Open Access Article
Open Access ArticleDevelopment of improved dual-diazonium reagents for faster crosslinking of tobacco mosaic virus to form hydrogels†
Dejun Ma‡
 b,
Zhuoyue Chen‡a,
Long Yi
b,
Zhuoyue Chen‡a,
Long Yi *a and
Zhen Xi*bc
*a and
Zhen Xi*bc
aState Key Laboratory of Organic-Inorganic Composites, Beijing University of Chemical Technology, Beijing 100029, China. E-mail: yilong@mail.buct.edu.cn
bState Key Laboratory of Elemento-Organic Chemistry, Department of Chemical Biology, National Pesticide Engineering Research Center (Tianjin), Nankai University, Tianjin, 300071, China. E-mail: zhenxi@nankai.edu.cn; Tel: +86 22 23504782
cCollaborative Innovation Center of Chemical Science and Engineering, Nankai University, Tianjin, 300071, China
First published on 17th September 2019
Abstract
New bench-stable reagents with two diazonium sites were designed and synthesized for protein crosslinking. Because of the faster diazonium-tyrosine coupling reaction, hydrogels from the crosslinking of tobacco mosaic virus and the reagent DDA-3 could be prepared within 1 min at room temperature. Furthermore, hydrogels with the introduction of disulfide bonds via DDA-4 could be chemically degraded by dithiothreitol. Our results provided a facile approach for the direct construction of virus-based hydrogels.
Introduction
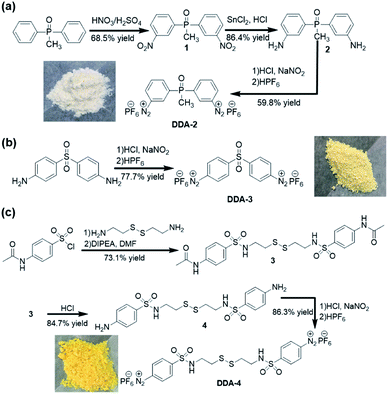
Hydrogels are widely used as biomedical materials in manufacturing cell culture matrices,1 tissue engineering scaffolds,2 drug delivery systems3 and wound dressings4 due to their unique properties, such as water retention capacity, softness, controllability and biocompatibility.5 One of the main approaches for preparing hydrogels is the chemical crosslinking of hydrophilic polymers (natural or synthetic) to provide desirable mechanical and chemical properties.6 As natural hydrophilic polymers, plant viruses like tobacco mosaic virus (TMV, 300 nm × 18 nm) exhibit a nanoscale size and abundant amino acid residues on their surface of capsid proteins, which facilitate facile chemical reactions to produce nanomaterials with multiple functions.7 Furthermore, plant viruses could be largely produced in gram-scale quantities and are also considered much safer to mammals than human-associated viruses like adenovirus and lentivirus in drug delivery and tissue engineering.8 Hence, using of plant viruses as building blocks to assemble virus-based hydrogels is attractive in biomedical applications.9Recently, there are mainly two approaches to utilizing plant viruses to construct hydrogels. Firstly, mixture of TMV with hydrogels alters their properties. As reported, the incorporation of TMV into the porous alginate hydrogels greatly reduced the immunogenicity in mice, and the covalent incorporation of tobacco mosaic virus into poly (ethylene glycol) diacrylate hydrogels increased the stiffness.10 Secondly, the hydrogels are directly made through chemically crosslinking TMV virus matrix, which was rarely explored. In our previous work, a TMV-based hydrogel was prepared by crosslinking TMV with a dual-diazonium reagent DDA-1 (Scheme 1).11 However, the virus-based hydrogels should be made within at least 30 min at 37 °C due to the relatively low diazonium-tyrosine (Tyr) coupling reaction12 between DDA-1 and Tyr residues.
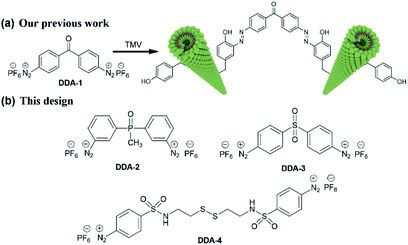 | ||
| Scheme 1 (a) Schematic illustration for crosslinking via TMV and DDA-1 in our previous work; (b) the chemical structure of new dual-diazonium reagents (DDA-2, DDA-3 and DDA-4). | ||
In this work, we designed and synthesized three new dual-diazonium reagents (DDA-2, DDA-3, DDA-4) to investigate the effects of substituents (CO, PO, SO2) on the crosslinking efficiency and gelation conditions. To our delight, DDA-3/DDA-4 could quickly crosslink TMV matrix as hydrogels within 1 min at room temperature. Moreover, the DDA-4-based hydrogel could be efficiently degraded with reducing chemicals like dithiothreitol (DTT), which enhanced the biocompatibility and biodegradability of such hydrogel.
