enhances biodegradation of phenol in high saline environments by the halophilic strain Oceanobacillus sp. PT-20†
Xiufeng Longa,
Denggang Wangb,
Yuqi Zoua,
Jiewei Tiana,
Yongqiang Tian *ac and
Xuepin Liaoac
*ac and
Xuepin Liaoac
aDepartment of Biomass and Leather Engineering, Sichuan University, Chengdu 610065, PR China. E-mail: yqtian@scu.edu.cn
bGuangdong Key Laboratory of Fermentation and Enzyme Engineering, South China University of Technology, Guangzhou 510006, PR China
cNational Engineering Laboratory for Clean Technology of Leather Manufacture, Sichuan University, Chengdu 610065, PR China
First published on 17th September 2019
Abstract
The halophilic bacterial strain PT-20, isolated from saline alkali soil samples and identified as a member of the genus Oceanobacillus, exhibited a robust ability to degrade phenol under high salt conditions. It was determined that strain PT-20 was capable of degrading 1000 mg L−1 phenol completely in the presence of 10% NaCl within 120 h. Under the optimal degradation conditions, pH 8.0, 3% NaCl and 30 °C, 1000 mg L−1 phenol could be completely degraded in 48 h. Interestingly, the biodegradation rate of phenol was dramatically improved in the presence of glycine betaine. When glycine betaine was added, the time required to degrade 1000 mg L−1 phenol completely was significantly reduced from 120 h to 72 h, and the corresponding average degradation rate increased from 8.43 to 14.28 mg L−1 h−1 with 10% NaCl. Furthermore, transcriptome analysis was performed to investigate the effects of phenol and glycine betaine on the transcriptional levels of strain PT-20. The results indicated that the addition of glycine betaine enhanced the resistance of cells to phenol, increased the growth rate of strain PT-20 and upregulated the expression of related enzyme genes. In addition, the results of enzyme activity assays indicated that strain PT-20 degraded phenol mainly through a meta-fission pathway.
1. Introduction
Phenol and phenolic compounds are widely distributed in the environment as contaminants because they are commonly found in wastewater from many industrial processes, including coal gasification, coking plants, oil refineries, pesticide factories, leather, textile and pharmaceutical manufacturers.1–4 Due to their toxicity and persistence, phenol and phenolic compounds might be bioaccumulated and biomagnified in the food chain.5 Moreover, they are toxic even at very low concentrations, and not only are harmful to aquatic organisms and plants but also induce cancer or cause neurotoxicity and acute toxicity to humans.6–8 Therefore, phenol is one of the most important pollutants in the environment and is considered as a priority pollutant worldwide.Conventional methods for treating wastewater containing phenol and phenolic compounds include physical, chemical and biological technologies, such as activated carbon adsorption, chlorination, ozonation, H2O2/UV treatment, membrane separation, solvent extraction and bioremediation.9 However, these strategies are costly and lead to secondary effluent problems. Bioremediation using microorganisms provides an attractive alternative treatment option because it is relatively cost-effective and environmentally compatible.
Numerous microorganisms, such as Alcaligenes, Rhodococcus, Acinetobacter, Gulosibacter, and Methylobacterium, have been used in the aerobic degradation of phenol.10–13 Jiang et al.10 reported that Alcaligenes faecalis could degrade 1600 mg L−1 phenol completely in 76 h. In Banerjee and Ghoshal's research,14 it was shown that strains Bacillus cereus MTCC 9817 and Bacillus cereus MTCC 9818 could completely degrade phenol up to 1000 mg L−1 within 60 h. However, these strains cannot tolerate high salt concentrations. In high saline conditions, the phenol pollutant degradation efficiency of microorganisms decreases dramatically. In fact, a high salt concentration can reduce cell viability as the cells dehydrate or disintegrate by osmotic differences across the cell membranes. Additionally, it would inhibit the oxygenase activity by decreasing the dissolved oxygen content in the water, and lead to a decrease in the biodegradation rate.15
Actually, salt is commonly contained in industrial wastewater.16 Currently, effective bioremediation of such contaminated environments cannot be achieved using conventional microorganisms because their growth and the bioavailability of hydrocarbons might be inhibited by high salt concentrations.17 Desalinization of wastewater is expensive and dilution with freshwater is a waste of this resource and may significantly increase wastewater volume. Thus, halophilic organisms are a promising alternative in the treatment of polluted wastewater with higher salt concentrations. The characteristic of some halophilic and halotolerant bacteria to degrade phenol has been reported in previous studies. Bastos et al. isolated two phenol-degrading strains Candida tropicalis and Alcaligenes faecalis from Amazonian rain forest soil, and the yeast could tolerate 1500 mg L−1 phenol in the presence of 15% (w/v) salt concentration 148 h, higher than the bacterium which could utilize 1100 mg L−1 phenol in the presence of 5.6% (w/v) salt with 200 h.18 Halomonas sp. can 100% degrade 100 mg L−1 phenol as its sole carbon source in the presence of 5% NaCl within 13 h.19 Similarly, the halotolerant strain Debaryomyces sp. JS4 could completely degrade 500 mg L−1 phenol in 3% (w/v) NaCl within 36 h.20 Haddadi and Shavandi2 also demonstrated that strain Halomonas sp. PH2-2 could degrade 1100 mg L−1 phenol completely in 7 days in the presence of 10% NaCl. However, these halophilic or halotolerant bacteria have some limitations that prevent the efficient degradation of high concentrations of phenol under high saline conditions. Therefore, it is still a challenge to isolate microorganisms that have the ability to degrade high phenol concentrations under high salt concentrations.
In this study, a halophilic strain PT-20 was isolated from saline-alkali soil samples, and identified as Oceanobacillus sp. PT-20. The phenol degradation ability of PT-20 was investigated, and the effects of different pH, culture temperatures and NaCl concentrations on phenol degradation were also determined. Furthermore, compatible solutes, as an important molecule used by microorganisms to resist osmotic pressure, were added to increase phenol biodegradation efficiency at high salt concentrations. How can the compatible solute (glycine betaine) enhance the biodegradation of phenol? By alleviating the environmental stress to cells? Or by inducing/regulating the expression of related genes? There is little research focused on this topic. Transcriptome analysis was carried out to investigate the effects of phenol and glycine betaine on the transcriptional levels of strain PT-20. In addition, the phenol degradation pathway of strain PT-20 was also explored.
