Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Hexanuclear Cu3O–3Cu triazole-based units as novel core motifs for high nuclearity copper(II) frameworks†
Sacramento Ferrer *a,
Javier Hernández-Gila,
Francisco Javier Valverde-Muñozb,
Francisco Lloretb and
Alfonso Castiñeiras
*a,
Javier Hernández-Gila,
Francisco Javier Valverde-Muñozb,
Francisco Lloretb and
Alfonso Castiñeiras c
c
aDepartament de Química Inorgànica, Universitat de València, Av. Vicent Andrés Estellés, s/n, 46100 Burjassot, Valencia, Spain. E-mail: Sacramento.Ferrer@uv.es
bInstitut de Ciència Molecular (ICMol), Universitat de València, C/Catedrático José Beltrán Martínez, 2, 46980 Paterna, Valencia, Spain
cDepartamento de Química Inorgánica, Facultad de Farmacia, Universidad de Santiago de Compostela, Praza do Seminario de Estudos Galegos, s/n, 15782 Santiago de Compostela, Spain
First published on 17th September 2019
Abstract
The asymmetric 3,5-disubstituted 1,2,4-triazole ligand H2V (5-amino-3-picolinamido-1,2,4-triazole) by reaction with an excess of Cu(II) perchlorate (Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2V being 12
H2V being 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) has produced a novel hexanuclear {Cu6(μ3-O/H)(HV/V)3} fragment, with one triangular Cu3(μ3-O/H) group connected to three peripheral single Cu(II) ions through a cis–cis–trans bridging mode of the ligand, which is the building block of the three structures described here: one hexanuclear, [Cu6(μ3-O)(HV)3(ClO4)7(H2O)9]·8H2O (1), one dodecanuclear, [Cu12(μ3-O)2(V)6(ClO4)5(H2O)18](ClO4)3·6H2O (2), and one tetradecanuclear 1D-polymer, {[Cu14(μ3-OH)2(V)6(HV)(ClO4)11(H2O)20](ClO4)2·14H2O}n (3), the last two containing hexanuclear subunits linked by perchlorato bridges. The Cu–Cu av. intra-triangle distance is 3.352(2) Å and the Cu(central)–Cu(bridged external) av. distance is 5.338(3) Å. The magnetic properties of the hexanuclear “Cu3O–3Cu” cluster have been studied, resulting as best fit parameters: g = 2.18(1), J(intra-triangle) = −247.0(1) cm−1 and j(central CuII – external CuII) = −5.15(2) cm−1.
1) has produced a novel hexanuclear {Cu6(μ3-O/H)(HV/V)3} fragment, with one triangular Cu3(μ3-O/H) group connected to three peripheral single Cu(II) ions through a cis–cis–trans bridging mode of the ligand, which is the building block of the three structures described here: one hexanuclear, [Cu6(μ3-O)(HV)3(ClO4)7(H2O)9]·8H2O (1), one dodecanuclear, [Cu12(μ3-O)2(V)6(ClO4)5(H2O)18](ClO4)3·6H2O (2), and one tetradecanuclear 1D-polymer, {[Cu14(μ3-OH)2(V)6(HV)(ClO4)11(H2O)20](ClO4)2·14H2O}n (3), the last two containing hexanuclear subunits linked by perchlorato bridges. The Cu–Cu av. intra-triangle distance is 3.352(2) Å and the Cu(central)–Cu(bridged external) av. distance is 5.338(3) Å. The magnetic properties of the hexanuclear “Cu3O–3Cu” cluster have been studied, resulting as best fit parameters: g = 2.18(1), J(intra-triangle) = −247.0(1) cm−1 and j(central CuII – external CuII) = −5.15(2) cm−1.
Introduction
Polynuclear Cu(II) compounds are of interest in biology and in magnetochemistry.1 The structural and electronic factors that govern exchange coupling have been well stablished in small di-, tri- and tetrametallic clusters.2 More recently, the pursuit of single molecule magnets has focused the attention on ever larger clusters with higher multiplicity ground states.3 In addition, the last few years have witnessed an impressive growth of the literature on 1D, 2D and 3D coordination polymers (CPs) with the aggregates of Cu(II) compounds emerging as a type of subunit whose (self)assemblies may form supramolecular structures.4The rich bridging chemistry of the 1,2,4-triazole ligands towards first-row transition metals has produced multiple nuclearities and topologies.5,6 We have previously reported a series of 3-substituted-1,2,4-triazole derivatives which are able to generate trinuclear copper-complexes containing the [Cu3(μ3-OH)(trz)3]n+ core.7,8 This specific class of tricopper clusters (with trigonal symmetry) are receiving great attention for two reasons: (1) because of their magnetic singularity, which involves geometric spin frustration effects, antisymmetric exchange coupling and unusual electron paramagnetic resonance (EPR) response,7–9 features which make them relevant as magnetic models to understand the mechanism of action of multicopper enzymatic systems;10 and (2) as subunits (secondary building units, SBU) of metal–organic frameworks (MOFs, also termed porous coordination polymers, PCPs) which combine magnetism and porosity.11,12
Literature on triangular Cu3O/OH arrays has basically been focused on oximato (N,O), pyrazole (N,N), 1,2,4-triazole (N,N) and more recently 1,2,3-triazole (N,N) and tetrazole (N,N) bridging groups.13 The assembly of triangular units in larger aggregates has been achieved (a) by dimerization of the trimeric units through H-bonds between μ3-O/μ3-OH centered triangles,14,15a (b) through bridging counteranions,15b,c (c) by means of bi-topic amines15d,e or carboxylate bridges/bis-carboxylato linkers,15f,g and (d) with the use of carboxylate-functionalized N,N ligands (construction of MOFs).16–19
In oximate systems, some hexanuclear systems20 and a few catena21 have been reported to date. In contrast, for the pyrazole family there are many examples of hexacopper complexes and of chains of Cu3O fragments grown with chlorido, sulfato, perchlorato, thiocyanato, carboxylato or bipyridine bridges.15
As for the 1,2,4-triazol systems, Cu3O/OH assembles are more scarce in number but more diverse than the pyrazole ones,13 and the corresponding works could be classified in three groups. (A) The most prolific one comprises a series of fascinating 2D and 3D structures mostly obtained from solvothermal synthesis by self-association of bifunctional triazole-carboxylate/carboxylic ligands,16–18 triazole-isophthalate ligands,19 or ternary triazolate-sulfoisophthalate systems.11,12 (B) A second group includes the studies which combine triazole ligands with polyoxometalates to render POM-based trinuclear clusters.22–24 (C) The third class contains a few examples of polymers made up with simple triazole ligands in which the triangular Cu3O motifs are bridged by ditopic anions such as sulfonate25 or nitrate,14 as well as one hexanuclear structure with an unusual μ6-Cl− anion.26 There are also two special cases with a Cu(II) center linking two Cu3O units.7b,25 Finally, it has been described one related tetrazole hexanuclear copper complex constructed with sulfato bridges.27
In this report we introduce the small polydentate 1,2,4-triazole ligand which not only affords triangular Cu3O cores but also chelates in trans one external Cu(II) center to give an unprecedented Cu3O–3Cu (Cu3 + Cu + Cu + Cu) hexanuclear compound with formula [Cu6(μ3-O)(HV)3(ClO4)7(H2O)9]·8H2O (1). Further on, the linkage of two hexanuclear motifs through μ2/μ3-perchlorate anions has yielded one dodecanuclear discrete compound and one tetradecanuclear 1D-coordination polymer formulated as [Cu12(μ3-O)2(V)6(ClO4)5(H2O)18](ClO4)3·6H2O (2) and {[Cu14(μ3-OH)2(V)6(HV)(ClO4)11(H2O)20](ClO4)2·14H2O}n (3), respectively. The three structures and the magnetic properties of this new type of hexanuclear cluster are presented.
