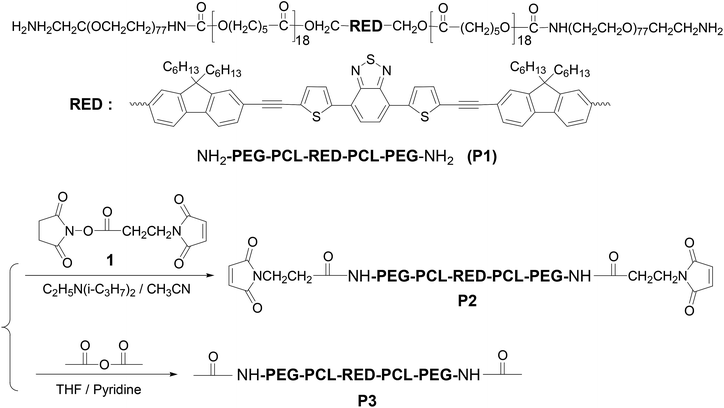
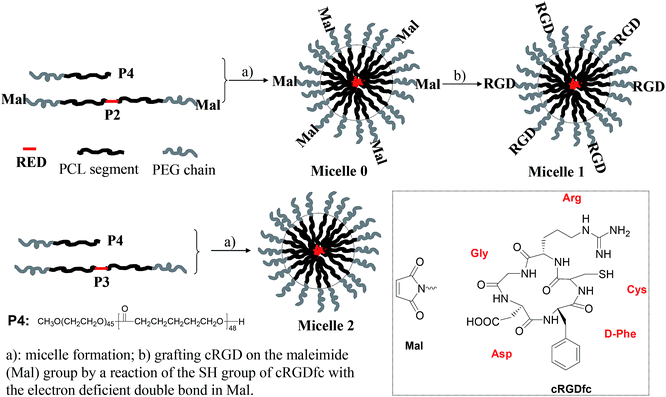
Open Access Article
Open Access ArticlecRGD functionalized 2,1,3-benzothiadiazole (BTD)-containing two-photon absorbing red-emitter-conjugated amphiphilic poly(ethylene glycol)-block-poly(ε-caprolactone) for targeted bioimaging†
Shanshan Wu *a,
Fengyu Sub,
Hansa Y. Mageec,
Deirdre R. Meldrumd and
Yanqing Tian
*a,
Fengyu Sub,
Hansa Y. Mageec,
Deirdre R. Meldrumd and
Yanqing Tian *e
*e
aGuangdong Industry Polytechnic, Foshan Municipality Anti-counterfeiting Engineering Research Center, Guangzhou, Guangdong 510300, China. E-mail: 2018090123@gdip.edu.cn
bAcademy for Advanced Interdisciplinary Studies, Southern University of Science and Technology, Shenzhen, Guangdong 518055, China
cKnowledge Enterprise, Arizona State University, Tempe, AZ 85287-5001, USA
dCenter for Biosignatures Discovery Automation, Biodesign Institute, Arizona State University, Tempe, AZ 85287-5001, USA
eDepartment of Materials Science and Engineering, Southern University of Science and Technology, Shenzhen, Guangdong 518055, China. E-mail: tianyq@sustech.edu.cn
First published on 23rd October 2019
Abstract
A two-photon absorbing (2PA) red emitter group was chemically conjugated onto amphiphilic poly(ethylene glycol)-block-poly(ε-caprolactone) (PEG-b-PCL) copolymers, and further grafted with cyclo(Arg-Gly-Asp) (cRGD) peptide to form micelle 1. Micelle 1 with cRGD targeting groups were used for targeted bioimaging. For comparison, micelle 2 without the cRGD targeting groups were also prepared and investigated. The micelles were characterized using dynamic light scattering (DLS), showing average diameters of around 77 nm. The cRGD targeting group is known to bind specifically with αvβ3 integrin in cancer cells. In this study, αvβ3 integrin overexpressed human glioblastoma U87MG cell line and αvβ3 integrin deficient human cervical cancer HeLa cell line were chosen. Results showed that the cRGD targeting group enhanced the cellular uptake efficiency of the micelles significantly in αvβ3 integrin rich U87MG cells. Higher temperature (37 °C versus 4 °C) and calcium ions (with 3 M calcium chloride in the cell culture medium versus no addition of calcium ions) enhanced the cellular uptake efficiency, suggesting that the uptake of the micelles is through the endocytosis pathway in cells. A 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) viability assay was used to evaluate the cytotoxicity of the micelles and no significant cytotoxicity was observed. The BTD-containing two-photon absorbing emitter in the micelles showed a two-photon absorbing cross-section of 236 GM (1 GM = 1 × 10−50 cm4 s per photonper molecule) at 820 nm, which is among the highest values reported for red 2PA emitters. Because of the two-photon absorbing characteristics, micelle 1 was successfully used for two-photon fluorescence imaging targeted to U87MG cells under a two-photon fluorescence microscope. This study is the first report regarding the targeted imaging of a specific cancer cell line (herein, U87MG) using the BTD-conjugated-fluorophore-containing block copolymers.