2. Materials and methods
2.1. Sample collection and culture conditions
Strain PT-20 was isolated from saline alkali soil samples collected from Shache County, Xinjiang Province, China (38° 42′ N 77° 23′ E). Soil samples were thoroughly shaken in PT media which containing (per liter): 10 g yeast extract, 7.5 g casein peptone, 100 g NaCl, 0.016 mol phenol and 1 mL MS mixture (glycine, proline, betaine, D-sorbitol and glutamate, each 0.5 g, added to 1000 mL H2O) at 30 °C for 7 days. The suspension was diluted in gradients of 10−1, 10−2, 10−3, 10−4, 10−5 and spread onto TSB plates (0.5% soya peptone, 1.5% tryptone, m/v, pH adjusted to 8.0) supplemented with 2% agar and 10% NaCl and then cultured at 30 °C. Individual colonies on the plates were inoculated and stored on slants. Strains were tested for the phenol degrading potential.2.2. Identification of strain PT-20
The genomic DNA of PT-20 was extracted as described by Li et al.21 PCR amplification of the 16S rRNA gene was performed with the primers 27F and 1492R. Sequencing reactions were performed by Shanghai Sangon (Shanghai, China). MEGA 5.2 (ref. 22) was used to construct phylogenetic trees under maximum-likelihood,23 minimum-evolution24 and neighbor-joining25 models with bootstrap values under 1000 replications.262.3. Analytical techniques
The cell concentration of the sample was determined by measuring the optical density (OD) at 600 nm using a UV-vis spectrophotometer. Samples were taken periodically and centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800g for 10 min at 4 °C to remove cell debris, and the supernatants were filtered through a 0.22 μm filter membrane. The filtrate was analyzed by high-performance liquid chromatography (HPLC, waters). The HPLC conditions for phenol concentration determination were as follows: C18 column (4.5 mm i.d. × 250 mm, waters) with a methanol–water (50
800g for 10 min at 4 °C to remove cell debris, and the supernatants were filtered through a 0.22 μm filter membrane. The filtrate was analyzed by high-performance liquid chromatography (HPLC, waters). The HPLC conditions for phenol concentration determination were as follows: C18 column (4.5 mm i.d. × 250 mm, waters) with a methanol–water (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v) mobile phase, using a UV detector at 270 nm. The flow rate was 0.8 mL min−1.
50, v/v) mobile phase, using a UV detector at 270 nm. The flow rate was 0.8 mL min−1.
2.4. Determination of the phenol degradation ability of PT-20
The ability of strain PT-20 to degrade different concentrations of phenol was confirmed by inoculating a series of TSB medium with different initial phenol concentrations ranging from 100 to 1200 mg L−1, which were then cultured at 30 °C and 200 rpm. Strain PT-20 grown in TSB medium without phenol and nonseeded samples were used as controls. The flasks were shaken in the dark to avoid phenol photodestruction. Samples were taken periodically for the determination of biomass and phenol concentration.2.5. Effects of pH, temperature and salt concentration on phenol degradation
The optimal pH, temperature and phenol concentration for phenol degradation were determined. The experiments were performed at various temperatures ranging from 25 to 37 °C, maintaining the initial phenol concentration at 1000 mg L−1, pH 8.0 and 5% NaCl. To determine the optimum pH, various pH values ranging from 6.5 to 9.0 with an were used; the initial phenol concentration, culture temperature and NaCl concentration were 1000 mg L−1, 30 °C and 5% respectively. TSB medium supplemented with different NaCl concentrations (3, 5, 8, 10, 12 and 15%, m/v) and 1000 mg L−1 phenol were used to confirm the effect of NaCl on phenol degradation. Other inorganic salts [NaCl, KCl, Na2SO4 and MgCl2, 5% (m/v)] were also used to determine the effect on phenol degradation, and the initial phenol concentration was 500 mg L−1.2.6. Effects of compatible solutes on phenol degradation
Glycine betaine, proline, and ectoine were used to investigate the effects of additional compatible solutes on phenol degradation by the halophilic strain PT-20. Several 250 mL flasks containing 50 mL of TSB medium with different initial compatible solute concentrations were used. Glycine betaine, proline, and ectoine were added to the medium after filter sterilization. All experiments were performed in triplicate, and error bars were used to denote the standard deviation.2.7. Transcriptome analysis
To investigate the effect of phenol and glycine betaine on the transcriptional levels of strain PT-20, cells of PT-20, PT-20 supplemented with 1000 mg L−1 phenol, and PT-20 supplemented with 1000 mg L−1 phenol and 500 mg L−1 glycine betaine were used. Phenol and glycine betaine were added to TSB medium (10% NaCl) after filter sterilization at the exponential growth stage. After 45 min, bacterial cells were harvested by centrifugation at 1500g at for 10 min 4 °C. The cells were immediately frozen in liquid nitrogen to stop cellular metabolism.Total RNA was extracted using Trizol reagent (Invitrogen Life Technologies). Reverse transcriptase and random primers were used to synthesize the first-strand cDNA. dUTP was subsequently used for second-strand cDNA synthesis. The remaining overhangs were converted to blunt ends by enzymes exonuclease/polymerase. Illumina PE adapter oligonucleotides were ligated to prepare for hybridization after adenylation of the 3′ ends of the DNA fragments. UNG enzyme was used to digest the second-strand cDNA for constructing strand-specific libraries. DNA fragments with ligated adaptor molecules on both ends were selectively enriched using the Illumina PCR Primer Cocktail in a 15 cycles of PCR. Products were purified (AMPure XP beads) and quantified using Agilent high sensitivity DNA assay. The sequencing library was then sequenced on the Illumina HiSeq platform by Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China).
2.8. Enzyme activity
Enzyme activities were determined by spectrophotometry in cell-free extracts at 25 °C using quartz cuvettes. Cells were harvested by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800g at 4 °C for 10 min. Cells were resuspended in 0.1 M phosphate sodium buffer (pH 7.2) after being washed twice. The bacterial suspensions were disrupted by ultrasonication treatment (65 W, 4 × 4 s) (Y92-IIN; Scientz Biotechnology Co, Ningbo, China) for 30 min, and supernatant were obtained by centrifugation at 13
800g at 4 °C for 10 min. Cells were resuspended in 0.1 M phosphate sodium buffer (pH 7.2) after being washed twice. The bacterial suspensions were disrupted by ultrasonication treatment (65 W, 4 × 4 s) (Y92-IIN; Scientz Biotechnology Co, Ningbo, China) for 30 min, and supernatant were obtained by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800g at 4 °C for 20 min. The activities of phenol hydroxylase (PH), catechol 1,2-dioxygenase (C12O) and catechol 2,3-dioxygenase (C23O) were investigated according to Jiang et al.10 One unit of hydroxylase activity was defined as the amount of enzyme that caused the oxidation of 1 μM NADPH/min in the present of phenol. And one unit of catechol 1,2 (or 2,3)-dioxygenase activity was defined as the amount of enzyme that produced 1 μM of cis,cis-muconate or 2-hydroxymuconic semialdehyde per minute at 25 °C.
800g at 4 °C for 20 min. The activities of phenol hydroxylase (PH), catechol 1,2-dioxygenase (C12O) and catechol 2,3-dioxygenase (C23O) were investigated according to Jiang et al.10 One unit of hydroxylase activity was defined as the amount of enzyme that caused the oxidation of 1 μM NADPH/min in the present of phenol. And one unit of catechol 1,2 (or 2,3)-dioxygenase activity was defined as the amount of enzyme that produced 1 μM of cis,cis-muconate or 2-hydroxymuconic semialdehyde per minute at 25 °C.