Experimental
Synthesis of 1, 2 and 3
Synthesis of the ligand H2V was performed as previously reported.28A methanolic suspension (20 mL) of H2V (0.25 mmol, 0.052 g) was stirred for 20 min and then heated with stirring at 50 °C for 10 min (ligand only partially solved). At that point a methanolic solution (5 mL) of Cu(ClO4)2·6H2O (3 mmol, 1.111 g) was added dropwise and the mixture stirred for 1 h. The resulting green suspension was filtered off. Additional methanol (2.5 mL) was added to the filtered solution, which was deposited on a crystallizing dish (initial reactants ratio is H2V![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cu(II) = 1
Cu(II) = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12). Cubic-like green crystals of 1 suitable for X-ray analysis were obtained after ca. two months. Yield: ca. 60%. IR (ATR, cm−1): 3459(m), 3363(m), 1660(s, sharp), 1617(m), 1588(s, sharp), 1532(s, sharp), 1484(w), 1434(w), 1394(s, sharp), 1307(w), 1266–1241(w), 1054(vs), 985(w, shoulder), 929(w). Anal. calc. for C24H55Cl7Cu6N18O49 (1) (2009.25): C, 14.35; H, 2.76; N, 12.55, Cl, 12.35. Found: C, 14.00; H, 2.60; N, 11.96; Cl, 11.98. The synthesis was reproduced several times. Some of the crystallizations (the most concentrated, approx. 1 out of 5) rendered crystals of different shapes. The X-ray study revealed that the most abundant, the cubic-like ones, correspond to compound 1 (approx. 55% yield). A few of them, hexagonal-prismatic shaped, correspond to compound 2 (approx. 4% yield). Finally, a third type, big long-prismatic shaped, corresponds to compound 3 (approx. 1% yield).
12). Cubic-like green crystals of 1 suitable for X-ray analysis were obtained after ca. two months. Yield: ca. 60%. IR (ATR, cm−1): 3459(m), 3363(m), 1660(s, sharp), 1617(m), 1588(s, sharp), 1532(s, sharp), 1484(w), 1434(w), 1394(s, sharp), 1307(w), 1266–1241(w), 1054(vs), 985(w, shoulder), 929(w). Anal. calc. for C24H55Cl7Cu6N18O49 (1) (2009.25): C, 14.35; H, 2.76; N, 12.55, Cl, 12.35. Found: C, 14.00; H, 2.60; N, 11.96; Cl, 11.98. The synthesis was reproduced several times. Some of the crystallizations (the most concentrated, approx. 1 out of 5) rendered crystals of different shapes. The X-ray study revealed that the most abundant, the cubic-like ones, correspond to compound 1 (approx. 55% yield). A few of them, hexagonal-prismatic shaped, correspond to compound 2 (approx. 4% yield). Finally, a third type, big long-prismatic shaped, corresponds to compound 3 (approx. 1% yield).
Crystal structure determination
Crystal data, data collection and structure refinement details for 1, 2 and 3 are summarized in Table 1. Diffraction data were obtained at 100(1) K using a Bruker SMART CCD 1000 (1 and 2) or a Bruker X8 Kappa APEXII (3) diffractometer from crystals mounted on glass fibers. Data were corrected for Lorentz and polarization effects and for absorption following multi-scan type.29 The structures were solved by direct methods and subsequent difference Fourier maps30 and refined on F2 by a full-matrix least-squares procedure using anisotropic displacement parameters.30 Hydrogen atoms attached to carbon and nitrogen atoms were placed in geometrically idealized positions and were refined with isotropic displacement parameters constrained to 1.2/1.5 Ueq of the carrier atoms. Molecular graphics were generated with DIAMOND.31 For each structure peculiarities of the refinement are indicated in ESI (S1–S3).†| 1 | 2 | 3 | |
|---|---|---|---|
| Chem. formula | C24H55Cl7Cu6N18O49 | C48H84Cl8Cu12N36O64 | C56H113Cl13Cu14N42O95 |
| Mr | 2009.25 | 3235.59 | 4245.29 |
| Cryst. size (mm) | 0.260 × 0.230 × 0.220 | 0.240 × 0.150 × 0.100 | 0.320 × 0.260 × 0.120 |
| Cryst. syst. | Trigonal | Hexagonal | Monoclinic |
| Space group | R![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) :H :H |
P63/m | C2/c |
| a (Å) | 20.496(3) | 20.811(3) | 35.294(3) |
| b (Å) | 20.496(3) | 20.811(3) | 20.8571(17) |
| c (Å) | 33.675(6) | 22.620(3) | 23.370(3) |
| α (°) | 90 | 90 | 90 |
| β (°) | 90 | 90 | 98.149(5) |
| γ (°) | 120 | 120 | 90 |
| V (Å3) | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 251(4) 251(4) |
8484(3) | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 030(3) 030(3) |
| Z, Dc (g cm−3) | 6, 1.634 | 2, 1.267 | 4, 1.656 |
| h/k/l | −25/12, 0/25, 0/42 | −18/0, 0/21, 0/23 | −39/38, 0/23, 0/25 |
| F(000) | 6060 | 3240 | 8520 |
| Absorption coeff. (mm−1) | 1.865 | 1.673 | 2.019 |
| No. of collected/unique rflns | 33712/5630 | 47107/3580 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 143/12 143/12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 143 143 |
| Rint | 0.0498 | 0.0542 | 0.0516 |
| No. of data/restraints/params | 5630/67/318 | 3580/13/301 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 143/148/777 143/148/777 |
| R1/wR2 (I > 2σ(I)) | 0.1021/0.2846 | 0.0798/0.2308 | 0.1198/0.2663 |
| R1/wR2 (all data) | 0.1521/0.3377 | 0.1125/0.2547 | 0.1768/0.2873 |