Results and discussion
As shown in Scheme 2, DDA-2 was obtained by nitration, reduction, diazotization and hexafluorophosphoric acid precipitation from the inexpensive methyl biphenyl phosphine oxide. The final product DDA-2 (white solid) was obtained after mixing 2 with the concentrated HCl, NaNO2 and HPF6 in one-pot. DDA-3 (light yellow solid) could be prepared from 4,4′-diaminodiphenyl sulfone in one-pot synthesis. For DDA-4, cystamine was firstly reacted with 4-acetamidobenzenesulfonyl chloride to provide 3, which was then hydrolysed with the concentrated HCl to give 4. After treatment of the concentrated HCl, NaNO2 and HPF6 in one-pot, a yellow solid DDA-4 was obtained with good yield.These dual-diazonium reagents (DDA-2, DDA-3 and DDA-4) were well characterized by 1H NMR, 13C NMR, 31P NMR (see the ESI†). Compared with the chemical shift of 2 in 1H NMR (less than 8 ppm), the high chemical shift (9.19, 8.86, 8.63 and 8.17 ppm) of DDA-2 implied the existence of a positively charged diazonium moiety. Affected by phosphorus, the methyl group in DDA-2 was split into two peaks in 1H NMR and 13C NMR. The 31P NMR implied the existence of two kinds of phosphorus, in which the seven peaks were the result of the coupling of six fluorine nucleuses in hexafluorophosphate. Since the dual-diazonium salt was not successfully characterized by mass spectrum, we tried to react DDA-2 with 4,4′-methylenediphenol to get 5. The structure of 5 was clearly confirmed by 1H NMR, 13C NMR spectra and high-resolution mass spectra (HRMS), which was indirectly verified that the obtained diazonium salt was DDA-2 (see the ESI†). As for DDA-3 and DDA-4, the high chemical shift values in 1H NMR compared to their aromatic amine feedstock also implied the existence of positively charged diazo moieties. All dual-diazonium hexafluorophosphates were bench-stable and could be stored in the refrigerator (−20 °C) for at least two months.
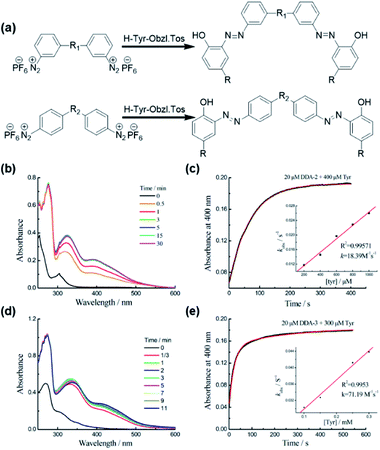
We firstly studied the reaction of DDA-2 or DDA-3 with Tyr using the UV-Vis spectrophotometry (Fig. 1). The time-dependent absorption changes at 400 nm implied the formation of azo bonds11,12 and enabled us to follow the reaction kinetics conveniently. When the dual-diazonium reagent and Tyr were mixed under pseudo-first-order conditions, the absorption peak at 400 nm was monitored with time. The pseudo-first-order rate kobs was determined by fitting the data with a single exponential function. The linear fitting between kobs and Tyr concentrations gave the reaction rate (k2). DDA-2 showed much smaller k2 (18.39 M−1 s−1) than that of DDA-3 (71.19 M−1 s−1), implying that the substitution of CO group with SO2 group greatly enhanced the reaction rate (Fig. 1, S1 and S2†). Substituents linked to aromatic cycles could affect the reactivity of diazonium with Tyr due to conjugate and other effects. Herein, the Hammett equation was further used to study the relationship between the para and meta substituents of benzene derivatives and the reaction rate. In this work, the Hammett parameter σ13 was obtained by using two kinds of similar structural substituents (Fig. S3†).14 By comparing the reactivity of the three diazonium salts, we found that the crosslinking rate of the diazonium cations and Tyr was higher with increasing electron withdrawing of the substituent: DDA-3 > DDA-1 > DDA-2. Remarkably, the slope of the linear fit was positive, demonstrating that the electron-withdrawing substituents accelerated the reaction, which was consistent with the kinetic results. This study might help us to construct more efficient multi-diazonium reagents in the future.
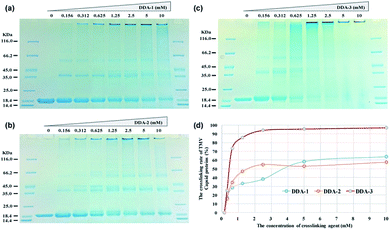
To compare the efficiency of dual-diazonium reagents (DDA-1, DDA-2, DDA-3) in protein labelling, we firstly used these reagents to crosslink the capsid proteins from tobacco mosaic virus (TMV CP) at 25 °C. The SDS-PAGE assay revealed that DDA-3 had higher crosslinking efficiency than that of DDA-1 and DDA-2 at room temperature. When the concentration of DDA-3 reached at least 1.25 mM, the percentage of crosslinked capsid proteins could be more than 85%. At such concentration, DDA-1 and DDA-2 could just achieve 30–50% protein crosslinking (Fig. 2).
Subsequently, we used these reagents to crosslink TMV virus at 25 °C and 37 °C. Based on SDS-PAGE analysis, we also found that the crosslinking efficiency of DDA-3 at 37 °C were slightly higher than that at 25 °C while the crosslinking efficiency of DDA-1 at 25 °C was significantly lower than that at 37 °C (Fig. S4–S6†). Furthermore, time-dependent crosslinking of TMV also suggested that the reaction could finish within 10 min for DDA-3 (Fig. S7†) and not finish until 60 min for DDA-1 (Fig. S8†) at 25 °C. These results clearly demonstrated that the substitution of CO group with SO2 group in the linker reagent greatly enhanced the speed and efficiency of protein crosslinking and TMV crosslinking at room temperature.
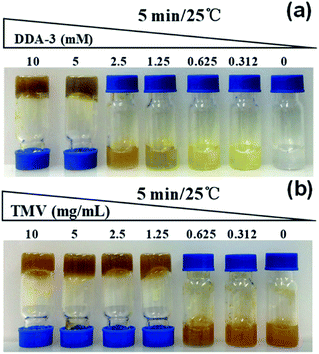
Encouraging with the above results, we then employed these reagents to crosslink TMV virus matrix to construct hydrogels. We firstly performed the time- and dose-dependent gelation tests. The results showed that 10 mM DDA-3 could completely solidify TMV matrix (2.5 mg mL−1) in 1 min at 25 °C. While for DDA-2, the complete gelation should cost at least 15 min under similar conditions (Fig. S9†). Therefore, we further used DDA-3 in following studies. The dose-dependent tests also suggested that 1.25 mM DDA-3 and 1.25 mg mL−1 TMV matrix were the minimum concentration for gelation within 30 min (Fig. 3, S10 and S11†). The high concentration of crosslinker and TMV matrix like 10 mM DDA-3 and 5 mg mL−1 TMV matrix also provided the high solidification. These results implied that improved dual-diazonium reagents could be used for faster crosslinking of tobacco mosaic virus as hydrogels, which further simplify the gelation method.