2.9. Statistical analysis
All measurements were performed in triplicate, and error bars denote the standard deviation.3. Results and discussion
3.1. Identification of strain PT-20
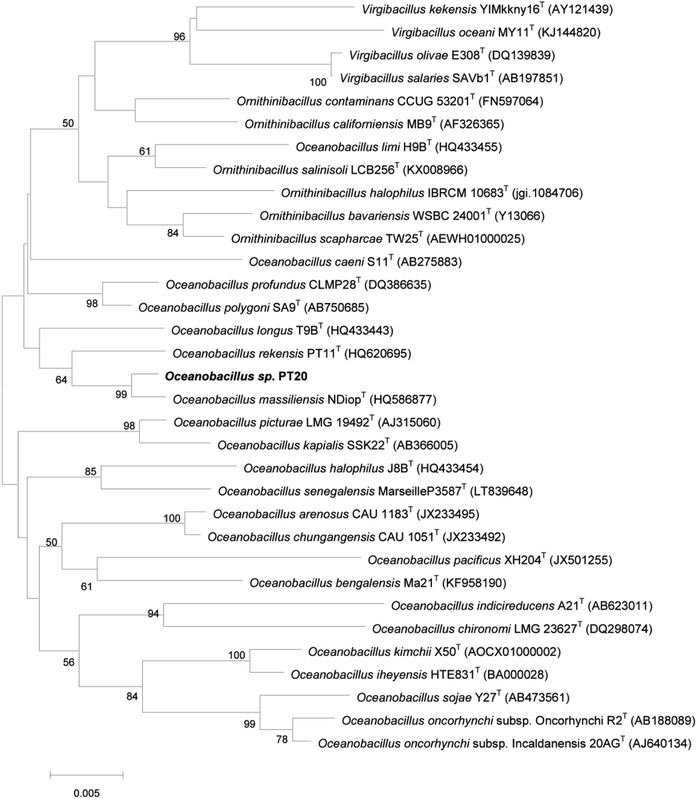
Strain PT-20 was Gram-positive, aerobic, motile and rod-shaped bacterium. Growth of strain PT-20 was found to occur at pH 6.5–10.0 (optimum 7.5–9.0), 15–40 °C (optimum 30–37 °C) and 3–15% (w/v) NaCl (optimum 5–10%). According to the comparisons of pairwise 16S rDNA sequence, PT-20 was 98.76% identical to Oceanobacillus massiliensis N'DiopT. Based on the phylogenetic analysis (Fig. 1 and S1†), strain PT-20 was found to be a typical member of the genus Oceanobacillus and was named Oceanobacillus sp. PT-20 (Genbank accession number was HQ620704).3.2. Phenol biodegradation by Oceanobacillus sp. PT-20
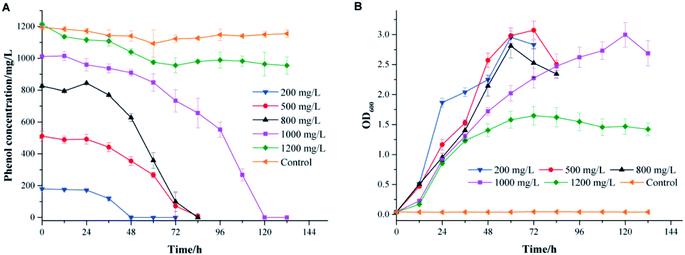
The effects of the initial phenol concentration on phenol degradation behaviors and cell growth are shown in Fig. 2. It was impressive that strain PT-20 could completely degrade phenol in 120 h when the phenol concentration was as high as 1000 mg L−1 with 10% NaCl.As shown in Fig. 2A, the time required to completely degrade phenol was prolonged as the initial phenol concentration increased. At an initial phenol concentration of 200 mg L−1, the time to degrade phenol completely was 48 h, and when the phenol concentration increased to 800 mg L−1, 84 h was needed. The corresponding average degradation rate increased from 3.74 to 9.84 mg L−1 h−1 as the initial phenol concentration increased from 200 to 800 mg L−1. However, a period of 120 h was needed to degrade 1000 mg L−1 of phenol completely, with an average phenol degradation rate of 8.43 mg L−1 h−1. In addition, as shown in Fig. 2B, the cell growth curves illustrated an increase in the bacterial cell numbers, while the lag phase prolonged when the initial phenol concentration increased. This phenomenon was similar to previous studies showing that the toxicity of phenol might affect the integrity of the cytoplasmic membrane.27–29
When the phenol concentration increased to 1200 mg L−1, only 20% of phenol was degraded, and the average degradation rate of phenol was reduced to only 1.36 mg L−1 h−1. It was reported that an obvious inhibitory effect was observed when the initial phenol concentration was higher than 1000 mg L−1.7 The same phenomenon was found with a salt-tolerant fungi that can degrade 500 mg L−1 phenol completely in 34 h, while the removal efficiency apparently reduced when the phenol concentration was higher than 800 mg L−1.30 The bacterium strain Halomonas sp. PH2-2 was able to degrade up to 1100 mg L−1 of phenol in 10% (w/v) NaCl concentration within 7 day but cell growth was inhibited at higher phenol concentrations.2 The above results indicated that strain PT-20 was a highly effective phenol degrading bacteria in high salinity.