Introduction
Micelles, formed from amphiphilic block copolymers (ABCs) with hydrophobic cores and hydrophilic coronas have been demonstrated as a powerful tool for cell imaging, disease diagnosis, disease therapy and delivery of various water insoluble materials (including nanoparticles and drugs etc.) into cells.1–8 In order to understand the internalization of block copolymers with cells and clarify the payload delivery mechanism, it is extremely important to directly monitor the subcellular distribution of micelles. Thus, fluorophore (fluorescence probe)-conjugated block copolymers have been synthesized and investigated, which include rhodamine derivative-conjugated poly(ε-caprolactone)-block-poly(ethylene oxide) (TMRCA-PCL-b-PEO),9 fluorescein-conjugated poly(ethylene oxide)-block-poly(β-benzyl L-aspartate) (PEO-b-PBLA-FITC),10 rhodamine-conjugated poly(ethylene glycol)-block-phosphatidylethanolamine (rhodamine-PEG-b-PE),11 and fluorescein-conjugated poly(ethylene glycol)-block-poly(lactic acid) (FITC-PEG-b-PLA)12 etc. Cellular internalization of these block copolymers was found to be either through a non-specific endocytosis9,11,13 or a receptor mediated endocytosis12 process, which depends on the cell lines and polymer structures with or without targeting groups on the micelles. In early studies, Savić et al.9 studied the cellular internalization of TMRAC-PCL-b-PEG by rat pheochromocytoma cells (P12) and mouse embryonic fibroblast cells (NIH 3T3) and concluded that the micelles were internalized through a non-specific endocytosis. However, it was later argued that the micelles were not endocytosed but taken up by a strong interaction of the cationic fluorescent probes (TMRACs) with the cell's membrane.14 Nevertheless, Tang et al.11 reported that a cationic rhodamine-conjugated PEG-b-PE was not able to stain human lung cancer cells (A549). These discrepancies indicate the complexes of the cell internalization of the block copolymers.On the other hand, two-photon absorbing (2PA) materials that can be excited in the near infrared (NIR) spectral region (700–1000 nm) have been developed and applied in frequency upconversion lasing, high-density optical storage, two-photon fluorescence microscopy, and photodynamic therapy.15,16 However, commercially available hydrophilic fluorescent molecular probes, such as fluoresceins, rhodamines, and green fluorescence protein usually have less than 200 GM 2PA cross-sections17–19 (δ, expressed in GM = 1 × 10−50 cm4 s per photon per molecule−1). Recently, 2PA dyes based on the D–π–D, D–π–A–π–D, D–π–A, and A–π–A (D: donor; π: conjugates; A: acceptor) structures have been shown to exhibit d values higher than 2000 GM.20–23 Nevertheless, most of these 2PA materials are hydrophobic. Only a few water-soluble 2PA green emitters have been reported with high 2PA cross-sections (2050 GM).24–26 In addition to the commonly used blue and green emitters, red emitters are highly desirable for bioimaging because of the reduced autofluorescence of biosubstrates within the red spectral window.27 Until now, studies of the syntheses of efficient red 2PA emitters are quite limited. Red emission can be obtained either by direct excitation of chromophores28 or indirect excitation using fluorescence resonance energy transfer (FRET) mechanism.29 Among the red 2PA emitters reported, the 2,1,3-benzothiadiazole (BTD)-containing hydrophobic D–π–A–π–D type dyes have been shown to exhibit high d values of about 800 GM at 780 nm.28 However, their poor solubility in water still limited their biological applications. As most of the efficient 2PA chromophores are hydrophobic, the application of these materials in biological environment is a major challenge. The approach of using dye-doped silica particles30–32 has been demonstrated as an efficient solution to disperse 2PA materials in aqueous solutions. These particles have been used for bioimaging,30 pH sensing,31 and photodynamic therapy.32 Another potential method for the delivery of 2PA compounds is to use in micelles formed from ABCs, either through a physical incorporation of the hydrophobic 2PA fluorophores33,34 or chemically grafted the 2PA fluorophores into a block copolymer35,36 The hydrophobic cores of the micelles provide compatible environment for hydrophobic 2PA materials to retain their high two-photon absorbing efficiencies and the hydrophilic shells of the micelles stabilize the micelles.26 This approach enables the applications of hydrophobic 2PA materials in aqueous conditions for bioapplications and retains the high two-photon absorbing efficiencies of the hydrophobic 2PA materials.