| Max, min transmission | 1.0000, 0.7645 | 1.0000, 0.7555 | 1.0000, 0.8926 |
| GOF on F2 | 1.049 | 1.041 | 1.096 |
| Δρmax, Δρmin (e Å−3) | 1.939, −1.352 | 1.407, −0.503 | 3.268, −1.283 |
| CCDC number | 1935979 | 1935980 | 1935981 |
Magnetic measurements
Magnetic susceptibility measurements on polycrystalline samples were carried out with a Superconducting Quantum Interference Design (SQUID) magnetometer in the temperature range of 1.9–300 K under magnetic fields of 250–5000 Gauss. Diamagnetic corrections of the constituent atoms were estimated from Pascal's constants. Experimental susceptibilities were also corrected for the temperature independent paramagnetism, χTIP = 60 × 10−6 cm3 mol−1 per copper(II), and for the magnetization of the sample holder. X-Band EPR spectra of polycrystalline samples were recorded at different temperatures with a Bruker ER 200 spectrometer equipped with a helium continuous-flow cryostat.Results
The ligand
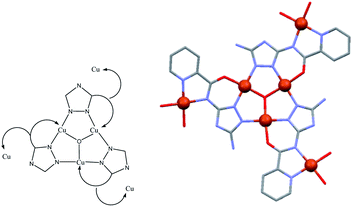
The ligand of the present study, H2V [5-amino-3(pyridine-2-yl-acetamido)-1,2,4-triazole![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) 5-amino-3-picolinamido-1,2,4-triazole], was obtained from mono-acetylation of the guanazole with picolinic acid.28 In a previous work we reported on a similar ligand, the H3diV [3,5-bis(picolinamido)-1,2,4-triazole], synthetized by di-acetylation of the guanazole (Scheme 1). From the previous H3diV ligand and with an excess of copper(II) perchlorate a hexanuclear cationic complex of formula [Cu6(HdiV)2(ClO4)6(H2O)14]2+ was formed, which could be described as a cluster of the “1 + 1 + 2 + 1 + 1” type.32 The ligand H2V, a 1,2,4-triazole ligand bearing only one chelating arm, studied here as such for the first time, in the presence of Cu(II) forms the Cu3N6 nine-membered ring of the Cu3O(H) triangular family.7,8 The feature which makes H2V unique is the bis-chelating nature of the triazole substituent which links in trans one additional atom centre, thus resulting in the “3 + 1 + 1 + 1” hexanuclear copper(II) core-motif (Scheme 2). From this building block three different structures have been formed which we present here: one hexanuclear Cu6 (1), one di-hexanuclear (2 × Cu6=Cu12) (2), and one 1D-polymer made of dimers linked by pairs of hexamers (Cu2–Cu6–Cu6–Cu2′, Cu14) (3).
5-amino-3-picolinamido-1,2,4-triazole], was obtained from mono-acetylation of the guanazole with picolinic acid.28 In a previous work we reported on a similar ligand, the H3diV [3,5-bis(picolinamido)-1,2,4-triazole], synthetized by di-acetylation of the guanazole (Scheme 1). From the previous H3diV ligand and with an excess of copper(II) perchlorate a hexanuclear cationic complex of formula [Cu6(HdiV)2(ClO4)6(H2O)14]2+ was formed, which could be described as a cluster of the “1 + 1 + 2 + 1 + 1” type.32 The ligand H2V, a 1,2,4-triazole ligand bearing only one chelating arm, studied here as such for the first time, in the presence of Cu(II) forms the Cu3N6 nine-membered ring of the Cu3O(H) triangular family.7,8 The feature which makes H2V unique is the bis-chelating nature of the triazole substituent which links in trans one additional atom centre, thus resulting in the “3 + 1 + 1 + 1” hexanuclear copper(II) core-motif (Scheme 2). From this building block three different structures have been formed which we present here: one hexanuclear Cu6 (1), one di-hexanuclear (2 × Cu6=Cu12) (2), and one 1D-polymer made of dimers linked by pairs of hexamers (Cu2–Cu6–Cu6–Cu2′, Cu14) (3).
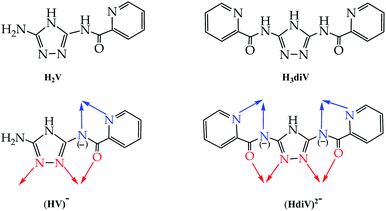 | ||
| Scheme 1 Comparing ligands H2V and H3diV: bridging mode of the mono-deprotonated (HV)− (or the di-deprotonated V2−) (this work) and the di-deprotonated (HdiV)2− ligands.32 | ||
Description of structures
| 1 | 2 | 3 | |
|---|---|---|---|
| a For 1: symmetry transformations used to generate equivalent atoms: #1 −y + 1, x − y + 1, z, #2 −x + y, −x + 1, z. For 2: symmetry transformations used to generate equivalent atoms: &3 x, y, −z + 1/2, &4 −x + y + 1, −x + 1, −z + 1/2.b Labels referred to compound 1 (Fig. 1a and b); for compounds 2 and 3 the distances listed correspond to the equivalent bonds. In italic, labels specific of compound 2; in bold labels specific of compound 3. For complete tables of distances and angles see S4–S6. | |||
| Cu(1)–N(20) | 1.909(6) | 1.903(6) | 1.912(10), 1.910(10), 1.895(10) |
| Cu(1)–N(21) | 1.957(6) | 1.947(6) | 1.945(10), 1.935(10), 1.955(11) |
| Cu(1)–O(17) | 1.969(6) | 1.964(5) | 1.968(9), 1.968(9), 1.972(9) |
| Cu(1)–O(1) | 1.995(3) | 2.009(3) | 1.986(8), 1.994(9), 2.012(8) |
| Cu(1)–O(31)/O(21)/O(w) | 2.501(11) | 2.417(7) | 2.403(13), 2.385(11), 2.387(17) |
| Cu(1)–O(11) | 2.634(14) | 2.506(8) | 2.450(12), 2.852(19), 2.546(15) |
| Cu(1)#1–O(1)–Cu(1) | 113.8(3) | 113.6(2) | 114.8(4), 113.1(4), 114.1(4) |
| Cu(1)–Cu(1)#1 | 3.344(2) | 3.362(2) | 3.335(2), 3.353(2) |
| O(1)–O(1)′ | 5.208(9) | 5.648(18) | 6.816(9) |