3.3. Effect of incubation pH, temperature and NaCl concentration on phenol degradation
The phenol degradation performance of strain PT-20 with variations in temperature, pH and NaCl concentration was investigated since cell growth and enzyme activities can be markedly affected by these parameters.31As shown in Fig. 3A, the cells had high degradation rates of phenol at 28 °C, 30 °C, and 34 °C. The optimal temperature was found to be 30 °C, at which the phenol degradation efficiency in 24 h, 36 h, and 48 h was 11.7%, 85.7% and 98.9%, respectively. When the temperature dropped to 25 °C or rose to 37 °C, there was only a slight decrease in the residual concentration of phenol. This phenomenon was similar to many previous studies that the removal performance of pollutants by isolated microorganisms worsened at temperatures that were too low or too high.32–34 Regarding pH, Fig. 3B shows satisfactory performance when the pH was 8.0, at which 1000 mg L−1 phenol was completely degraded within 60 h at 5% NaCl. 94.4% and 88.6% phenol was degraded at pH 8.5 and 9.0, respectively. When the pH was lower than 7.5, the phenol degradation efficiency was significantly decreased. Extremely high or low pH values usually lead to the inactivation of most enzymes.9,35
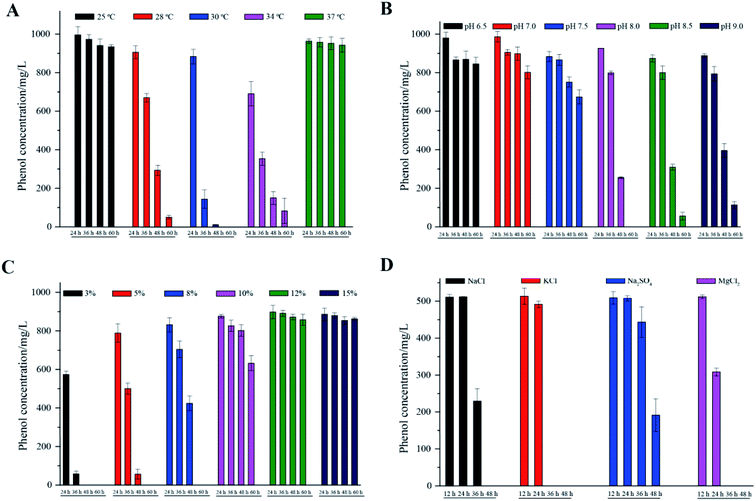 | ||
| Fig. 3 The effects of different temperatures (A), pH (B), NaCl concentrations (C) and inorganic salts (D) on phenol degradation. All experiments were performed in triplicate. | ||
Different NaCl concentrations (3, 5, 8, 10, 12 and 15%, m/v) were used to investigate the effect on phenol degradation by halophilic strain PT-20. In general, strain PT-20 exhibited relatively strong salt tolerance in terms of phenol degradation and cell growth. It was observed that the required time to completely degrade phenol (1000 mg L−1) was 60 h and 120 h when the NaCl concentration was 5% and 10%, respectively. However, as shown in Fig. 3C, the phenol-degrading ability of strain PT-20 was dramatically inhibited when the concentration of NaCl was higher than 10%, at which the high salinity and high phenol concentration likely inhibited cell metabolism.17 It was reported2 that the halophilic bacterium Halomonas sp. can effectively degrade more than 95% of 400 mg L−1 phenol under a 7% NaCl concentration, while only 88% of phenol was degraded at 12% NaCl. As the salt concentration increased from 1 M to 2 M, the time required for Acidobacterium sp. to completely degrade 120 mg L−1 phenol increased form 32 h to 72 h.36 And in the media containing 10% (w/v) total salts, halophilic bacteria Halomonas organivorans isolated from saline soil was able to degrade only 280 mg L−1 phenol within 120 h.37 From the above results, it can be suggested that high salinity can significantly decrease the biodegradation rate of most microorganisms. Strain PT-20 has high phenol degradation efficiency under high salt conditions, indicating that it is an excellent strain for the effective degradation of phenol in high salt concentration environments.
Since other inorganic salts often exist in phenol-containing wastewater, the biodegradation behavior of phenol by strain PT-20 with other inorganic salts was further investigated. The phenol degradation results of PT-20 in the presence of NaCl, KCl, Na2SO4 and MgCl2 are summarized in Fig. 3D. It was found that PT-20 can completely degrade 500 mg L−1 phenol in the presence of three inorganic salts in 48 h, but there was a little difference in the phenol biodegradation rate. In the presence of KCl and MgCl2, the phenol degradation rate was significantly faster than that of NaCl and Na2SO4, and a similar phenomenon was observed for cell growth. It was suggested that KCl and MgCl2 can contribute to the increase in the cell viability and metabolic rate;38 therefore, the phenol biodegradation rate was also increased. According to the results, PT-20 exhibited broad salt tolerance in phenol degradation.
3.4. Effects of compatible solutes on phenol degradation
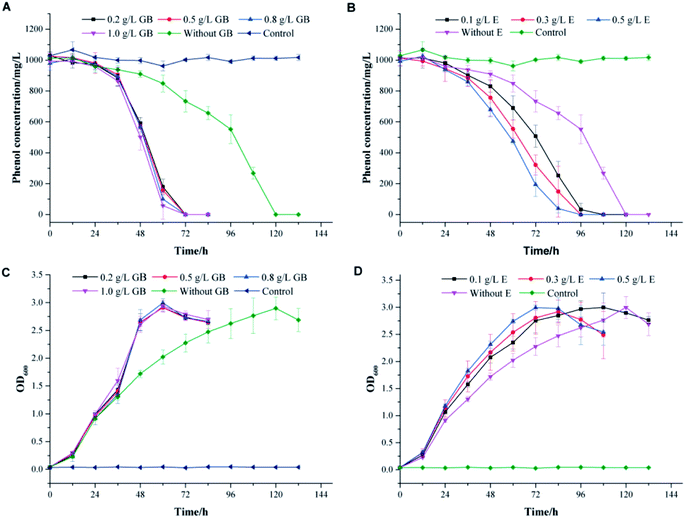
To thrive in a high-salinity environment, most of prokaryotes response to the increasing osmotic pressure through the organic-osmolyte mechanism, which control the synthesis or uptake of compatible solutes.39,40 They can achieve high intracellular concentrations without interfering with important cellular functions.41–43 In high salt conditions, the toxicity of phenol might break the physiological balance of cells and lead to the readjustment of the osmotic equilibrium. The addition of compatible solutes might effectively alleviate the stress effect of high salinity on cells. Glycine betaine, ectoine and proline are compatible solutes commonly used by microorganisms to resist salt stress.44–46 In this work, these compounds were used to investigate the effects of additional compatible solutes on phenol degradation behaviors in high salinity by strain PT-20.The profiles of phenol degradation and cell growth by PT-20 at different concentrations of compatible solutes are shown in Fig. 4. At an initial phenol concentration of 1000 mg L−1 in TSB medium with 10% NaCl, approximately 120 h was required to completely degrade phenol without the addition of any compatible solutes. It was impressive that the time was significantly reduced when glycine betaine and ectoine were added to the system (Fig. 4A and B). However, this effect was not observed for proline (data not shown). In Fig. 4A, the complete degradation of 1000 mg L−1 phenol only took 72 h in TSB medium supplemented with glycine betaine. When the concentration of glycine betaine (GB) was increased from 0.1 to 1.0 g L−1, the degradation efficiency of phenol was slightly improved, suggesting that only a small dose of glycine betaine is sufficient to resist salt stress. As shown in Fig. 4B, TSB medium supplemented with ectoine can also improve the phenol degradation efficiency. When the concentration of ectoine (E) was increased from 0.1 to 0.5 g L−1, the degradation efficiency correspondingly increased, indicating the dose-dependent effect of ectoine on salt stress resistance. In addition, the cell growth rate also increased with glycine betaine and ectoine, as shown in Fig. 4C and D, leading to rapid degradation of phenol.
To confirm the effect of glycine betaine on phenol degradation by PT-20, the degradation performances of phenol with various concentrations of phenol and salt were investigated, as shown in the ESI (Fig. S3†). A similar phenomenon was observed in the samples, and it was impressively that 1200 mg L−1 phenol was completely degraded after 120 h when glycine betaine was added, which was only degraded by 20% after 132 h in the glycine betaine-free system.
