Open Access Article
Open Access ArticleVisual detection of Fusarium proliferatum based on asymmetric recombinase polymerase amplification and hemin/G-quadruplex DNAzyme†
Ying Wang *a,
Xiangdong Lib,
Dongmei Xia and
Xiaoqiang Wangc
*a,
Xiangdong Lib,
Dongmei Xia and
Xiaoqiang Wangc
aCollege of Life Science, Shandong Provincial Key Laboratory of Detection Technology for Tumor Markers, School of Chemistry and Chemical Engineering, Linyi University, Linyi 276005, People's Republic of China. E-mail: wangying@lyu.edu.cn
bShandong Province Key Laboratory of Agricultural Microbiology, College of Plant Protection, Shandong Agricultural University, Tai'an, Shandong 271018, People's Republic of China
cPlant Protection Research Center, Tobacco Research Institute of Chinese Academy of Agricultural Sciences, Qingdao 266101, People's Republic of China
First published on 13th November 2019
Abstract
A one-step and instrument-free visual method was established based on asymmetric recombinase polymerase amplification coupled with hemin/G-quadruplex DNAzyme for the detection of Fusarium proliferatum.
Fusarium proliferatum causes rot disease, which is difficult to control worldwide. The vascular systems of F. proliferatum-infected crops are destroyed. It causes rot of the stems, stalks, roots, flowers, and ears of maize1–3 and decreases its yield and quality remarkably. In addition, the mycotoxins of fumonisin B1, fumonisin B2, beauvericin, enniatins, fusaproliferin, and moniliformin are produced by F. proliferatum during infection processes.4–6 When these grains are used as food or feedstuffs, the health of consumers or livestock is exposed to danger.7 There are many reports on the illnesses of livestock and humans caused by Fusarium mycotoxins.8–10 Methods based on qPCR have been developed for F. proliferatum detection.11 Although they show high accuracy, these methods require expensive instruments and skilled operators; they are also limited by many factors, such as electric power, high cost, and long testing times, which results in inability to apply these methods outside the lab. Therefore, sensitive and simple detection of F. proliferatum is still needed in field testing of crops and their byproducts.
Bio-sensing has become important in recent decades because it provides alternative methods to solve some of the above problems.12–14 Recombinase polymerase amplification (RPA)15 is a conventional and isothermal method to obtain double-stranded DNA (dsDNA). There are many reports of rapid detection based on RPA,16 such as monitoring of viruses, plasmodium, mycoplasma, fungi, and other causative agents. End-point detection is usually performed using lateral flow strips.17–19 The greatest strengths of the lateral flow strip method are that it is rapid and straightforward. However, its cost is quite high because of the primer labeling and preparing of the strips. In addition, the strip is disposable, which is not suitable for high throughput detection. The product of RPA can also be observed with the aid of SYBR Green I in UV light.20 However, there is no selectivity of binding between SYBR Green I and dsDNA, which results in false positive results of primer dimers and nonspecific amplification of other dsDNA.
Methods of visual detection are greatly valued due to the intuitiveness of the results.21 Gold nanoparticles are widely used for colorimetric detection of various targets.22 However, preparation of gold nanoparticles with specific sizes is laborious and difficult. Proteins, genomic DNA, salts, et al. are present in the crude liquids of biological samples used in point-of-care detection, which may give rise to nonspecific aggregation of gold nanoparticles. Hemin/G-quadruplex-based visual methods are used to detect targets of nucleotides, proteins, and other signal molecules.23 In the presence of hemin/G-quadruplex DNAzyme, H2O2 and ABTS2− react, and a specific green color is observed; this method is very simple, cost-effective, and straightforward compared with lateral flow strips or gold nanoparticles.
Asymmetric RPA is a type of amplification that involves different concentration ratios of primer-F/primer-R. First, the dsDNA product is obtained with the primer pair. As the reaction continues, one primer in a small amount is exhausted, and single-stranded DNA (ssDNA) is produced by the other primer using the newly synthesized dsDNA as a template. In this paper, an one-step and instrument-free strategy for visual monitoring of F. proliferatum is established based on asymmetric recombinase polymerase amplification coupled with hemin/G-quadruplex DNAzyme.