| Cu(2)–O(3) | 1.972(8) | 1.933(18)AA/2.014(16)BB | 1.988(12), 1.965(12), 1.995(11) |
| Cu(2)–N(11) | 1.968(8) | 2.009(9)A/1.971(8)B | 1.974(12), 1.983(11), 1.960(12) |
| Cu(2)–O(2) | 1.994(8) | 2.005(18)AA/2.005(19)BB | 1.989(12), 2.003(11), 1.94(15) |
| Cu(2)–N(18) | 2.008(6) | 2.011(8)A/2.010(8)B | 2.009(11), 2.003(10), 2.007(12) |
| Cu(2)–O(21)/O(x1)/O(w) | 2.470(11) | 2.265(18), 2.422(15), 2.258(19) | |
| Cu(2)–O(4)/O(y1) | 2.69(3) | 2.21(4)AA/2.39(6)BB | 2.469(13), 2.456(17), 2.423(14) |
| Cu(1)–Cu(2) | 5.345(2) | 5.256(3)A/5.365(3)B | 5.361(2), 5.367(2), 5.352(3) |
| Cu(1)–Cu(1)&3 | 6.687(2) | ||
| Cu(1)–Cu(1)&4 | 7.485(2) | ||
| Cu(7)–O(77)/Cu(8)–O(103) | 1.76(3)/1.88(3) | ||
| Cu(7)–N(80)/Cu(8)–N(71) | 1.85(3)/1.964(16) | ||
| Cu(7)–O(101)/Cu(8)–N(78) | 1.94(5)/1.95(3) | ||
| Cu(7)–O(102)/Cu(8)–O(104) | 2.11(6)/2.00(3) | ||
| Cu(7)–O(52)/Cu(8)–O(14) | 2.67(2)/2.498(18) | ||
| Cu(7)–Cu(8) | 5.272(9) | ||
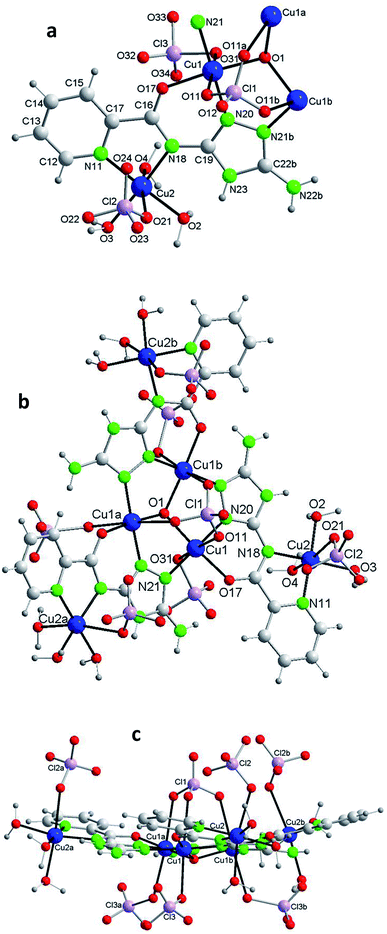
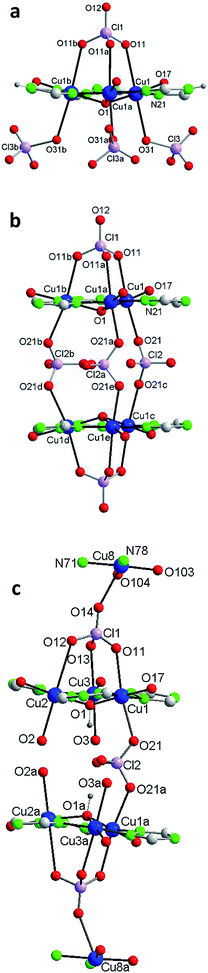
Each of the 3 (HV)− ligands bridges two Cu(II) ions of the Cu3O group while chelating one of them (bite angle of 86.5(2)°). Besides, each ligand chelates in trans another Cu(II) center, Cu2 (bite angle of 82.5(2)°) (Scheme 2, Fig. 1a and b). So, in whole, there are 3 external single Cu(II) atoms (Cu2, Cu2a, Cu2b). This second type of copper center exhibits a very distorted tetragonally octahedral stereochemistry, in which one N-amido, one N-pyridine and two O-water atoms occupy the basal sites and one O-water molecule and one O-perchlorate atom (at 2.47(2) Å) the axial ones.
In the triangular array, the Cu(1)–O(1) distances are of 1.995(3)Å and the Cu(1)–O(1)–Cu(1a) angles of 113.8(2)°. The central oxygen lies 0.51(1) Å below the plane defined by the three copper atoms. In spite of this deviation of the Cu3O moiety from planarity, the lack of H-bonding together with the charge balance (and the symmetry of the molecule) lead us to exclude the existence of the central oxygen as OH species (as found in related triangular triazole compounds)7,8 instead of O. The intra-triangle Cu1–Cu1 distance, 3.344(2) Å, is in the range observed for related systems.7a,8 The Cu1–Cu2 distance is 5.345(2) Å.
A view perpendicular to the c axis shows the {Cu6(μ3-O)(HV)3} fragment defining a rough plane (Fig. 1c). The hexanuclear unit is chiral although, since in the network there are inversion centers, the bulk solid is racemic (i.e. the compound is achiral). Above the plane there are 4 perchlorate anions, three monodentate (towards each of the external Cu(II) centers) and the mentioned tridentate (which bridges the three central Cu(II) atoms); below the plane another 3 monodentate perchlorato ligands complete the octahedral coordination of the three Cu3O copper atoms. The internal perchlorates together with the axially coordinated water molecules connect one {Cu6(μ3-O)(HV)3} unit with an upside-down second {Cu6(μ3-O)(HV)3} unit, of opposite quirality, through H-bonds (Fig. S7†). The O1⋯O1′ distance is 5.208(9) Å. In turn, the external perchlorate anions bind each pair of hexamers with another pair via additional H-bonds thus giving chains of pairs of hexanuclear compounds (Fig. S8†).
The structure of 2, as that of 1, is based on the hexanuclear {Cu6(μ3-O)(L)3} block, the main difference between both being that in 2, two hexanuclear units related by one reflection plane (Fig. 2c and S7†) are linked by 3 perchlorate bidentate anions to give a dodenuclear Cu(II) compound (Fig. 2b and d). The O(1)⋯O(1)′ distance in 2, of 5.648(18) Å, is somewhat longer than the equivalent O(1)⋯O(1)′ distance in paired units of 1 (5.208(9) Å). The deviation of O(1) from the Cu3 plane is similar (0.52(1) Å). The schematic view of Fig. 2d shows the position of the 8 ClO4− groups: 2 tridentate external (on a 6-fold axis) and 6 interleaved, 3 of which are bidentate and connect central Cu3–O atoms from two opposite triangular units, and the other 3 being non-coordinating but contributing to stabilize the dodenuclear species through H-bonds. The 6 enclosed perchlorate anions are placed on a mirror plane which contains the Cl atom and two out of the four O atoms of each perchlorate.
A second difference with 1 stands on the lack of any perchlorate on the apical coordination positions of the Cu2 peripheral centers. To be noticed the disorder on these Cu2 atoms (Fig. 2a and S9†). The fragment O(2)–Cu(2)–O(3) of the metal coordination sphere has been assumed to occupy two alternative orientations inclined at an angle of 37(1)° and given an occupancy of 0.5 for each position. Fig. 2b–d display only one out of the two positions for clarity purposes. While the geometry of the central Cu1 atoms is distorted octahedral, Cu(N2O2 + O2) (like in 1), that of the outer Cu2 centers is Cu(N2O2 + O) penta-coordinated. Table 2 compares selected bond angles and distances in the structure of 1 and 2.