These results lead us to infer that the addition of glycine betaine can effectively alleviate the stress effect of high salinity on cells, enabling the cells to cope with phenol toxicity. Glycine betaine is a simple but useful osmoprotectant for many prokaryotic organisms. This versatile solute is efficiently taken up by many bacteria that use it to cope with salt stress.46 Glycine betaine has also been shown to enhance the physiological activity of cyanobacterium Synechococcus sp. and Listeria monocytogenes at cold temperatures.47,48 Acting as an important osmoprotectant, glycine betaine has great potential to help cells resist stress under high salinity, low temperature or other harsh environment.
3.5. Transcriptome analysis
To investigate the effects of phenol and glycine betaine on the transcriptional levels of strain PT-20, transcriptome analysis of three groups (PT-20, PT-20 supplemented with 1000 mg L−1 phenol and PT-20 supplemented with 1000 mg L−1 phenol and 500 mg L−1 glycine betaine) was performed. Illumina HiSeq sequencing platform was used for transcriptome analysis. Reads were mapped onto the reference genome, and TPM values were used for comparison. Differentially expressed genes (DEGs) were identified in the case of p-adjust <0.001 and |log2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FC| > 1.
FC| > 1.
Based on transcriptome analysis, we performed hierarchical clustering of the significantly DEGs of PT-20 cultured in different conditions, the heat map exhibited significant differences in gene expression with addition of phenol and glycine betaine (Fig. 5, Table S1†). The transcriptome analysis in response to 1000 mg L−1 phenol stress revealed the induction of 464 genes and repression of 1782 genes after 45 min. In previous research, the protective mechanism under phenolic compound stress was studied, and the results indicated that phenol caused the activation of heat-shock regulons and oxidative stress response.49,50 In our work, the HrcA, σB and CtsR heat-shock regulons were significantly upregulated (Table 1), while no obvious response was found in oxidative stress proteins, which was different from previous reports.50,51 Ten general stress proteins were found to respond to phenol stress (upregulated 2.49–38.06-fold). Most membrane proteins were significantly downregulated, which was consistent with Putrinš et al.52 showing that phenol was toxic to cell membranes. The cell membrane acted as the first barrier against external stress, and to resist the toxicity of phenol, some membrane proteins were upregulated (2–26-fold) (Table 1), which was important for resisting phenol stress.
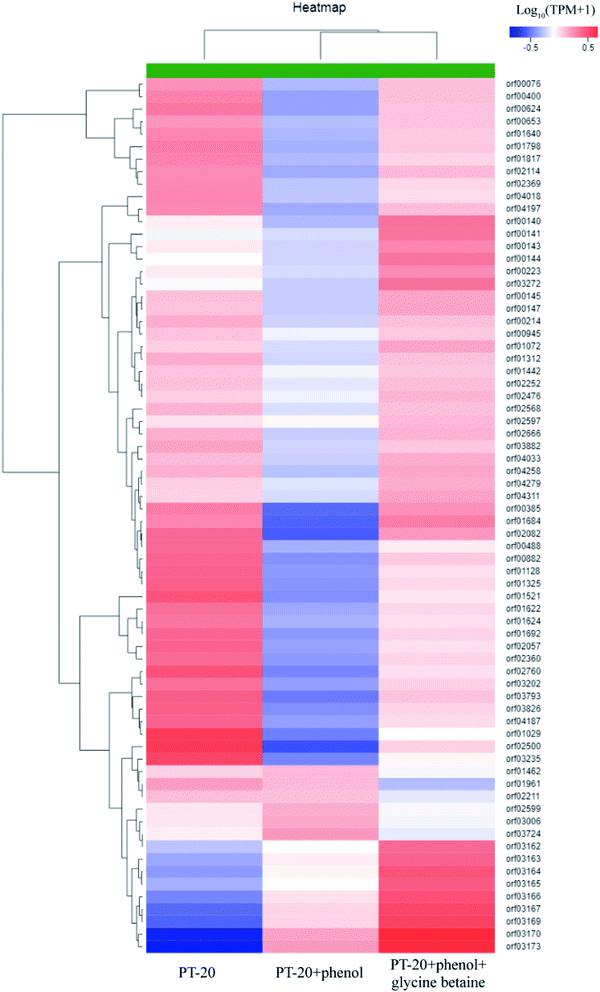 | ||
| Fig. 5 Hierarchical clustering of features of the whole transcriptome of PT-20 cultured in different conditions (PT-20 without phenol and glycine betaine; PT-20 supplemented with 1000 mg L−1 phenol; PT-20 supplemented with 1000 mg L−1 phenol and 500 mg L−1 glycine betaine). The heat map represented the expression of part genes in Table S1.† | ||
| Regulon | Gene | Gene id | Gene description | FC (PT20P/PT20) | FC (PT20PG/PT20P) |
|---|---|---|---|---|---|
| orf01045 | Phenol hydroxylase | 5.09 | 1.25 | ||
| dmpB | orf01044 | Catechol 1,2-dioxygenase | 2.42 | 1.33 | |
| dmpB | orf01046 | Catechol 2,3-dioxygenase | 7.15 | 1.30 | |
| HrcA | dnaK | orf01732 | Molecular chaperone DnaK | 1.87 | 0.83 |
| hrcA | orf01730 | HrcA family transcriptional regulator | 2.86 | 0.69 | |
| groES | orf01396 | Molecular chaperone GroES | 3.37 | 0.70 | |
| groEL | orf01397 | Molecular chaperone GroEL | 3.56 | 0.62 | |
| σ | csbD | orf00540 | Stress response protein CsbD | 14.75 | 0.79 |
| CtsR | clpE | orf02939 | ATP-dependent Clp protease ATP-binding protein | 2.01 | 0.56 |
| clpC | orf03313 | ATP-dependent Clp protease ATP-binding protein | 3.81 | 0.81 | |
| ctsR | orf03310 | CtsR family transcriptional regulator | 7.05 | 0.58 | |
| SPX | mecA1 2 | orf03550 | Adaptor protein | 3.48 | 0.67 |
| Others | Fnr | orf02527 | Crp/Fnr family transcriptional regulator | 0.22 | 0.95 |
| K06884 | orf01488 | General stress protein B | 8.69 | 0.95 | |
| orf03645 | General stress protein 39 | 38.06 | 0.78 | ||
| pfpI | orf04081 | General stress protein 18 | 13.58 | 0.89 | |
| orf02394 | General stress protein 17M | 19.18 | 0.70 | ||
| orf03489 | General stress protein | 25.39 | 0.86 | ||
| orf04007 | General stress protein | 4.59 | 1.09 | ||
| orf04006 | General stress protein | 4.48 | 1.06 | ||
| orf01907 | General stress protein | 2.56 | 1.14 | ||
| orf01906 | Multispecies: General stress protein | 2.49 | 1.15 | ||
| E1.2.1.3 | orf01060 | Aldehyde dehydrogenase | 30.38 | 0.77 | |
| murA | orf02845 | UDP-N-acetylglucosamine | 12.93 | 0.80 | |
| 1-Carboxyvinyltransferase | |||||
| treC | orf00661 | Trehalose-6-phosphate hydrolase | 6.56 | 0.92 | |
| scrK | orf01802 | Sugar kinase | 10.76 | 0.81 | |
| sacA | orf02140 | Sucrase-6-phosphate hydrolase (gene sacA) | 7.21 | 0.68 | |
| gabD | orf01878 | Succinate-semialdehyde dehydrogenase | 7.47 | 0.67 | |
| rpe | orf02203 | Ribulose-phosphate 3-epimerase | 3.63 | 0.75 | |