The strategy is shown in Scheme 1; it includes extraction of genomic DNA of F. proliferatum DSM62267 (F120), asymmetric RPA, and visual detection. First, genomic DNA was released from F120 cells with NaOH solution. It was used directly as a template for asymmetric RPA with primer-F and primer-R in a certain ratio. The trans-complementary sequence of the G-quadruplex was designed at the 5′-terminal of primer-F. The RPA product was obtained in the presence of a TwistDx/TwistAmp® Liquid Basic kit. The product was a mixture of dsDNA and ssDNA with the G-quadruplex sequence at their 3′-ends. In the presence of NH4+ and hemin, a DNAzyme of hemin/G-quadruplex was constructed at the end of the ssDNA. When ABTS2− and H2O2 were added, the resulting solution turned green. However, no color changes occurred in the final solution if no F120 genomic DNA was present, no target DNA was extracted, or no ssDNA was amplified by asymmetric RPA.
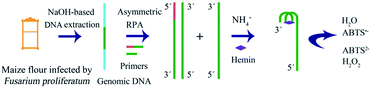 | ||
| Scheme 1 Schematic of the asymmetric recombinase polymerase amplification and hemin/G-quadruplex DNAzyme-based visual detection of F. proliferatum. | ||
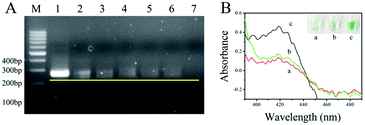
A genomic DNA mixture of maize and F120 was extracted from maize flour using NaOH solution. The feasibility of the asymmetric RPA reaction24 was evaluated using this genomic DNA as a template. The ratios of primer-F and primer-R were set as 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 1
20, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, 1
50, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75, and 1
75, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100. The products were analyzed by 2.5% agarose gel electrophoresis and stained with ethidium bromide. As shown in Fig. 1A, the bands of ssDNA were all in front of the bands of dsDNA, compared with the band of the 1
100. The products were analyzed by 2.5% agarose gel electrophoresis and stained with ethidium bromide. As shown in Fig. 1A, the bands of ssDNA were all in front of the bands of dsDNA, compared with the band of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of the primer pair (1). Faint bands of ssDNA were observed when the ratio of primer-F and primer-R was 1
1 ratio of the primer pair (1). Faint bands of ssDNA were observed when the ratio of primer-F and primer-R was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75 (5) or 1
75 (5) or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (6). When the ratio was 1
100 (6). When the ratio was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (3), the band of ssDNA was brighter than those of 1
20 (3), the band of ssDNA was brighter than those of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (2) and 1
10 (2) and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 (4). The yellow line represents the leading edge of the ssDNA bands. At the same time, the bands of dsDNA tapered off with the appearance of ssDNA and the down-ratio of primer-F/primer-R. These data show that genomic DNA of F120 could be extracted by NaOH solution, and the unpurified template of F120 was available for asymmetric RPA. In addition, more ssDNA was produced by the primer pair ratio of 1
50 (4). The yellow line represents the leading edge of the ssDNA bands. At the same time, the bands of dsDNA tapered off with the appearance of ssDNA and the down-ratio of primer-F/primer-R. These data show that genomic DNA of F120 could be extracted by NaOH solution, and the unpurified template of F120 was available for asymmetric RPA. In addition, more ssDNA was produced by the primer pair ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 than the other ratios; thus, this ratio was used in the following tests.
20 than the other ratios; thus, this ratio was used in the following tests.
To demonstrate the feasibility of the asymmetric RPA-hemin/G-quadruplex assay, the absorbance of the resulting solutions was measured by UV-Vis absorption (390 to 490 nm) of NanoDrop 2000. As shown in Fig. 1B, no absorption signal was observed in solutions containing hemin, H2O2 and ABTS2− (a) or the assay without genomic F120 DNA near 420 nm (b). An obvious increase of absorbance was observed in the presence of the target DNA (c) at 420 nm. A specific green color appeared in tube c (inserted in the top right corner of Fig. 1B). This result was in keeping with that of the UV-Vis absorption, demonstrating that the asymmetric RPA coupled with hemin/G-quadruplex DNAzyme method is viable. The visual biosensor could monitor F. proliferatum in maize flour.
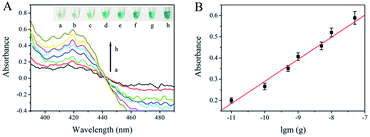
The intensity of UV-Vis absorption generated by the asymmetric RPA-hemin/G-quadruplex assay depended on the temperature gradient and time of the RPA reaction, as well as the different monovalent cations in the final solution (Fig. S1–S3†). To assess the analytical performance of the asymmetric RPA-hemin/G-quadruplex assay, various amounts of F120 genomic DNA were monitored under the optimal conditions. The UV-Vis absorption increased with the amount of F120 DNA from 0 to 50 ng (Fig. 2A). A larger amount of F120 DNA was used as the template, and more ssDNA was produced by the asymmetric RPA reaction. More hemin/G-quadruplex DNAzyme was constructed, causing an increase of the UV-Vis absorption of the resulting solution. A good linear dependence was observed between the absorbance and the amount of genomic DNA of F120 in a range from 0.01 ng to 50 ng at 420 nm (Fig. 2B and S4†). The regression equation was A = 0.1042![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) m + 1.331, with a correlation coefficient of 0.989, in which A and m represent the absorbance and quantity of F120 genomic DNA. The detection limit was calculated to be 0.01 ng by three times the standard deviation of the blank. The type of reaction vessel, reaction temperature, and sensitivity of the proposed strategy were compared with RPA-related colorimetric detection methods (ESI Table 1†).