Targeted imaging using disease-specific probes is very important for disease and cancer detection, especially at the early stage. Further, materials integrating targeted imaging probes with drugs can be used to diagnose and treat cancers simultaneously with high detection accuracy and treatment efficiency. To achieve these goals, various targeting groups were chemically grafted onto different non-invasive fluorescence imaging probes and magnetic resonance imaging probes, to specifically target tumors through molecular recognition of unique cancer-specific biomarkers. Cyclo-Arg-Gly-Asp (cRGD) peptide is one type of highly efficient targeting groups.37–40 The cRGD moiety is known to bind strongly and selectively with αvβ3 integrin containing cancer cells.41 cRGD modified micelles were reported and used for targeted delivery of drugs,37 magnetic nanoparticles,38 quantum dots,39 and carbon nanotubes.40
Along the line of our studies, herein, we for the first time use the cRGD as a targeting group to deliver new BTD-containing 2PA red emitter-conjugated block copolymer derived from P142 (Scheme 1) to αvβ3 integrin-overexpressed human glioblastoma tumor U87MG cell line (Fig. 1). The BTD-conjugated fluorophore (RED) in P1 shows large Stokes shift (∼120 nm) and emits in the red spectral window (550–750 nm with a maximum at 630 nm).43 Further, although the fluorophore (herein, RED) is extremely hydrophobic, by conjugating with amphiphilic block copolymer and assembling into the micellar cores (Fig. 1), the hydrophobic materials can be applied in biological environment as fluorescent probes for bioimaging.44,45 To demonstrate that the micellar probe is for targeted imaging of U87 cell line, αvβ3 integrin deficient human cervical cancer HeLa cell line46 was used as a counterpart. The cellular uptake efficiency of the micelles with and without cRGD peptides was compared. Influence of temperature and addition of calcium ion on the cellular uptake efficiency was also evaluated. The micelles were demonstrated to be applied for bioimaging under a two-photon fluorescent microscope. This study provides direct information not only for further understanding cellular internalization of the block copolymers, especially the two-photon absorbing block copolymers, but also for the development of new cancer diagnosis and detection materials.