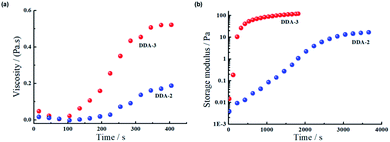
To further probe the crosslinking efficiency, we subsequently determined the rheological characters of the hydrogels formed by TMV and DDA-2 or DDA-3 from the solution phase to gel phase. As shown in Fig. 4a, the viscosity of both hydrogels started to increase after 100 s, while the hydrogel with DDA-3 showed higher viscosity (about 2.5-fold) than that with DDA-2, implying that DDA-3 had the higher gelation efficiency. Both the hydrogels were solidified after 400 s. Moreover, we also analysed the storage modulus of TMV hydrogels (Fig. 4b) and found that the hydrogel based on DDA-3 had much faster gelation speed with a relatively higher storage modulus than that based on DDA-2. These results indicate that the improved reagent DDA-3 could be used for faster crosslinking of TMV as hydrogels than DDA-2.
We further used scanning electron microscope (SEM) to analyze the structure of TMV hydrogels from 10 mM DDA-2 or DDA-3 and 5 mg mL−1 TMV. SEM results showed multiple virus rods in DDA-2 constructed hydrogel was crosslinked to form the 3D net structure, while a large number of sheets and less net structure were observed in DDA-3 constructed hydrogel (Fig. 5).
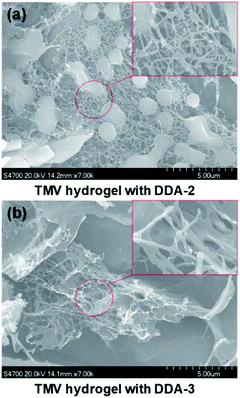 | ||
| Fig. 5 Characterization of the TMV hydrogel with DDA-2 (a) and DDA-3 (b) by SEM. Scale bar, 500 μm for full view and 500 nm for the red rectangular box. | ||
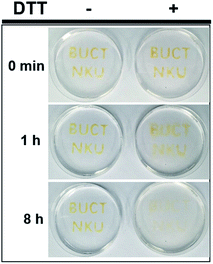
Considering the importance of biocompatibility and degradability of hydrogels in applications, we also synthesized DDA-4 through introducing a disulfide bond on the basis of DDA-3. The DDA-4 has similar ability as DDA-3 for TMV gelation (Fig. S9†). We then used the TMV hydrogels with DDA-4 to make the capital “BUCT” and “NKU” in the dish (Fig. 6), and determined whether the hydrogels could be degraded by reducing chemicals like DTT. After the gelation, we immersed the gel in the DTT solution (500 mM) and observed the degradation of the gel within 8 h, suggesting the disulfide bond was gradually cleaved by DTT (Fig. 6 and S12†). On the contrary, the same hydrogel in the dish was not affected when no DTT was added. We believe that this kind of disulfide bond-based hydrogel could be disrupted by reducing molecules in vivo, which might add its biocompatibility for chemically controlled disassembly.15
Conclusions
To improve the reaction rate and gelation efficiency, we developed new bench-stable reagents with different substituents (CO, PO, SO2) between two diazonium sites. Through comparing the reaction rate constant and protein crosslinking efficiency, we found that DDA-3 greatly improved the crosslinking efficiency and also shortened the gelation time even at room temperate, which was a great improvement compared with DDA-1.11 The SDS-PAGE and SEM characterizations also confirmed the virus crosslinking via the dual-diazonium reagents. Furthermore, hydrogels with the introduction of disulfide bonds via DDA-4 could be chemically degraded by biocompatible reducing molecules. We propose that the new dual-diazonium reagents may provide a general approach to preparing diverse functional hydrogels from other biocompatible viruses for future biomedical applications.Experimental
General
All chemicals and solvents used for synthesis were purchased from commercial suppliers and applied directly in the experiments without further purification. The progress of the reaction was monitored by TLC on pre-coated silica plates (Merck 60F-254, 250 μm in thickness), and spots were visualized by UV light (254 nm). Merck silica gel (100–200 mesh) was used for general column chromatography purification. 1H NMR and 13C NMR spectra were recorded on a Bruker 400 spectrometer. Chemical shifts are reported in parts per million relative to internal standard tetramethylsilane (Si(CH3)4 = 0.00 ppm) or residual solvent peaks (DMSO-d6 = 2.50 ppm, 1H; 39.52 ppm, 13C). 1H NMR coupling constants (J) are reported in hertz (Hz), and multiplicity is indicated as the following: s (singlet), d (doublet), t (triplet), q (quartet), td (triplet doublet), dd (doublet of doublets), m (multiple). High-resolution mass spectra (HRMS) were obtained on an Agilent 6540 UHD Accurate-Mass Q-TOFLC/MS. The UV-Visible spectra were recorded on a UV-6000 UV-VIS-NIR-spectrophotometer (METASH, China).Synthesis of DDA-2
Methyl diphenyl phosphine oxide (4.89 g, 22.6 mmol) was dissolved in concentrated sulfuric acid (20 mL), and stirred under ice-cooling to dissolve the solid. A mixture of concentrated sulfuric acid (20 mL) and concentrated nitric acid (20 mL) was slowly added dropwise with a dropper. After the addition was completed, the ice bath was removed and the mixture was stirred at room temperature overnight. The reaction was monitored by TLC (CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 19
MeOH = 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The reaction mixture was poured into ice water and then the precipitated solid were collected by filtration. After being washed with saturated aqueous NaHCO3 until neutral pH, the product was recrystallized with absolute ethanol, yielding a light-yellow solid of 1 (4.74 g, 68.5%). 1H NMR (400 MHz, DMSO-d6) δ 8.61 (d, J = 12.0 Hz, 2H), 8.41 (d, J = 8.1 Hz, 2H), 8.29 (dd, 2H), 7.85 (td, J = 7.9, 2.6 Hz, 2H), 2.31 (d, J = 14.1 Hz, 3H); 13C NMR (101 MHz, DMSO-d6) δ 148.0, 147.8, 137.0, 136.7, 136.6, 136.0, 130.8, 130.7, 126.7, 126.7, 125.0, 124.9, 15.6, 14.9.