A last remarkable feature of the structure of 2 refers to the packing. A view projected on plane ab reveals the existence of wide channels in the network (a 18.631(9) Å void between opposite N4-triazole atoms) (Fig. 2e).
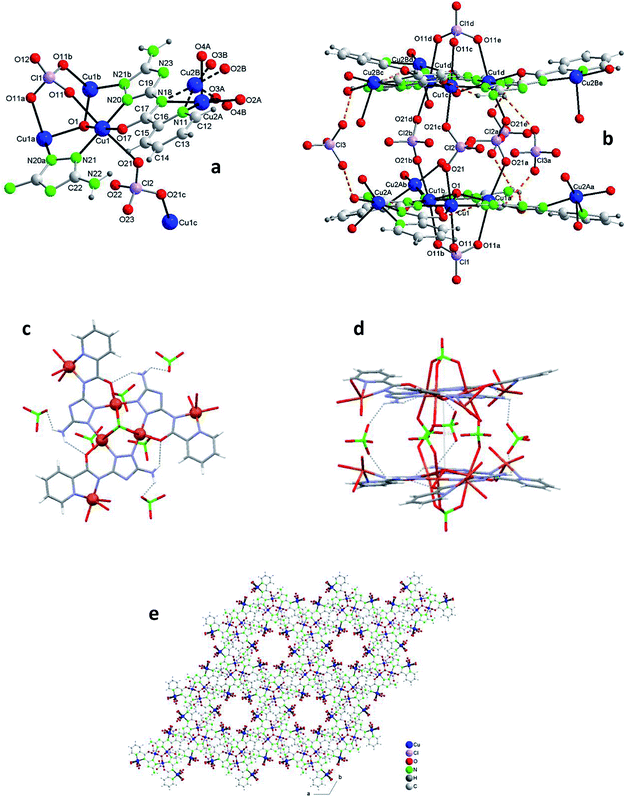
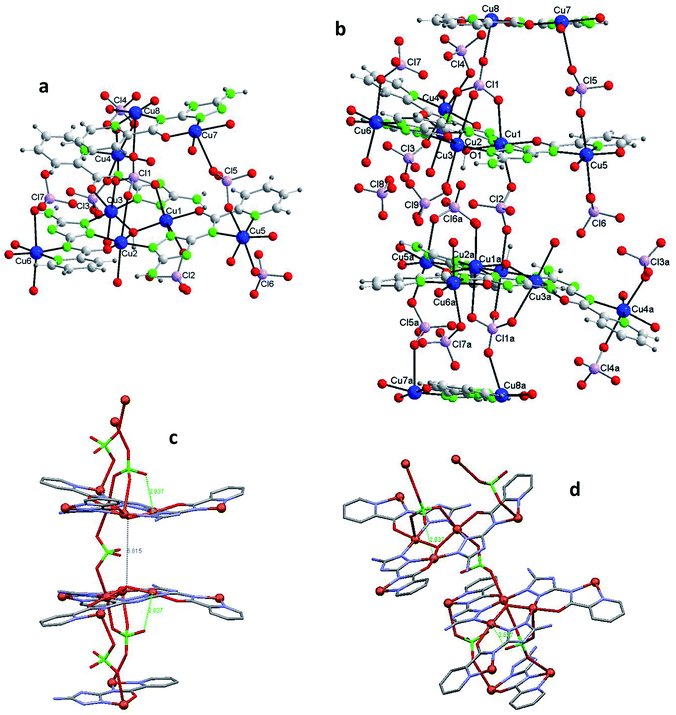
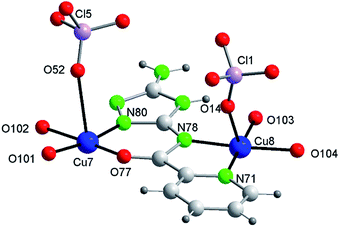
Fig. 3b shows one of those 14Cu species. Fig. 3c and d are alleviated pictures in which water molecules (coordinating/crystallization) and non-coordinating/monodentate perchlorate anions are not displayed to facilitate visualization of the 14Cu centers and the different bridges that connect them (note: Fig. 3b–d contain 14 + 2 Cu).
The 14Cu aggregate contains 8 different (symmetry independent) Cu(II) centers, 9 different (symmetry independent) perchlorate anions and 7 (6V2− + 1(HV)−) ligands. Cu1, Cu2, Cu3, Cu4, Cu5 and Cu6, together with 3 ligands and one μ3-hidroxo ligand, produce each of the two hexanuclear units, which are symmetry related and of different quirality (see 1 and 2) (Fig. S10†). The 7th (HV)− ligand, which acts as trans-bichelate, bridges Cu7 and Cu8 to form the dimeric group (Fig. 4). The two hexamers are linked via one μ2-perchlorato ligand (Cl2) [bonds Cu1–O21(Cl2)O21a–Cu1a], with the O1⋯O1a distance being 6.816(9) Å, far longer than the equivalent distance in 1 and 2, as expected since in 3 the two hexanuclear units are not properly paired but rather shifted towards each other. One μ4-perchlorato ligand (Cl1) which coordinates on apical positions simultaneously Cu1, Cu2 (semi-coordination, 2.852(19) Å, in this case), Cu3 and Cu8 links one hexamer with the dinuclear moiety (Fig. 3a; see also S10†). There is one additional bridge, a second μ2-perchlorato ligand (Cl5) which connects Cu7 with Cu5 [Cu5–O51(Cl5)O52–Cu7]. 4 monodentate perchlorate anions coordinate axially to Cu4 (Cl3, Cl4), Cu5 (Cl6) and Cu6 (Cl7) (Fig. 3c and d). The other 2 perchlorate anions (Cl8 and Cl9) are not coordinating.
The chain grows through (a) another μ4-perchlorato ligand (Cl1a) which coordinates the Cu3-triangular copper centers of the second hexamer (Cu1a, Cu2a, Cu3a), with the Cu8a copper atom of another dimeric unit (Cu8a, Cu7a), and (b) a new μ2-perchlorato ligand (Cl5a) which links Cu5a and Cu7a (Fig. 3b).
In the hexanuclear unit the three ligand planes are visibly deviated from the plane defined by Cu1, Cu2 and Cu3. The large distortion is produced by the polydentate character of the coordinating perchlorate anions. Deviation of the central μ3-O1(H) atom toward the triangular Cu3 plane is 0.498(10)Å (slightly lower than in 1 and 2).
As for the coordination geometry of the copper sites, the 6Cu(II) of the hexanuclear arrays display analogous distorted octahedral N2O2 + O2 environment but with different ligands on apical positions (Fig. 3b). On axial positions Cu1 presents two polydentate perchlorato ligands; Cu2 and Cu3 one (the) tetradentate perchlorate anion and one water molecule; Cu4 and Cu5, two perchlorate anions; Cu6 one perchlorate and one water molecule. The 2Cu(II) centers of the dimeric moiety are NO3 + O penta-coordinated (Fig. 4). The basal positions of Cu7 are occupy by the N(triazole) and O(carbonyl) atoms of the ligand, and two O water atoms; the apical ones, by one O perchlorate atom (O52). To end, Cu8 is equatorially coordinated to the N(acetamido) and N(pyridine) atoms of the ligand and to two O water atoms, and apically to the O14 atom of the Cl1 perchlorate. Bond distances in the coordination polyhedron are listed in Table 2.