m + 1.331, with a correlation coefficient of 0.989, in which A and m represent the absorbance and quantity of F120 genomic DNA. The detection limit was calculated to be 0.01 ng by three times the standard deviation of the blank. The type of reaction vessel, reaction temperature, and sensitivity of the proposed strategy were compared with RPA-related colorimetric detection methods (ESI Table 1†).
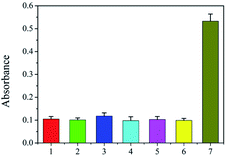
The selectivity of the proposed assay is critical because numerous DNA sequences belonging to the host and other living bodies are present in biological samples. The primer pair used in the strategy was based on the intergenic sequence of the ribosomal RNA gene cluster, which is specific to species of F. proliferatum. Three fungi, F. equiseti RD13 (F216), F. culmorum 3.37 dus Bomm (F109), and F. avenaceum borm (F112), are also species of Fusarium. Ralstonia solancearum and Puccinia sorghi are common causes of disease in corn. The three Fusarium species, R. solancearum, and P. sorghi were used as controls to check the selectivity of the proposed strategy. As shown in Fig. 3, the absorbances of the final solutions of F. equiseti RD13 (2), F. culmorum 3.37 dus Bomm (3), F. avenaceum borm (4), R. solancearum (5), and P. sorghi (6) were as low as that of the blank control (1). However, the absorbance from genomic F120 DNA (7) was obviously high, showing the good selectivity of our strategy.
To demonstrate the feasibility of the proposed assay for real sample detection, 4 positive samples and 10 field-collected samples were successively tested by the RPA/G-quadruplex visual assay and PCR. The results show that 2 of the 8 field-collected samples were infected with F. proliferatum; also, the two methods were consistent, indicating that our RPA/G-quadruplex visual strategy is plausible and can be used in field tests (Table 1).
| Sample | PCR-based detection | RPA/G-quadruple-based detection | ||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| 4 | 4 | 0 | 4 | 0 |
| 10 | 2 | 8 | 2 | 8 |
This strategy has obvious advantages. It employs specific primers of F. proliferatum, guaranteeing good sensitivity and selectivity of the biosensor over other species of Fusarium. Asymmetric RPA and hemin/G-quadruplex strategies were combined to achieve visual detection of F. proliferatum; the method is very simple, straightforward and cost-effective. The strategy is instrument-free, and only two label-free oligonucleotides were used in the assay, which was performed in a foam box with hot water; this implies that the proposed assay can be applied in field tests.
Therefore, the one-step and instrument-free visual detection of F. proliferatum species based on asymmetric RPA coupled with hemin/G quadruplex DNAzyme was established. The asymmetric RPA reaction was performed in a foam box with hot water, and the results of a specific green color could be observed by the naked eye. The method is label-free and extremely simple, and it can be operated by general staff; thus, it has great potential for field tests. The proposed strategy has good sensitivity, with a detection limit of 0.01 ng by absorption spectroscopy. It shows satisfactory selectivity for F. proliferatum species over other Fusarium species. It also has great potential to be applied to detect other DNA-based samples, such as viruses, bacteria, fungi, plants and animals, by changing the corresponding specific primers.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to acknowledge the assistance of Professor Petr Karlovsky from the University of Goettingen, Germany, for kindly providing strains of F. proliferatum DSM62267, F. equiseti RD13, F. culmorum 3.37 dus Bomm, F. avenaceum borman, and maize flour contaminated with F. proliferatum DSM62267. This work was financially supported by the National Natural Science Foundation of China (No. 31701760, 31901937), Shandong Provincial Major Application Technology Innovation Project, and Shandong Provincial National Science Foundation (ZR2018BC037).Notes and references
- A. Logrieco, A. Rizzo, R. Ferracane and A. Ritieni, Appl. Environ. Microbiol., 2002, 68, 82–85 CrossRef CAS PubMed.