Results and discussion
Synthesis, photophysical property, and characterization of the RED-fluorophore-containing micelles with or without cRGD targeting groups
P1 with amino-terminal groups on PEG-b-PCL was synthesized according to our procedure published previously.42 The fluorophore RED exhibited an absorption maximum at 509 nm and red emission with a maximum at 630 nm. Critical micelle concentration (CMC) was measured to be 0.022 mg mL−1 (Fig. S1†). Thus for all studies herein, the polymer concentration is higher than its CMC. P1 was firstly conjugated to a heterobifunctional material, 4-maleimidopropionic acid N-hydroxysuccinimide ester (compound 1), yielding a maleimide modified block copolymer P2 (Scheme 1). Typical 1H NMR of P2 was given in Fig. S2A.† The inset figure in Fig. S2A† shows the magnified area from 4.8 to 8.8 ppm. The observation of chemical shift at 6.72 ppm (peak g) indicates the successful grafting of the maleimide moiety. Basing on the integration ratio of peak g to peak f (–RED–CH2O–), maleimide group on P2 was calculated to be 65%.Micelles were prepared using P2 and a commercially available PEG-b-PCL (P4) to reduce the amount of P2 for micelle preparation. The micelles were stable in refrigerator at 4 °C for at least two months. The maleimide group on micelle surface was used to link the αvβ3 integrin-targeting group of cRGD through a chemical addition reaction of the thio-maleimide conjugation at pH 7.2 in HEPES buffer. Excess cRGD in the reaction mixture was removed through an intense dialysis using a regenerated cellulose membrane with a Mw cut off 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 for three days to form cRGD-functionalized micelles (called micelle 1). To confirm the cRGD grafting degree on the micelles, the micelles were freeze-dried and suspended in D2O with 10% DMSO-d6 by volume to measure the 1H NMR spectrum. The spectrum was given in Fig. S2B.† The insert figure in Fig. S2B† shows the magnified area from 5.8 to 8.8 ppm. Clearly, the chemical shifts at aromatic area around 6.7–7.9 ppm from the RED segment were not observed, indicating the RED part was located inside the solid PCL cores of the micelles due to its insufficient mobility in the mixture of D2O and DMSO-d6. Successful conjugation of cRGDfc onto the solvated PEG shells were confirmed by the observation of the chemical shifts of the phenyl protons of cRGDfc around 7.2 ppm and complete disappearance of the maleimide signal at δ = 6.72 ppm. Thus, basing on the 65% maleimide percentages on P2, and the molar ratio of P4 to P2 (2
000 for three days to form cRGD-functionalized micelles (called micelle 1). To confirm the cRGD grafting degree on the micelles, the micelles were freeze-dried and suspended in D2O with 10% DMSO-d6 by volume to measure the 1H NMR spectrum. The spectrum was given in Fig. S2B.† The insert figure in Fig. S2B† shows the magnified area from 5.8 to 8.8 ppm. Clearly, the chemical shifts at aromatic area around 6.7–7.9 ppm from the RED segment were not observed, indicating the RED part was located inside the solid PCL cores of the micelles due to its insufficient mobility in the mixture of D2O and DMSO-d6. Successful conjugation of cRGDfc onto the solvated PEG shells were confirmed by the observation of the chemical shifts of the phenyl protons of cRGDfc around 7.2 ppm and complete disappearance of the maleimide signal at δ = 6.72 ppm. Thus, basing on the 65% maleimide percentages on P2, and the molar ratio of P4 to P2 (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) used for micelle preparation, the cRGD percentage on the micellar shells was calculated to be ∼22%.
1) used for micelle preparation, the cRGD percentage on the micellar shells was calculated to be ∼22%.
For understanding whether the cRGD moiety can achieve the targeted delivery of the RED-fluorophore-conjugated block copolymer, P3 were prepared through an end capping of the amino groups of P1. Micelles made from P3 without cRGD moieties were named as micelle 2. Micelles nanostructures were characterized using DLS (Fig. S3†). Results showed the average diameters of the micelles before grafting cRGD (named as micelle 0) and after grafting cRGD (micelle 1) were 77.3 and 76.8 nm with polydispersites of 0.24 and 0.25, which are almost the same as those of micelles 2 with size of 77.2 nm and polydispersity of 0.26. This indicates the surface modification of cRGD did not change the micelle structures significantly.
The 2PA cross-sections of RED in micelle-1 were measured by using the two-photon-induced fluorescence method with fluorescein in 1 N NaOH aqueous solution as the reference. The plots of the 2PA cross-sections against the excitation wavelength from 760 nm to 840 nm were given in Fig. S4.† It was found, at the most popularly used wavelengths of 800 to 820 nm, the 2PA cross-sections were around 240 GM, which is among the highest reported values of water-soluble red 2PA emitters (for example, the commonly used rhodamine B has a maximum 2PA cross-sections of ∼210 GM18).