Finally to be indicated that in the dimeric unit the Cu7⋯Cu8 distance is 5.272(9) Å [the average Cu(central)–Cu(bridged-external) distance is 5.360(2) Å in the hexanuclear units of 3]. The Cu8⋯Cu8a distance in two consecutive Cu14 units of the chain is 20.387(9) Å whereas the Cu7⋯Cu7a distance is 22.117(9) Å.
Fig. 5 shows the Cu3O cores of 1, 2 and 3 with the corresponding coordinating perchlorates (the Cu peripheral centers are not included) to emphasize the different association mediated by the perchlorate anions.
Magnetic studies
The thermal dependence of the χMT product for 1 (χM being the magnetic susceptibility per Cu6 unit) is shown in Fig. 6.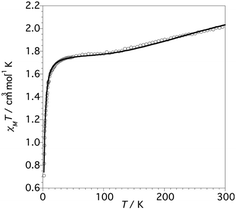 | ||
| Fig. 6 χMT vs. T plot for complex 1 (values per Cu6 unit). The solid line corresponds to the best-fit parameters obtained from the Hamiltonian (eqn 1). | ||
At 300 K the χMT value is 2.05 cm3 K mol−1, which is lower than the value expected for six non-interacting CuII S = 1/2 ions (∼2.4 cm3 K mol−1) with a reasonable g-value.2,33 Upon cooling, the χMT product decreases slowly reaching a plateau at 1.75 cm3 K mol−1 between 90 and 60 K. Below ca. 20 K, χMT decreases sharply to achieve a value of 0.70 cm3 K mol−1 at 2 K.
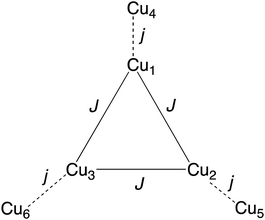
From structural analysis (Schemes 2 and 3) this behavior can be rationalized considering two steps. Firstly, the high-temperature region (300–100 K) reveals the presence of a moderate-to-strong antiferromagnetic interaction within the “triangular unit” between the Cu1, Cu2 and Cu3 centers, doubly bridged via the central oxo ligand and the peripheral N–N triazole atoms. This {Cu3(μ3-O/OH)(N–N)3} bridging system has already been described as leading to significant antiferromagnetic coupling.7–9,33 In previous studies we reported a series of trinuclear compounds of this type built with triazole ligands which exhibited χMT values of 0.37–0.40 cm3 K mol−1 in the range 90–60 K.7,8 The plateau value of 1.75 cm3 K mol−1 for 1 would roughly correspond to the presence of the Cu3 triangle plus three almost isolated CuII additional centers (0.40 + 1.30).
Secondly, at lower temperatures (60–2 K), a weaker antiferromagnetic exchange involving the pairs Cu1–Cu4, Cu2–Cu5 and Cu3–Cu6 occurs. This weak antiferromagnetic interaction takes place through the double “NCN + NCO” bridge (Scheme 2), in a trans-bridging mode (following literature terminology),34 as observed previously in the “2 + 1 + 1 + 1 + 1” hexanuclear compound [Cu6(HdiV)2(ClO4)6(H2O)14](ClO4)2·10H2O (A).32
Considering all of these features and the symmetry of the structure (Cu3 is a perfect triangle) the magnetic data were simulated by means of the following C3-symmetric Hamiltonian (eqn 1):2
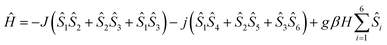 | (1) |
Fitting of the magnetic properties of CuII triangular clusters, example of spin frustrate systems, requires the use of both isotropic and ASE Hamiltonian terms to account for the low temperatures behavior.7–9,33 For 1, however, the experimental data were well reproduced without incorporation of the ASE term into the magneto-chemical analysis. Presumably the ASE is shadowed by the dominant antiferromagnetic interaction within the 3 dimeric Cu(II) units. The resulting best-fit parameters were:
| g = 2.18(1), J = −247.0(1) cm−1 and j = −5.15(2) cm−1. |
The high symmetry of the structure of 1 must be responsible of the high value of J (−247 cm−1), constant of the AF exchange within the tri-copper triangle, when compared with the values exhibited by the reported analogous triazole trimers (177–195 cm−1).8 In contrast, the j constant (−5.15 cm−1), corresponding to the AF exchange between the coupled “internal Cu-peripheral Cu” pairs is significantly lower than the one observed for a similar pathway in the hexanuclear compound A (−35 cm−1).32 The polydentate character of the perchlorate ligands on axial positions, which induces significant distortion on the equatorial planes of Cu1 (central) and Cu(2) (peripheral), could account for the lower value of j.35
EPR spectra at low T have been recorded in search of the ASE signature. The X-band powder spectrum of 1 at 4 K (Fig. S11†) displays signals at g‖ = 2.30 and g⊥ = 1.61, the last one being indeed indicative of the existence of ASE (g < 2.0).8,9
Conclusions
We have prepared and tested a new simple 1,2,4-triazole ligand, H2V, capable of yielding with Cu(II) a new building block consisting of hexanuclear Cu(II) clusters with Cu3–O(H) core, that is, of the triangular type, but with expanded nuclearity: Cu3O(H)–3Cu. The hexanuclear units can aggregate in different forms. In the presence of perchlorate anions we have isolated and described three different structures with 6Cu (compound 1), 12Cu (compound 2) and 14Cu (compound 3) of nuclearity. The magnetic properties of the hexanuclear cluster have been studied with 1 and rationalized. Compound 2 combines high nuclearity and a porous 3D structure. Compound 3 is a mixed system (hexanuclear units and dimeric units) in which the dinuclear units link dodecanuclear assembles to afford a one-dimensional coordination polymer. While assembles of triangular Cu3O groups have been extensively described the introduction of the Cu3O–Cu3 fragment offers a straightforward way to enhance the magnetism of the resulting frameworks/CPs. We plan to explore the possibilities of this hexanuclear Cu(II) scaffold with different anions and anionic linkers.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Spanish Ministerio de Economía y Competitividad and FEDER (CTQ2016-78341-P), Unidad de Excelencia María de Maeztu (MDM-2015-0538) and Generalitat Valenciana through PROMETEO/2016/147. F. J. V.-M. thanks MINECO for a predoctoral (FPI) grant.References
- (a) D. Gatteschi, A. Caneschi, L. Pardi and R. Sessoli, Science, 1994, 265, 1054–1058 CrossRef CAS PubMed; (b) A. P. Cole, D. E. Root, P. Mukherjee, E. I. Solomon and T. D. P. Stack, Science, 1996, 273, 1848–1850 CrossRef PubMed; (c) D. Gatteschi and R. Sessoli, Angew. Chem., Int. Ed., 2003, 42, 268–297 CrossRef CAS.