| exaA | orf00623 | Quinonprotein alcohol dehydrogenase | 4.16 | 0.78 | |
| crr | orf02473 | PTS glucose transporter subunit IIA | 2.34 | 1.14 | |
| treB | orf00662 | PTS beta-glucoside transporter subunit IIABC | 7.38 | 0.91 | |
| Pta | orf02791 | Phosphotransacetylase | 3.48 | 0.81 | |
| deoB | orf02646 | Phosphopentomutase | 10.83 | 0.82 | |
| nanE | orf03905 | N-acetylmannosamine-6-phosphate 2-epimerase | 5.29 | 0.71 | |
| sqd1 | orf02123 | Multispecies: NAD-dependent dehydratase | 2.11 | 1.26 | |
| prpB | orf00566 | Methylisocitrate lyase | 2.87 | 0.69 | |
| E3.2.1.41 | orf00889 | Glycogen debranching enzyme | 9.26 | 0.63 | |
| GPI | orf03085 | Glucose-6-phosphate isomerase | 3.92 | 0.85 | |
| glmS | orf00912 | Glucosamine-fructose-6-phosphate | 5.31 | 0.71 | |
| Aminotransferase | |||||
| frmA | orf00622 | Dehydrogenase | 22.25 | 0.82 | |
| gltA | orf00564 | Citrate synthase 3 | 3.77 | 0.64 | |
| katE | orf00869 | Catalase | 3.10 | 1.10 | |
| mhpD | orf01416 | Alcohol dehydrogenase | 2.63 | 0.81 | |
| ACSS | orf01063 | Acetyl-CoA synthetase | 3.09 | 1.05 | |
| accB | orf00586 | Acetyl-CoA carboxylase | 7.12 | 0.66 | |
| hxlA | orf02204 | 3-Hexulose-6-phosphate synthase | 4.08 | 0.70 | |
| prpD | orf00565 | 2-Methylcitrate dehydratase | 3.15 | 0.66 | |
| katE | orf02549 | Hydroperoxidase II | 15.77 | 0.79 | |
| butA | orf00125 | Hypothetical protein | 18.09 | 1.00 | |
| E3.2.1.10 | orf00659 | Hypothetical protein | 2.98 | 1.01 | |
| mmsA | orf00569 | Hypothetical protein | 2.32 | 0.72 | |
| IMPA | orf04266 | Hypothetical protein | 2.02 | 0.98 | |
| orf02489 | Glycine/betaine ABC transporter permease | 3.10 | 1.09 | ||
| orf02720 | Glycine/betaine ABC transporter permease | 48.09 | 0.86 | ||
| orf00513 | Glyoxalase | 9.76 | 2.31 | ||
| orf04003 | N-acetylmuramoyl-L-alanine amidase | 2.75 | 2.08 | ||
| orf01320 | Unknown | 9.15 | 2.47 | ||
| orf02933 | Unknown | 2.75 | 2.30 | ||
| orf02358 | Hypothetical protein | 3.18 | 2.34 | ||
| orf02994 | Hypothetical protein | 1.83 | 2.96 | ||
| orf03162 | Hypothetical protein | 2.15 | 3.28 | ||
| orf03163 | Hypothetical protein | 3.07 | 2.98 | ||
| orf03164 | Type II secretion system protein E | 3.35 | 3.64 | ||
| orf03165 | Hypothetical protein, partial | 2.64 | 3.84 | ||
| orf03166 | Hypothetical protein | 4.95 | 3.39 | ||
| orf03167 | Hypothetical protein | 9.73 | 3.42 | ||
| orf03169 | Hypothetical protein | 11.89 | 3.65 | ||
| orf03170 | Hypothetical protein | 45.75 | 3.08 | ||
| orf03171 | Hypothetical protein | 10.98 | 2.10 | ||
| orf03172 | Hypothetical protein | 12.63 | 2.26 | ||
| orf03173 | Hypothetical protein | 17.59 | 2.68 | ||
| orf03187 | Hypothetical protein | 1.83 | 2.96 |
Similar to other studies, most of the carbon metabolism was significantly inhibited50 while the metabolic pathways associated with phenol metabolism were correspondingly upregulated. Of 149 genes associated with carbohydrates according to KEGG enrichment analyses, 31 and 118 genes were significantly upregulated and downregulated, respectively (Table 1). Succinic acid, oxaloacetate and acetyl-CoA, as the intermediates in phenol degradation, eventually enter the TCA cycle. Transcriptome analysis indicated that the metabolism of acetyl-CoA and acetyl compounds was significantly upregulated (2.63–30.38-fold) under phenol stress.
PH, C23O and C12O, the key enzymes in phenol biodegradation, were located in the same operon and upregulated 5.09-, 2.42- and 7.15-fold, respectively, under phenol stress. Both C12O and C23O showed significant upregulation, indicating that the two pathways were induced in this period. Notably, although the expression of most genes downregulated, the expression of some glycine/betaine ABC transporter permease genes was significantly upregulated under the stress of phenol, and the highest was 48-fold relative to that of the phenol-free system. These results indicated that glycine betaine played an important role in resisting phenol stress. Glycine betaine is an important osmoprotectant for Gram-positive bacteria, and osmotic induced the expression of glycine/betaine ABC transporter permease has been reported in many reports.53,54 Strain PT-20 degraded phenol under high salt concentrations, the toxicity of phenol might break the physiological balance of the cells and lead to the readjustment of the osmotic equilibrium. This is consistent with the inhibition of cell growth.
The addition of glycine betaine caused an induction of 272 genes. Among them, 16 genes were upregulated with the addition of phenol and continued to be upregulated after the addition of glycine betaine (Table 1). Of the 16 genes, 10 genes with unknown functions (mainly flp-family type proteins) were close to each other in position, which might be related to the resistance of phenol stress. The addition of glycine betaine further increased their expression and accelerated the degradation of phenol. Of the remaining 6 genes, two of which were an N-acetylmuramoyl-L-alanine amidase and a glyoxalase involved in the metabolism of the phenol degradation process, while the functions of the other four genes were unknown.