1). The reaction mixture was poured into ice water and then the precipitated solid were collected by filtration. After being washed with saturated aqueous NaHCO3 until neutral pH, the product was recrystallized with absolute ethanol, yielding a light-yellow solid of 1 (4.74 g, 68.5%). 1H NMR (400 MHz, DMSO-d6) δ 8.61 (d, J = 12.0 Hz, 2H), 8.41 (d, J = 8.1 Hz, 2H), 8.29 (dd, 2H), 7.85 (td, J = 7.9, 2.6 Hz, 2H), 2.31 (d, J = 14.1 Hz, 3H); 13C NMR (101 MHz, DMSO-d6) δ 148.0, 147.8, 137.0, 136.7, 136.6, 136.0, 130.8, 130.7, 126.7, 126.7, 125.0, 124.9, 15.6, 14.9.
Tin II chloride (14.48 g, 64.2 mmol) was dissolved in 40 mL concentrated HCl. 1 (3.11 g, 10.1 mmol) was added portionwise with stirring and the reaction mixture was refluxed at 80 °C for 2 h. After the mixture had cooled to room temperature, the pH of the mixture was adjusted to 9–10 by the saturated solution of NaOH at 0 °C. The mixture was extracted with EtOAc. The organic phase was washed by brine and dried over anhydrous Na2SO4. After removing the solvent under reduced pressure, the product was purified by silica gel column chromatography to give 2 (2.15 g, 86.4%). 1H NMR (400 MHz, DMSO-d6) δ 7.11 (td, J = 7.7, 3.6 Hz, 2H), 6.89 (d, J = 13.1 Hz, 2H), 6.78 (dd, J = 11.1, 7.6 Hz, 2H), 6.67 (d, J = 7.9 Hz, 2H), 5.31 (s, 4H), 1.81 (d, J = 13.1 Hz, 3H); 13C NMR (101 MHz, DMSO-d6) δ 148.8, 148.6, 136.2, 135.2, 129.1, 128.9, 117.1, 117.0, 116.4, 116.4, 115.2, 115.1, 16.4, 15.6; 31P NMR (162 MHz, DMSO-d6) δ 28.63 (s).
2 (998 mg, 4.0 mmol) was dissolved in 30 mL cold concentrated HCl and cooled down to 0 °C. The water solution of NaNO2 (1.82 g, 26.4 mmol) was slowly added to the mixture at 0 °C. After reaction for 30 min, 4 mL 60% HPF6 was added and the mixture was stirred for 1 h. The product was collected by filtration and washed by ice-cold water, yielding a creamy-white solid of DDA-2 (1.34 g, 59.8%). 1H NMR (400 MHz, DMSO-d6) δ 9.19 (d, J = 11.5 Hz, 2H), 8.86 (d, J = 8.2 Hz, 2H), 8.63 (t, 2H), 8.17 (t, J = 7.6 Hz, 2H), 2.36 (d, J = 14.2 Hz, 3H); 13C NMR (101 MHz, DMSO-d6) δ 141.7, 141.6, 137.3, 136.4, 135.9, 135.1, 135.0, 132.0, 131.9, 118.0, 117.8, 15.5, 14.8; 31P NMR (162 MHz, DMSO-d6) δ 26.52 (s), −144.20 (h, JP-F = 711.3 Hz).
Synthesis of DDA-3
4,4′-Diaminodiphenyl sulfone (1.16 g, 4.6 mmol) was dissolved in 20 mL concentrated HCl and 40 mL H2O and cooled down to 0 °C. The water solution of NaNO2 (2.07 g, 29.6 mmol) was slowly added to the mixture at 0 °C. After reaction for 1 h, 4 mL 60% HPF6 was added and the mixture was stirred for 1 h. The product was collected by filtration and washed with methanol and dichloromethane, yielding a yellow solid of DDA-3 (2.01 g, 77.7%). 1H NMR (400 MHz, DMSO-d6) δ 8.94 (d, J = 8.8 Hz, 4H), 8.65 (d, J = 8.8 Hz, 4H); 13C NMR (101 MHz, DMSO-d6) δ 147.6, 134.3, 130.6, 122.9; 31P NMR (162 MHz, DMSO-d6) δ −144.19 (h, JP-F = 711.3 Hz).Synthesis of DDA-4
Cystamine dihydrochloride (0.99 g, 4.4 mmol) and 4-acetamidobenzenesulfonyl chloride (2.48 g, 10.6 mmol) was dissolved in 10 mL DMF. The mixture was stirred at 0 °C; DIPEA (3.4 mL, 20.0 mmol) was added into 10 mL DMF and add in batches with a syringe in portions at 0 °C. The mixture was stirred at room temperature overnight. The solvent was evaporated under reduced pressure and the crude was diluted with 50 mL dichloromethane. The product was collected by filtration and washed with dichloromethane to give 3 (1.76 g, 73.1%). 1H NMR (400 MHz, DMSO-d6) δ 10.32 (s, 1H), 7.78–7.67 (m, 2H), 2.98 (q, J = 13.1, 6.5 Hz, 1H), 2.66 (t, J = 6.9 Hz, 1H), 2.08 (s, 1H); 13C NMR (101 MHz, DMSO-d6) δ 169.0, 142.8, 133.9, 127.7, 118.7, 41.7, 37.0, 24.1.3 (1.16 g, 2.1 mmol) was dissolved in 40 mL concentrated HCl, and the mixture was stirred at 60 °C for 4 h. Then it was collected by filtration and the saturated solution of NaHCO3 was added until pH 7–8. Next, the mixture was extracted with EtOAc, and the combined organic extracts were washed with brine, dried over anhydrous Na2SO4, and evaporated under reduced pressure. The product was purified by silica gel column chromatography (CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 95
MeOH = 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) to give 4 (823 mg, 84.7%). 1H NMR (400 MHz, DMSO-d6) δ 7.41 (t, J = 5.6 Hz, 1H), 7.29 (t, J = 5.9 Hz, 1H), 6.63–6.59 (m, 1H), 5.93 (s, 1H), 2.92 (dd, J = 13.7, 6.2 Hz, 1H), 2.65 (t, J = 7.0 Hz, 1H); 13C NMR (101 MHz, DMSO-d6) δ 152.6, 128.5, 125.2, 112.7, 41.8, 37.0. HRMS (ESI): m/z [M + H]+ calculated for C16H23N4O4S4+: 463.0597; found: 463.0597.