- (a) O. Kahn, Molecular Magnetism, VCH Publishers, New York, 1993 Search PubMed; (b) P. J. Hay, J. C. Thibeault and R. offmann, J. Am. Chem. Soc., 1975, 97, 4884–4899 CrossRef CAS.
- E. M. Zueva, M. M. Petrova, R. Herchel, Z. Trávnícek, R. G. Raptis, L. Mathivathananc and J. E. McGrady, Dalton Trans., 2009, 5924–5932 RSC.
- N. Stock and S. Biswas, Chem. Rev., 2012, 112, 933–969 CrossRef CAS PubMed.
- (a) J. Haasnoot, Coord. Chem. Rev., 2000, 200–202, 131–185 CrossRef CAS; (b) G. Aromí, L. Barrios, O. Roubeuau and P. Gámez, Coord. Chem. Rev., 2011, 255, 485–546 CrossRef.
- (a) M. H. Klingele and S. Brooker, Coord. Chem. Rev., 2003, 241, 119–132 CrossRef CAS; (b) U. Beckmann and S. Brooker, Coord. Chem. Rev., 2003, 245, 17–29 CrossRef CAS; (c) J. Olguín, M. Kalisz, R. Clerac and S. Brooker, Inorg. Chem., 2012, 51, 5058–5069 CrossRef PubMed.
- (a) S. Ferrer, J. G. Haasnoot, J. Reedijk, E. Müller, M. Biagini-Cingi, M. Lanfranchi, A. M. Manotti Lanfredi and J. Ribas, Inorg. Chem., 2000, 39, 1859–1867 CrossRef CAS PubMed; (b) S. Ferrer, E. Aznar, F. Lloret, A. Castiñeiras, M. Liu-González and J. Borrás, Inorg. Chem., 2007, 46, 372–374 CrossRef CAS PubMed.
- (a) S. Ferrer, F. Lloret, I. Bertomeu, G. Alzuet, J. Borrás, S. García-Granda, M. Liu-González and J. G. Haasnoot, Inorg. Chem., 2002, 41, 5821–5830 CrossRef CAS PubMed; (b) S. Ferrer, F. Lloret, E. Pardo, J. M. Clemente-Juan, M. Liu-González and S. García-Granda, Inorg. Chem., 2012, 51, 985–1001 CrossRef CAS PubMed.
- (a) T. Moriya, Phys. Rev. Lett., 1960, 4, 228–230 CrossRef CAS; (b) B. S. Tsukerblat, B. Y. Kuyavskaya, M. I. Belinskii, A. V. Ablov, V. M. Novotortsev and V. T. Kalinnikov, Theor. Chim. Acta, 1975, 38, 131–138 CrossRef CAS.
- (a) S. I. Chan, V. C.-C. Wang, J. C.-H. Lai, S. S.-F. Yu, P. P.-Y. Chen, K. H.-C. Chen, C. L. Chen and M. K. Chan, Angew. Chem., Int. Ed., 2007, 46, 1992–1994 CrossRef CAS PubMed; (b) P. P.-Y. Chen, R. B.-G. Yang, J. C.-M. Lee and S. I. Chan, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 14570–14575 CrossRef CAS PubMed; (c) J. Yoon and E. I. Solomon, Coord. Chem. Rev., 2007, 251, 379–400 CrossRef CAS.
- M. Deng, P. Yang, X. Liu, B. Xia, Z. Chen, Y. Ling, L. Weng, Y. Zhou and J. Sun, Cryst. Growth Des., 2015, 15, 1526–1534 CrossRef CAS.
- E.-C. Yang, Y.-Y. Zhang, Z.-Y. Liu and X.-J. Zhao, Inorg. Chem., 2014, 53, 327–335 CrossRef CAS PubMed.
- ConQuest Interface, The Cambridge Structural Database, release 5.40; Cambridge, U.K., accessed May 2019 Search PubMed.
- C.-Y. Su, Y.-Z. Tong, F.-C. Yuan, Q.-L. Wang, Y. Ma, G.-M. Yang and D.-Z. Liao, Inorg. Chim. Acta, 2014, 423, 545–549 CrossRef CAS.
- (a) L. Mathivathanan, M. Rivera-Carrillo and R. G. Raptis, Inorg. Chim. Acta, 2012, 391, 201–205 CrossRef CAS; (b) L.-L. Zheng, J.-D. Leng, S.-L. Zheng, Y.-C. Zhaxi, W.-X. Zhang and M.-L. Tong, CrystEngComm, 2008, 10, 1467–1473 RSC; (c) C. Di Nicola, F. Garau, M. Gazzano, M. Monari, L. Pandolfo, C. Pettinari and R. Pettinari, Cryst. Growth Des., 2010, 10, 3120–3131 CrossRef CAS; (d) F. Condello, F. Garau, A. Lanza, M. Monari, F. Nestola, L. Pandolfo and C. Pettinari, Cryst. Growth Des., 2015, 15, 4854–4862 CrossRef CAS; (e) C. Di Nicola, F. Garau, M. Gazzano, M. F. C. Guedes da Silva, A. Lanza, M. Monari, F. Nestola, L. Pandolfo, C. Pettinari and A. J. L. Pombeiro, Cryst. Growth Des., 2012, 12, 2890–2901 CrossRef CAS; (f) S. Massignani, R. Scatena, A. Lanza, M. Monari, F. Condello, F. Nestola, C. Pettinari, F. Zorzi and L. Pandolfo, Inorg. Chim. Acta, 2017, 455, 618–626 CrossRef CAS; (g) S. Contaldi, C. Di Nicola, F. Garau, Y. Y. Karabach, L. M. D. Martins, M. Monari, L. Pandolfo, C. Pettinari and A. J. L. Pombeiro, Dalton Trans., 2009, 4928–4941 RSC.
- S. I. Vasylevs'kyy, G. A. Senchyk, A. B. Lysenko, E. B. Rusanov, A. N. Chernega, J. Jezierska, H. Krautscheid, K. V. Domasevitch and A. Ozarowski, Inorg. Chem., 2014, 53, 3642–3654 CrossRef PubMed.
- D.-M. Chen, X.-P. Zhang, W. Shi and P. Cheng, Inorg. Chem., 2015, 54, 5512–5518 CrossRef CAS PubMed.
- N. N. Adarsh, M. M. Dirtu, A. D. Naik, A. F. Leonard, N. Campagnol, K. Robeyns, J. Snauwaert, J. Fransaer, B. L. Su and Y. Garcia, Chem. - Eur. J., 2015, 21, 4300–4307 CrossRef CAS PubMed.