Thirty-seven genes did not discernibly change when phenol was added but were significantly upregulated after the addition of phenol and glycine betaine. A total of 216 genes were significantly downregulated after the addition of phenol and upregulated after the addition of glycine betaine. They were mostly related to amino acid metabolism, cell division and cell wall, indicating that the addition of glycine betaine can effectively reduce the toxicity of phenol to cells. Compared with the glycine betaine-free system, the general stress protein expression was upregulated, and the carbon source metabolism was restored to some extent. These results indicated that glycine betaine relieved the pressure of phenol and reduced the toxicity of phenol to cells.
Among the 272 upregulated genes, 25 were involved in transport. Transporters are important for the uptake and utilization of phenol53,55 and the upregulation of transporters after the addition of glycine betaine is an important reason for the accelerated degradation of phenol. The remaining genes were involved in energy metabolism, cell division, carbon source metabolism and some genes of unknown function. In addition, the expression of PH, C12O and C23O was upregulated to some extent after the addition of glycine betaine, which played an essential role in improving the degradation efficiency of phenol. In brief, the addition of glycine betaine enhanced the resistance of cells to phenol, increased the growth rate of strain PT-20 and increased the expression of related enzyme genes, thereby improving the degradation efficiency of phenol.
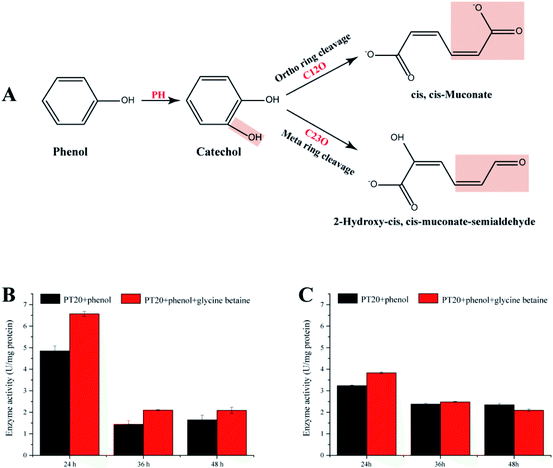
3.6. Enzyme activities
The present study demonstrates that strain PT-20 has a high phenol degradation ability at high salt concentrations and that the addition of glycine betaine could significantly improve the degradation efficiency. Therefore, it is significant to study the phenol degradation process of PT-20. As shown in Fig. 6A, aerobic phenol degradation was usually first catalyzed by PH to catechol, followed C12O (ortho pathway) or C23O (meta pathway) catalyzed the catechol ring fission.56,57 These two steps are crucial for typical microbial degradation of phenol. To understand the phenol degradation mechanism of PT-20, the relatively enzyme activities were determined.Both PH and C23O were detected (Fig. 6B and C), whereas almost no C12O activity was observed in cell-free suspensions, indicating that phenol was mainly degraded by meta pathway in strain PT-20. According to transcriptome analysis, the expression of C12O and C23O were both upregulated under phenol stress after 45 min, indicating that strain PT-20 activated both two pathways during the stress period, while the strain degrade phenol through meta pathway in the late degradation phase. After the addition of glycine betaine, the activities of PH and C23O were improved, which were consistent with the results of degradation efficiency and transcriptome analysis. Certainly, it cannot not be excluded that the faster cell growth rate in the present of glycine betaine also slightly raised the phenol degradation rate by PT-20.
4. Conclusions
In this study, we isolated a moderately halophilic strain Oceanobacillus sp. PT-20 and investigated its phenol biodegradation performances. It was found that this strain has robust ability to degrade phenol with high salts stress. At the optimal degradation condition, pH 8.0, 3% NaCl and 30 °C, 1000 mg L−1 phenol was completely degraded in 48 h. The biodegradation rate of phenol could be obviously improved in the presence of glycine betaine. The addition of glycine betaine enhanced the resistance of cells to phenol, increased the growth rate of strain PT-20 and upregulated the expression of related enzyme genes, thereby improving the degradation efficiency of phenol. In summary, this halophilic strain showed great potential to apply into high-salt phenol contained wastewater treatment, and glycine betaine also exhibited excellent application potential in harsh environment.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFC1802201).References
- B. S. D. Araujo, B. V. Charlwood and M. Pletsch, Environ. Pollut., 2002, 117, 329–335 CrossRef PubMed.
- A. Haddadi and M. Shavandi, Int. Biodeterior. Biodegrad., 2013, 85, 29–34 CrossRef CAS.
- W. Jiang, J. Yang, X. Wang, H. Han, Y. Yang, J. Tang and Q. Li, Bioresour. Technol., 2018, 247, 1246–1248 CrossRef CAS PubMed.
- O. Lefebvre, N. Vasudevan, M. Torrijos, K. Thanasekaran and R. Moletta, Water Res., 2005, 39, 1471–1480 CrossRef CAS PubMed.
- Z. Z. Huang, P. Wang, H. Li, K. F. Lin, Z. Y. Lu, X. J. Guo and Y. D. Liu, Int. Biodeterior. Biodegrad., 2014, 94, 115–120 CrossRef CAS.
- F. A. Banat, B. Al-Bashir, S. Al-Asheh and O. Hayajneh, Environ. Pollut., 2000, 107, 391–398 CrossRef CAS PubMed.
- Y. Jiang, Y. Shang, K. Yang and H. Wang, Appl. Microbiol. Biotechnol., 2016, 100, 1883–1890 CrossRef CAS PubMed.
- O. Lefebvre and R. Moletta, Water Res., 2006, 40, 3671–3682 CrossRef CAS PubMed.
- A. Banerjee and A. K. Ghoshal, J. Hazard. Mater., 2010a, 176, 85–91 Search PubMed.
- Y. Jiang, J. Wen, J. Bai, X. Jia and Z. Hu, J. Hazard. Mater., 2007, 147, 672–676 CrossRef CAS PubMed.
- P. Khongkhaem, A. Intasiri and E. Luepromchai, Lett. Appl. Microbiol., 2011, 52, 448–455 CrossRef CAS PubMed.
- L. Křiklavová, M. Truhlář, P. Škodová, T. Lederer and V. Jirků, Bioresour. Technol., 2014, 167, 510–513 CrossRef PubMed.
- Z. Zhai, H. Wang, S. Yan and J. Yao, J. Chem. Technol. Biotechnol., 2012, 87, 105–111 CrossRef CAS.
- A. Banerjee and A. K. Ghoshal, Bioresour. Technol., 2010b, 101, 5501–5507 Search PubMed.
- R. J. Von Wedel, J. F. Mosquera, C. D. Goldsmith, G. R. Hater, A. Wong, T. A. Fox, W. T. Hunt, M. S. Paulies, J. M. Quiros and J. W. Wiegand, Water Sci. Technol., 1988, 20, 501–503 CrossRef CAS.
- S. Le Borgne, D. Paniagua and R. Vazquez-Duhalt, J. Mol. Microbiol. Biotechnol., 2008, 15, 74–92 CrossRef CAS PubMed.