5) to give 4 (823 mg, 84.7%). 1H NMR (400 MHz, DMSO-d6) δ 7.41 (t, J = 5.6 Hz, 1H), 7.29 (t, J = 5.9 Hz, 1H), 6.63–6.59 (m, 1H), 5.93 (s, 1H), 2.92 (dd, J = 13.7, 6.2 Hz, 1H), 2.65 (t, J = 7.0 Hz, 1H); 13C NMR (101 MHz, DMSO-d6) δ 152.6, 128.5, 125.2, 112.7, 41.8, 37.0. HRMS (ESI): m/z [M + H]+ calculated for C16H23N4O4S4+: 463.0597; found: 463.0597.
4 (523 mg, 1.1 mmol) was dissolved in 20 mL concentrated HCl and 20 mL H2O and cooled down to 0 °C. The water solution of NaNO2 (483 mg, 7.0 mmol) was slowly added to the mixture at 0 °C. After reaction for 1 h, 2 mL 60% HPF6 was added and the mixture was stirred for 1 h. The product was collected by filtration and washed with ice-cold water, yielding a yellow solid of DDA-4 (741 mg, 86.3%). 1H NMR (400 MHz, DMSO-d6) δ 8.87 (d, 4H), 8.60 (t, J = 5.6 Hz, 2H), 8.33 (d, 4H), 3.16 (dd, J = 12.6, 6.4 Hz, 4H), 2.75 (t, J = 6.7 Hz, 4H); 13C NMR (101 MHz, DMSO-d6) δ 150.3, 134.1, 128.7, 120.0, 41.7, 37.1; 31P NMR (162 MHz, DMSO-d6) δ −144.19 (h, JP-F = 711.3 Hz).
UV-Vis spectrophotometry assay
All spectroscopic measurements were performed in phosphate-buffered saline buffer (50 mM, pH 7.4, containing 5% DMSO) in a 3 mL corvette with 2 mL solution. Compounds were dissolved into DMSO to prepare the stock solutions with a concentration of 200 mM Tyr or 20 mM diazonium reagent.Reaction of the diazonium reagent (DDA-2 or DDA-3) and Tyr-containing molecule (H-Tyr-OBzl Tos): 2 μL DDA-2 or DDA-3 and 4 μL Tyr was added to 1994 μL buffer. The progress of the reaction was monitored by UV-Vis spectrophotometry from 250 to 600 nm.
Determination of the kinetic parameters of the reaction between the diazonium reagent (DDA-2 or DDA-3) and tyrosine: the reaction rate was measured under pseudo-first order conditions, where 20 μM DDA-2 or DDA-3 was mixed with an excess of tyrosine (0.1–1 mM) in PBS buffer (50 mM, pH 7.4, containing 5% DMSO) and the absorption at 400 nm was recorded per second.
TMV and TMV CP purification
According to our previous procedure, TMV virus and TMV capsid proteins were purified.12aTMV purification: TMV-inoculated tobacco leaves were ground fully and mixed in 25 mL 0.2 M phosphate buffer (pH 7.0) including 250 μL β-mercaptoethanol (1%, v/v) on ice. After the filtration with two-layer screen cloth, the clear lysates were mixed with n-butyl alcohol (8%, v/v) for 15 min. After centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min, the precipitate was dissolved again with 2 mL 0.01 M phosphate buffer (pH 7.0) on ice and stirred for 6 h. After centrifugation at 10
000 × g for 20 min, the precipitate was dissolved again with 2 mL 0.01 M phosphate buffer (pH 7.0) on ice and stirred for 6 h. After centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min, the pellet was resuspended in 5 mL 0.01 M phosphate buffer (pH 7.0) for 2 h. After centrifugation at 10
000 × g for 20 min, the pellet was resuspended in 5 mL 0.01 M phosphate buffer (pH 7.0) for 2 h. After centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min, the supernatant was then added with NaCl (4%, w/v) and PEG6000 (4%, w/v) for overnight precipitation. After centrifugation at 10
000 × g for 20 min, the supernatant was then added with NaCl (4%, w/v) and PEG6000 (4%, w/v) for overnight precipitation. After centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min, the precipitate was resuspended in 2 mL 0.01 M phosphate buffer (pH 7.0) for 6 h. After centrifugation at 10
000 × g for 20 min, the precipitate was resuspended in 2 mL 0.01 M phosphate buffer (pH 7.0) for 6 h. After centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 5 min, the final virus suspension was obtained and the virus purity was evaluated according to the indicated ratio of A280/A260 (0.84) and A260/A248 (1.09). TMV concentration was calculated according to C (mg mL−1) = A260/3.1. TMV solution was stored at 4 °C for the following test.