- (a) D. Lässig, J. Lincke, J. Moellmer, C. Reichenbach, A. Moeller, R. Gläser, G. Kalies, K. A. Cychosz, M. Thommes, R. Staudt and H. Krautscheid, Angew. Chem., Int. Ed., 2011, 50, 10344–10348 CrossRef PubMed; (b) J. Lincke, D. Lassig, M. Kobalz, J. Bergmann, M. Handke, J. Mollmer, M. Lange, C. Roth, A. Moller, R. Staudt and H. Krautscheid, Inorg. Chem., 2012, 51, 7579–7586 CrossRef CAS PubMed; (c) J. Lincke, D. Lässig, K. Stein, J. Moellmer, A. V. Kuttatheyil, C. Reichenbach, A. Moeller, R. Staudt, G. Kalies, M. Bertmer and H. Krautscheid, Dalton Trans., 2012, 41, 817–824 RSC.
- (a) A. Escuer, G. Vlahopoulou, F. Lloret and F. A. F. Mautner, Eur. J. Inorg. Chem., 2014, 83–92 CrossRef CAS; (b) A. Escuer, B. Cordero, M. Font-Bardia and T. Calvet, Inorg. Chem., 2010, 49, 9752–9754 CrossRef CAS PubMed; (c) A. Chakraborty, A. Escuer, J. Ribas and T. K. Maji, Dalton Trans., 2016, 45, 15523–15531 RSC; (d) M. U. Anwar, L. K. Thompson and L. N. Dawe, Dalton Trans., 2011, 40, 1437–1440 RSC; (e) A. Tarushi, C. P. Raptopoulou, V. Psycharis, C. K. Kontos, D. P. Kessissoglou, A. Scorilas, V. Tangoulis and G. Psomas, Eur. J. Inorg. Chem., 2016, 219–231 CrossRef CAS; (f) J. P. Naskar, B. Guhathakurta, P. Basu, N. Bandyopadhyay, G. S. Kumar, M. Zhu and L. Lu, Inorg. Chim. Acta, 2017, 462, 158–166 CrossRef CAS; (g) M. Holynska, J. Mol. Struct., 2015, 1098, 175–180 CrossRef CAS; (h) L. K. Das, M. G. B. Drew, C. Diaz and A. Ghosh, Dalton Trans., 2014, 43, 7589–7598 RSC; (i) S. Karmakar, O. Das, S. Ghosh, E. Zangrando, M. Johann, E. Rentschler, T. Weyhermuller, S. Khanra and T. K. Paine, Dalton Trans., 2010, 39, 10920–10927 RSC; (j) H.-X. Li, X.-Y. Zhang, Z.-Y. Liu, E.-C. Yang and X.-J. Zhao, Inorg. Nano-Met. Chem., 2017, 47, 50–59 CrossRef CAS; (k) D. Maity, P. Mukherjee, A. Ghosh, M. G. B. Drew, C. Diaz and G. A. Mukhopadhyay, Eur. J. Inorg. Chem., 2010, 807–813 CrossRef CAS.
- (a) A. Chakraborty, K. L. Gurunatha, A. Muthulakshmi, S. Dutta, S. K. Pati and T. K. Maji, Dalton Trans., 2012, 41, 5879–5888 RSC; (b) L. Croitor, E. B. Coropceanu, O. Petuhov, K. W. Krämer, S. G. Baca, S.-X. Liu, S. Decurtins and M. S. Fonari, Dalton Trans., 2015, 44, 7896–7902 RSC.
- A.-X. Tian, Y.-L. Ning, J. Ying, G.-C. Liu, X. Hou, T.-J. Li and X.-L. Wang, CrystEngComm, 2015, 17, 5569–5578 RSC.
- X.-Y. Yang, M.-T. Li, N. Sheng, J.-S. Li, G.-D. Liu, J.-Q. Sha and J. Jiang, Cryst. Growth Des., 2018, 18, 5564–5572 CrossRef CAS.
- Z. Weng, Y. Ren, M. Gu, B. Yue and H. He, Dalton Trans., 2018, 47, 233–239 RSC.
- E. C. Yang, X. G. Wang, C. H. Zhang, N. Yang, Z. Y. Liu and X. J. Zhao, Sci. China: Chem., 2013, 56, 465–474 CrossRef CAS.
- E. V. Lider, E. V. Peresypkina, A. I. Smolentsev, V. N. Elokhina, T. I. Yaroshenko, A. V. Virovets, V. N. Ikorskii and L. G. Lavrenova, Polyhedron, 2007, 26, 1612–1618 CrossRef CAS.
- S.-R. Zheng, L. Zhang, J.-E. He, J. Fan and W.-G. Zhang, Inorg. Chem. Commun., 2016, 66, 19–23 CrossRef CAS.
- J. Hernández-Gil, S. Ferrer, R. Ballesteros and A. Castiñeiras, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2013, 69, o227–o228 CrossRef PubMed.
- G. M. Sheldrick, SADABS. Program for Empirical Absorption Correction of Area Detector Data, University of Gottingen, Germany, 2001 Search PubMed.
- G. M. Sheldrick, Crystal structure refinement with SHELXL, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- H. Putz and K. Brandenburg, DIAMOND Version 4.5.3, Crystal Impact GbR, Bonn, Germany, 2018 Search PubMed.
- J. Hernández-Gil, N. Ovèjak, S. Ferrer, F. Lloret and A. Castiñeiras, Inorg. Chem., 2013, 52, 2289–2291 CrossRef PubMed.
- (a) M. Angaroni, G. A. Ardizzoia, T. Beringhelli, G. La Monica, D. Gatteschi, N. Masciocchi and M. Moret, J. Chem. Soc., Dalton Trans., 1990, 3305–3309 RSC; (b) X. Liu, M. P. de Miranda, E. J. L. McInnes, C. A. Kilner and M. A. Halcrow, Dalton Trans., 2004, 59–64 RSC; (c) P. A. Angaridis, P. Baran, R. Boca, F. Cervantes-Lee, W. Haase, G. Mezei, R. G. Raptis and R. Werner, Inorg. Chem., 2002, 41, 2219–2228 CrossRef CAS PubMed.
- (a) J.-P. Zhang, Y.-Y. Lin, X.-C. Huang and X.-M. Chen, J. Am. Chem. Soc., 2005, 127, 5495–5506 CrossRef CAS PubMed; (b) J.-P. Zhang, Y.-Y. Lin, X.-C. Huang and X.-M. Chen, Chem. Commun., 2005, 1258–1260 RSC.
- (a) W. M. E. Koomen-Van Oudenniel, R. A. G. de Graaff, J. G. Haasnoot, R. Prins and J. Reedijk, Inorg. Chem., 1989, 28, 1128–1133 CrossRef CAS; (b) S. Ferrer, P. J. van Koningsbruggen, J. G. Haasnoot, J. Reedijk, H. Kooijman, A. L. Spek, L. Lezama, A. Arif and J. S. Miller, J. Chem. Soc., Dalton Trans., 1999, 4269–4276 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Additional crystallographic information, tables and figures (S1–S11), as indicated along the text. CCDC 1935979–1935981. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ra05922a |
| This journal is © The Royal Society of Chemistry 2019 |