- L. C. Castillo-Carvajal, J. L. Sanz-Martín and B. E. Barragán-Huerta, Environ. Sci. Pollut. Res., 2014, 21, 9578–9588 CrossRef CAS PubMed.
- A. E. R. Bastos, D. H. Moon, A. Rossi, J. T. Trevors and S. M. Tsai, Arch. Microbiol., 2000, 174, 346–352 CrossRef CAS PubMed.
- C. Hinteregger and F. Streichsbier, Biotechnol. Lett., 1997, 19, 1099–1102 CrossRef CAS.
- Y. Jiang, Y. Shang, K. Yang and H. Wang, Appl. Microbiol. Biotechnol., 2016, 100, 1883–1890 CrossRef CAS PubMed.
- W. J. Li, P. Xu, P. Schumann, Y. Q. Zhang, R. Pukall, L. H. Xu, E. Stackebrandt and C. L. Jiang, Int. J. Syst. Evol. Microbiol., 2007, 57, 1424–1428 CrossRef PubMed.
- K. Tamura, D. Peterson, N. Peterson, G. Stecher, M. Nei and S. Kumar, Mol. Biol. Evol., 2011, 28, 2731–2739 CrossRef CAS PubMed.
- J. Felsenstein, J. Mol. Evol., 1981, 17, 368–376 CrossRef CAS PubMed.
- A. Rzhetsky and M. Nei, Mol. Biol. Evol., 1992, 9, 945–967 CAS.
- N. Saitou and M. Nei, Mol. Biol. Evol., 1987, 4, 406–425 CAS.
- J. Felsenstein, Evolution, 1985, 39, 783–791 CrossRef PubMed.
- H. J. Heipieper, H. Keweloh and H. J. Rehm, Appl. Environ. Microbiol., 1991, 57, 1213–1217 CAS.
- H. Keweloh, G. Weyrauch and H. J. Rehm, Appl. Microbiol. Biotechnol., 1990, 33, 66–71 CrossRef CAS PubMed.
- Y. Wang, H. Chen, Y. X. Liu, R. P. Ren and Y. K. Lv, Bioresour. Technol., 2016, 211, 711–719 CrossRef CAS PubMed.
- Y. Jiang, K. Yang, H. Wang, Y. Shang and X. Yang, Mar. Pollut. Bull., 2015, 99, 230–234 CrossRef CAS PubMed.
- B. Bhunia, B. Basak, T. Mandal, P. Bhattacharya and A. Dey, Int. J. Biol. Macromol., 2013, 54, 1–8 CrossRef CAS PubMed.
- S. S. Meena, R. S. Sharma, P. Gupta, S. Karmakar and K. K. Aggarwal, J. Basic Microbiol., 2016, 56, 369–378 CrossRef CAS PubMed.
- P. Satapute and B. Kaliwal, 3 Biotech., 2016, 6, 1–10 CrossRef PubMed.
- T. Deng, H. Wang and K. Yang, Phenol biodegradation by isolated Citrobacter strain under hypersaline conditions, Water Sci. Technol., 2018, 77, 504–510 CrossRef CAS PubMed.
- B. J. Tibbles and A. A. W. Baecker, Environ. Pollut., 1989, 59, 227–239 CrossRef CAS PubMed.
- Z. Z. Huang, P. Wang, H. Li, K. F. Lin, Z. Y. Lu, X. J. Guo and Y. D. Liu, Int. Biodeterior. Biodegrad., 2014, 94, 115–120 CrossRef CAS.
- M. R. L. Bonfá, M. J. Grossman, F. Piubeli, E. Mellado and L. R. Durrant, Biodegradation, 2013, 24, 699–709 CrossRef PubMed.
- D. Roy, Lett. Appl. Microbiol., 1991, 12, 207–211 CrossRef CAS.
- X. Long, J. Tian, X. Liao and Y. Tian, RSC Adv., 2018, 8, 27525–27536 RSC.
- A. R. Strom and I. Kaasen, Mol. Microbiol., 1993, 8, 205–210 CrossRef CAS PubMed.
- K. Strange, Cellular and molecular physiology of cell volumeregulation, CRC Press, 1993 Search PubMed.
- M. T. Record Jr, E. S. Courtenay, D. S. Cayley and H. J. Guttman, Trends Biochem. Sci., 1998, 23, 143–148 CrossRef CAS.
- P. Lamosa, L. O. Martins, M. S. da Costa and H. Santos, Appl. Environ. Microbiol., 1998, 64, 3591–3598 CAS.
- K. Gouffi and C. Blanco, Int. J. Food Microbiol., 2000, 55, 171–174 CrossRef CAS PubMed.
- P. Shivanand and G. Mugeraya, Curr. Sci., 2011, 1516–1521 CAS.
- N. Empadinhas and M. S. Da Costa, Contributions to Science, 2009, 95–105 Search PubMed.
- P. Deshnium, Z. Gombos, Y. Nishiyama and N. Murata, J. Bacteriol., 1997, 179, 339–344 CrossRef CAS PubMed.
- A. S. Angelidis and G. M. Smith, Appl. Environ. Microbiol., 2003, 69, 7492–7498 CrossRef CAS PubMed.
- A. Petersohn, M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker and M. Hecker, J. Bacteriol., 2001, 183, 5617–5631 CrossRef CAS PubMed.
- T. Tamle, C. Eymann, D. Albrecht, R. Sietmann, F. Schauer, M. Hecker and H. Antelmann, Environ. Microbiol., 2010, 8, 1408–1427 Search PubMed.
- M. G. Giuffrida, E. Pessione, R. Mazzoli, G. Dellavalle, C. Barello, A. Conti and C. Giunta, Electrophoresis, 2015, 22, 1705–1711 CrossRef.
- M. Putrinš, H. Ilves, L. Lilje, M. Kivisaar and R. Hõrak, BMC Microbiol., 2010, 10, 110 CrossRef PubMed.
- A. S. Yaakop, K. G. Chan, R. Ee, Y. L. Lim, S. K. Lee, F. A. Manan and K. M. Goh, Sci. Rep., 2016, 6, 33660 CrossRef CAS PubMed.
- L. Liu, L. Si, X. Meng and L. Luo, J. Ind. Microbiol. Biotechnol., 2015, 42, 601–616 CrossRef CAS PubMed.
- W. Wu, H. Huang, Z. Ling, Z. Yu, Y. Jiang, P. Liu and X. Li, Ecotoxicology, 2016, 25, 234–247 CrossRef CAS PubMed.
- A. Gaal and H. Y. Neujahr, J. Bacteriol., 1979, 137, 13–21 CAS.
- X. Wei, T. Gilevska, F. Wetzig, C. Dorer, H. H. Richnow and C. Vogt, Environ. Pollut., 2016, 210, 166–173 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra05163e |
| This journal is © The Royal Society of Chemistry 2019 |