000 × g for 5 min, the final virus suspension was obtained and the virus purity was evaluated according to the indicated ratio of A280/A260 (0.84) and A260/A248 (1.09). TMV concentration was calculated according to C (mg mL−1) = A260/3.1. TMV solution was stored at 4 °C for the following test.
Purification of TMV capsid proteins (TMV CP): TMV solution was firstly diluted to 5 mg mL−1 and then stirred with two volumes of glacial acetic acid for 2 h at 4 °C. Subsequently, an equal volume of sterile water was added and all the liquid was then transferred into the dialysis bag. After three to five times of water exchange, TMV CP precipitated. After centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min, TMV CP precipitate was obtained and dissolved in 50 μL 0.1 M phosphate buffer (pH 7.0). The concentration of TMV CP was calculated according to C (mg mL−1) = A282/1.27. TMV CP was stored at 4 °C for the following test.
000 rpm for 30 min, TMV CP precipitate was obtained and dissolved in 50 μL 0.1 M phosphate buffer (pH 7.0). The concentration of TMV CP was calculated according to C (mg mL−1) = A282/1.27. TMV CP was stored at 4 °C for the following test.
Crosslinking TMV CP and TMV virus with the dual-diazonium reagent
TMV CP crosslinking: TMV CP crosslinking was performed by mixing TMV CP with various concentrations of the dual-diazonium reagent (DDA-1, DDA-2, DDA-3). The dual-diazonium reagent was diluted to a gradient concentration (100, 50, 25, 12.5, 6.25, 3.12, 1.56, mM). The reaction was performed by mixing 9 μL TMV CP (2 μg μL−1) and 1 μL the dual-diazonium reagent (100, 50, 25, 12.5, 6.25, 3.12, 1.56, mM). The reaction was incubated at 25 °C for 30 min.TMV virus crosslinking: TMV virus crosslinking was performed by mixing TMV virus with various concentrations of the dual-diazonium reagent (DDA-1, DDA-2, DDA-3). The dual-diazonium reagent was diluted to a gradient concentration (100, 50, 25, 12.5, 6.25, 3.12, 1.56, mM). The reaction was performed by mixing 9 μL TMV virus (1 μg μL−1) and 1 μL the dual-diazonium reagent (100, 50, 25, 12.5, 6.25, 3.12, 1.56, mM). The reaction was incubated at 25 °C or 37 °C for 30 min or 60 min.
After reaction, all samples were mixed with 5× SDS loading buffer and used for 12% SDS-PAGE assay. The SDS-PAGE gel was stained with Coomassie blue R250 and imaged with Quantity One software (Bio-Rad).
Construction of TMV hydrogel with the dual-diazonium reagent
To optimize conditions for hydrogel preparation, we tried different concentrations of TMV matrix (0–10 mg mL−1) and DDA-3 (0–10 mM). TMV hydrogel was made by gently mixing different concentrations of DDA-3 and TMV solution in a glass vial at room temperature for 0–30 min to evaluate the gelation condition.For TMV hydrogel with 2.5 mg mL−1 TMV matrix and 10 mM DDA-2 or DDA-3, DDA-2 or DDA-3 was freshly prepared in DMSO as 100 mM stock solution. 50 μL of DDA-2 or DDA-3 (100 mM) was added to 325 μL of 0.1 M phosphate buffer (pH 7.0) to prepare solution A; 62.5 μL of TMV solution (20 mg mL−1) was added to 62.5 μL of phosphate buffer to prepare solution B. The reaction was performed by mixing solution A and solution B. The gelation of TMV hydrogel was observed within 30 min at room temperature. It was the same for DDA-4 constructed hydrogel except that the concentration of DDA-4 and TMV matrix were 2 mM and 2.5 mg mL−1, respectively.
Scanning electron microscopy (SEM) characterization
TMV hydrogel was made by 5 mg mL−1 TMV and 10 mM DDA-2 or DDA-3 at room temperature for 1 h. Then, the hydrogel was firstly cold-dried and loaded onto the sample vessel. The sample was imaged using a Hitachi S-4700 field emission SEM.Rheology
All rheological measurements were performed using an ar2000ex Rheometer from TA Instruments at 25 °C. When the crosslinking reagent and the TMV solution were mixed, the mixture was quickly added to the mold to start testing before the hydrogel was completely solidified. TMV hydrogel was made by 2.5 mg mL−1 TMV and 10 mM DDA-2 or DDA-3. Due to the faster rate of DDA-3 crosslinking, we formulated this hydrogel in ice-water bath conditions.TMV hydrogel degradation
To degrade virus hydrogel with chemical agents, TMV hydrogel was made according to the above procedure with DDA-4. The TMV hydrogel was degraded by dithiothreitol (DTT). We firstly carved the word “BUCT” and “NKU” on the Petri dish. The hydrogel was made by transferring the mixture of 2.5 mg mL−1 TMV and 2 mM DDA-4 on the word groove for gelation at room temperature. After gelation, 20 mL DTT (500 mM) as the test group and 20 mL deionized water as the control group were transferred into the Petri dish to immerse the TMV hydrogel. The gel degradation was imaged at different time (0–12 h).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by NSFC (21572019) and Beijing Municipal Natural Science Foundation (2192038).Notes and references
- (a) S. R. Caliari and J. A. Burdick, Nat. Methods, 2016, 13, 405–414 CrossRef CAS PubMed; (b) K. T. Dicker, J. Song, A. C. Moore, H. Zhang, Y. Li, D. L. Burris, X. Jia and J. M. Fox, Chem. Sci., 2018, 9, 5394–5404 RSC; (c) O. Chaudhuri, Biomater. Sci., 2017, 5, 1480–1490 RSC; (d) S. J. Kim, J. Park, H. Byun, Y. W. Park, L. G. Major, D. Y. Lee, Y. S. Choi and H. Shin, Biomaterials, 2019, 188, 198–212 CrossRef CAS PubMed; (e) M. W. Tibbitt and K. S. Anseth, Biotechnol. Bioeng., 2009, 103, 655–663 CrossRef CAS PubMed; (f) T. Bai, A. Sinclair, F. Sun, P. Jain, H.-C. Hung, P. Zhang, J.-R. Ella-Menye, W. Liu and S. Jiang, Chem. Sci., 2016, 7, 333–338 RSC.