- J. H. Choi, S. Lee, J. Y. Nah, H. K. Kim, J. S. Paek, S. Lee, H. Ham, S. K. Hong, S. H. Yun and T. Lee, Int. J. Food Microbiol., 2018, 267, 62–69 CrossRef CAS PubMed.
- E. Cendoya, M. D. P. Monge, S. M. Chiacchiera, M. C. Farnochi and M. L. Ramirez, Int. J. Food Microbiol., 2018, 266, 158–166 CrossRef CAS PubMed.
- L. Morales, T. P. Marino, A. J. Wenndt, J. Q. Fouts, J. B. Holland and R. J. Nelson, Phytopathology, 2018, 108, 1475–1485 CrossRef PubMed.
- Q. Jian, T. Li, Y. Wang, Y. Zhang, Z. Zhao, X. Zhang, L. Gong and Y. Jiang, Food Res. Int., 2019, 116, 397–407 CrossRef CAS PubMed.
- M. Bryla, A. Waskiewicz, E. Ksieniewicz-Wozniak, K. Szymczyk and R. Jedrzejczak, Molecules, 2018, 23, 963–996 CrossRef PubMed.
- S. Yazar and G. Z. Omurtag, Int. J. Mol. Sci., 2008, 9, 2062–2090 CrossRef CAS PubMed.
- I. Zimmer, E. Usleber, H. Klaffke, R. Weber, P. Majerus, H. Otteneder, M. Gareis, R. Dietrich and E. Martlbauer, Mycotoxin Res., 2008, 24, 40–52 CrossRef CAS PubMed.
- H. K. Abbas, W. P. Williams, G. L. Windham, H. C. Pringle 3rd, W. Xie and W. T. Shier, J. Agric. Food Chem., 2002, 50, 5246–5254 CrossRef CAS PubMed.
- H. F. Vismer, G. S. Shephard, J. P. Rheeder, L. van der Westhuizen and R. Bandyopadhyay, Food Addit. Contam., Part A: Chem., Anal., Control, Exposure Risk Assess., 2015, 32, 1952–1958 CrossRef CAS PubMed.
- S. Nutz, K. Doll and P. Karlovsky, Anal. Bioanal. Chem., 2011, 401, 717–726 CrossRef CAS PubMed.
- Y. Wang, B. X. Li, J. Liu and H. Zhou, Sens. Actuators, B, 2018, 273, 649–655 CrossRef CAS.
- Y. Wang, B. Li, J. Liu and H. Zhou, Anal. Bioanal. Chem., 2019, 2915–2924 CrossRef PubMed.
- D. Xi, X. Wang, S. Ai and S. Zhang, Chem. Commun., 2014, 50, 9547–9549 RSC.
- S. Lutz, P. Weber, M. Focke, B. Faltin, J. Hoffmann, C. Muller, D. Mark, G. Roth, P. Munday, N. Armes, O. Piepenburg, R. Zengerle and F. von Stetten, Lab-on-a-Chip, 2010, 10, 887–893 RSC.
- J. Li, J. Macdonald and F. von Stetten, Analyst, 2019, 144, 31–67 RSC.
- S. H. Kim, J. Lee, B. H. Lee, C. S. Song and M. B. Gu, Biosens. Bioelectron., 2019, 134, 123–129 CrossRef CAS PubMed.
- X. Deng, C. Wang, Y. Gao, J. Li, W. Wen, X. Zhang and S. Wang, Biosens. Bioelectron., 2018, 105, 211–217 CrossRef PubMed.
- M. L. Powell, F. R. Bowler, A. J. Martinez, C. J. Greenwood, N. Armes and O. Piepenburg, Anal. Biochem., 2018, 543, 108–115 CrossRef CAS PubMed.
- R. Wang, F. Zhang, L. Wang, W. J. Qian, C. Qian, J. Wu and Y. B. Ying, Anal. Chem., 2017, 89, 4413–4418 CrossRef CAS PubMed.
- Y. Wang, J. Liu and H. Zhou, Sensors, 2019, 19, 1298 CrossRef PubMed.
- E. Priyadarshini and N. Pradhan, Sens. Actuators, B, 2017, 238, 888–902 CrossRef CAS.
- Z. Tang, H. Zhang, C. Ma, P. Gu, G. Zhang, K. Wu, M. Chen and K. Wang, Microchim. Acta, 2018, 185, 109 CrossRef PubMed.
- U. B. Gyllensten and H. A. Erlich, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 7652–7656 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra05709a |
| This journal is © The Royal Society of Chemistry 2019 |