- (a) J. Xing, M. Zheng and X. Duan, Chem. Soc. Rev., 2015, 44, 5031–5039 RSC; (b) X. Mao, G. Chen, Z. Wang, Y. Zhang, X. Zhu and G. Li, Chem. Sci., 2018, 9, 811–818 RSC; (c) B. Balakrishnan and R. Banerjee, Chem. Rev., 2011, 111, 4453–4474 CrossRef CAS PubMed; (d) S. Reakasame and A. R. Boccaccini, Biomacromolecules, 2018, 19, 3–21 CrossRef CAS PubMed; (e) M. Mehrali, A. Thakur, C. P. Pennisi, S. Talebian, A. Arpanaei, M. Nikkhah and A. Dolatshahi-Pirouz, Adv. Mater., 2017, 29, 1603612 CrossRef PubMed; (f) A. Brito, Y. M. Abul-Haija, D. S. da Costa, R. Novoa-Carballal, R. L. Reis, R. V. Ulijn, R. A. Pires and I. Pashkuleva, Chem. Sci., 2019, 10, 2385–2390 RSC; (g) Y. P. Singh, J. C. Moses, N. Bhardwaj and B. B. Madal, J. Mater. Chem. B, 2018, 6, 5499–5529 RSC.
- (a) S. Merino, C. Martin, K. Kostarelos, M. Prato and E. Vazquez, ACS Nano, 2015, 9, 4686–4697 CrossRef CAS PubMed; (b) J. Tsai, T. Zou, J. Liu, T. Chen, A. Chan, C. Yang, C. Lok and C. Che, Chem. Sci., 2015, 6, 3823–3830 RSC; (c) J. Xing, M. Zheng and X. Duan, Chem. Soc. Rev., 2015, 44, 5031–5039 RSC; (d) M. R. Saboktakin and R. M. Tabatabaei, Int. J. Biol. Macromol., 2015, 75, 426–436 CrossRef CAS PubMed; (e) R. Dimatteo, N. J. Darling and T. Segura, Adv. Drug Delivery Rev., 2018, 127, 167–184 CrossRef CAS PubMed; (f) T. Su, Z. Tang, H. He, W. Li, X. Wang, C. Liao, Y. Sun and Q. Wang, Chem. Sci., 2014, 5, 4204–4209 RSC; (g) B. D. Monnery and R. Hoogenboom, Polym. Chem., 2019, 10, 3480–3487 RSC.
- (a) D. Stern and H. Cui, Adv. Healthcare Mater., 2019, 8, e1900104 CrossRef PubMed; (b) G. M. Guebitz and G. S. Nyanhongo, Trends Biotechnol., 2018, 36, 1040–1053 CrossRef CAS PubMed; (c) Q. Xu, L. Guo, A. Sigen, Y. Gao, D. Zhou, U. Greiser, J. Creagh-Flynn, H. Zhang, Y. Dong, L. Cutlar, F. Wang, W. Liu, W. Wang and W. Wang, Chem. Sci., 2018, 9, 2179–2187 RSC; (d) A. H. Morris, H. Lee, H. Xing, D. K. Stamer, M. Tan and T. R. Kyriakides, ACS Appl. Mater. Interfaces, 2018, 10, 41892–41901 CrossRef CAS PubMed; (e) X. Chen, Z. Yue, P. C. Winberg, J. N. Dinoro, P. Hayes, S. Beirne and G. G. Wallace, Biomater. Sci., 2019, 10, 41892–41901 Search PubMed; (f) S. An, E. J. Jeon, J. Jeon and S. Cho, Mater. Horiz., 2019, 6, 1169–1178 RSC; (g) H. Jiang, M. Ochoa, J. F. Waimin, R. Rahimi and B. Ziaie, Lab Chip, 2019, 19, 2265–2274 RSC.
- (a) E. M. Ahmed, J. Adv. Res., 2015, 6, 105–121 CrossRef CAS PubMed; (b) H. Yuk, B. Lu and X. Zhao, Chem. Soc. Rev., 2019, 48, 1642–1667 RSC; (c) X. Le, W. Lu, J. Zheng, D. Tong, N. Zhao, C. Ma, H. Xiao, J. Zhang, Y. Huang and T. Chen, Chem. Sci., 2016, 7, 6715–6720 RSC; (d) T. E. Brown and K. S. Anseth, Chem. Soc. Rev., 2017, 46, 6532–6552 RSC; (e) S. An, E. J. Jeon, J. Jeon and S. Cho, Mater. Horiz., 2019, 6, 1169–1178 RSC.
- (a) Y. Zhu, Q. Zhang, X. Shi and D. Han, Adv. Mater., 2019, e1804950 CrossRef PubMed; (b) C. Chen, L. Wang, L. Deng, R. Hu and A. Dong, J. Biomed. Mater. Res., Part A, 2013, 101, 684–693 CrossRef PubMed; (c) S. De Koker, J. Cui, N. Vanparijs, L. Albertazzi, J. Grooten, F. Caruso and B. G. De Geest, Angew. Chem., Int. Ed., 2016, 55, 1334–1339 CrossRef CAS PubMed; (d) C. Wang, M. Fadeev, J. Zhang, M. Vázquez-González, G. Davidson-Rozenfeld, H. Tian and I. Willner, Chem. Sci., 2018, 9, 7145–7152 RSC; (e) Y. Wang, Y. Li, X. Yu, Q. Long and T. Zhang, RSC Adv., 2019, 9, 18394–18405 RSC; (f) J. Du, S. Xu, S. Feng, L. Yu, J. Wang and Y. Liu, Soft Matter, 2016, 12, 1649–1654 RSC.
- (a) A. M. Wen and N. F. Steinmetz, Chem. Soc. Rev., 2016, 45, 4074–4126 RSC; (b) T. L. Schlick, Z. Ding, E. W. Kovacs and M. B. Francis, J. Am. Chem. Soc., 2005, 127, 3718–3723 CrossRef CAS PubMed; (c) K. Mohan and G. A. Weiss, ACS Chem. Biol., 2016, 11, 1167–1179 CrossRef CAS PubMed; (d) Z. Liu, J. Qiao, Z. Niu and Q. Wang, Chem. Soc. Rev., 2012, 41, 6178–6194 RSC; (e) R. Farr, D. S. Choi and S. W. Lee, Acta Biomater., 2014, 10, 1741–1750 CrossRef CAS PubMed; (f) M. A. Bruckman, G. Kaur, L. A. Lee, F. Xie, J. Sepulveda, R. Breitenkamp, X. Zhang, M. Joralemon, T. P. Russell, T. Emrick and Q. Wang, ChemBioChem, 2008, 9, 519–523 CrossRef CAS PubMed.
- (a) S. Honarbakhsh, R. H. Guenther, J. A. Willoughby, S. A. Lommel and B. Pourdeyhimi, Adv. Healthcare Mater., 2013, 2, 1001–1007 CrossRef CAS PubMed; (b) K. L. Kozielski, Y. Rui and J. J. Green, Expert Opin. Drug Delivery, 2016, 13, 1475–1487 CrossRef CAS PubMed; (c) M. Wu, J. Shi, D. Fan, Q. Zhou, F. Wang, Z. Niu and Y. Huang, Biomacromolecules, 2013, 14, 4032–4037 CrossRef CAS PubMed; (d) N. M. Gulati, A. S. Pitek, A. E. Czapar, P. L. Stewart and N. F. Steinmetz, J. Mater. Chem. B, 2018, 6, 2204–2216 RSC; (e) P. Lam, R. D. Lin and N. F. Steinmetz, J. Mater. Chem. B, 2018, 6, 5888–5895 RSC; (f) A. S. Pitek, J. Park, Y. Wang, H. Gao, H. Hu, D. I. Simon and N. F. Steinmetz, Nanoscale, 2018, 10, 16547–16555 RSC.
- (a) N. F. Steinmetz and D. J. Evans, Org. Biomol. Chem., 2007, 5, 2891–2902 RSC; (b) B. D. B. Tiu, D. L. Kernan, S. B. Tiu, A. M. Wen, Y. Zheng, J. K. Pokorski, R. C. Advincula and N. F. Steinmetz, Nanoscale, 2017, 9, 1580–1590 RSC.
- (a) J. A. Luckanagul, L. A. Lee, S. J. You, X. M. Yang and Q. Wang, J. Biomed. Mater. Res., Part A, 2015, 103, 887–895 CrossRef PubMed; (b) A. Southan, T. Lang, M. Schweikert, G. E. M. Tovar, C. Wege and S. Eiben, RSC Adv., 2018, 8, 4686–4694 RSC; (c) L. Chen, X. Zhao, Y. Lin, Z. Su and Q. Wang, Polym. Chem., 2014, 5, 6754–6760 RSC; (d) X. Liu, J. Zhang, M. Fadeev, Z. Li, V. Wulf, H. Tian and I. Willner, Chem. Sci., 2019, 10, 1008–1016 RSC.
- D. Ma, J. Zhang, C. Zhang, Y. Men, H. Sun, L. Li, L. Yi and Z. Xi, Org. Biomol. Chem., 2018, 16, 3353–3357 RSC.
- (a) D. Ma, Y. Xie, J. Zhang, D. Ouyang, L. Yi and Z. Xi, Chem. Commun., 2014, 50, 15581 RSC; (b) J. Zhang, D. Ma, D. Du, Z. Xi and L. Yi, Org. Biomol. Chem., 2014, 12, 9528 RSC; (c) J. Zhang, Y. Men, S. Lv, L. Yi and J. Chen, Org. Biomol. Chem., 2015, 13, 11422 RSC; (d) D. Ma, X. Kang, Y. Gao, J. Zhu, L. Yi and Z. Xi, Tetrahedron, 2019, 75, 888–893 CrossRef CAS.
- (a) Y. Heider, N. E. Poitiers, P. Willmes, K. I. Leszczynska, V. Huch and D. Scheschkewitz, Chem. Sci., 2019, 10, 4523–4530 RSC; (b) L. P. Hammett, Chem. Rev., 1935, 17, 125–136 CrossRef CAS; (c) L. P. Hammett, J. Am. Chem. Soc., 1937, 59, 96–103 CrossRef CAS.
- C. Hansch, A. Leo and R. W. Taft, Chem. Rev., 1991, 91, 165–195 CrossRef CAS.
- (a) J. R. Fores, M. Criado-Gonzalez, M. Schmutz, C. Blanck, P. Schaaf, F. Boulmedais and L. Jierry, Chem. Sci., 2019, 10, 4761–4766 RSC; (b) H. G. Li, D. Buesen, R. Williams, J. Henig, S. Stapf, K. Mukherjee, E. Freier, W. Lubitz, M. Winkler, T. Happee and N. Plumeré, Chem. Sci., 2018, 9, 7596–7605 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra05630k |
| ‡ The authors contribute equally to the work. |
| This journal is © The Royal Society of Chemistry 2019 |