Cytotoxicity evaluation
Cytotoxicity of the micelles to the two cell lines were studied using the typical MTT viability assay. Greater than 85% of the cells were viable after stained for 16 h using micelles with RED fluorophore concentrations of 1.3–4.4 μM (Fig. S5†). These observations demonstrated the biocompatibility of the RED fluorophore-conjugated block copolymers.Fluorescence bioimaging using two-photon fluorescence microscopy
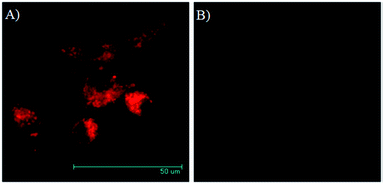
Fig. 2 gave the two-photon fluorescence images of U87MG and HeLa cells after internalization with a RED concentration of 2.2 μM in micelle 1 for 16 hours. Under the same experimental condition, strong emission was observed for the integrin rich U87MG cells, while almost no fluorescence was observed for integrin deficient HeLa cells. This result is consistent with that observed using confocal fluorescent microscopy. The observation of the fluorescence in U87MG cells under the two-photon excitation condition at 830 nm indicates the RED fluorophore is suitable for two-photon fluorescence imaging.Fluorescence bioimaging using confocal fluorescence microscopy
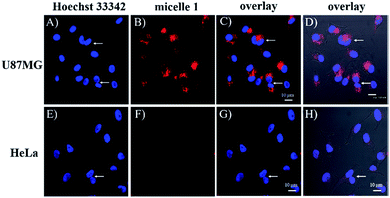
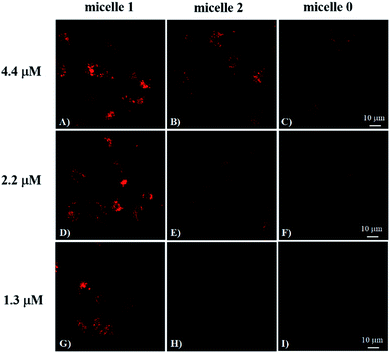
To investigate the cRGD influence on the cellular uptake of the micelles, live HeLa (human cervical cancer cell, αvβ3 integrin deficient) and U87MG (human glioblastoma, high αvβ3 integrin expression) cells were stained with micelle 0, micelle 1, and micelle 2 with 1.3, 2.2, and 4.4 μM of RED in the micelles and then examined under the confocal fluorescence microscope. The representative fluorescence images are shown in Fig. 3 and 4. Under the same experimental condition, micelle 1 slightly permeated the integrin-deficient cells (HeLa) and showed weak fluorescence signal under confocal fluorescence microscope (Fig. 3F). However, much stronger emission was observed for the integrin rich cells (U87MG) and showed the high cell permeability of micelle 1 (Fig. 3B). The difference between the two cell lines indicated the αvβ3 integrin on the cell membranes played a critical role for cellular internalization of the cRGD-containing materials. Co-staining of the micelles with nuclei staining Hoechst 33342 indicated the micelles were located in the cytoplasm regions of the cells. As indicated by the arrows in Fig. 3, two nuclei in one cell were observed, which means that under our experimental conditions, the cells were not only healthy, but also capable of dividing and proliferation.Fig. 4 shows the comparison of the micelle 0, micelle 1, and micelle 2 at different concentrations after 20 hours internalization with U87MG cells. Clearly, much brighter fluorescence was observed under the fluorescence microscope for micelle 1 than those of micelles 0 and 2 (Fig. 4). The results demonstrated that cRGD significantly enhanced the cellular uptake of the micelle 1 by U87MG cells through the receptor-mediated endocytosis mechanism, since U87MG cells are αvβ3 integrin over-expressed cells, which could selectively bind cRGD peptide. It should also be noted here, although non-specific endocytosis was observed, the cellular uptake efficiency through the non-specific endocytosis is much lower than those by the receptor-mediated endocytosis.
Quantitative comparison of the uptake efficiency of micelles using fluorescence spectrometer
To further quantitatively compare the cellular uptake of the micelles by U87MG and HeLa cells, the cells were lysed after the internalization with micelles. And then the micelles were extracted from the lysed cells using DMSO. The maximum fluorescence intensities at 650 nm were measured, which was excited using 488 nm excitation. The higher fluorescence intensity suggests more cellular uptake of the micelles. Results are given in Fig. S6.† Clear concentration dependent cellular uptake of micelle 1 was observed for U87MG cells. The internalized amount of micelle 1 possessing cRGD moieties in U87MG cells are much more than those of micelles 0 and 2 without the cRGD moieties. Weak fluorescence was observed using HeLa cells for cRGD-containing micelle 1. It means that certain amount micelles were taken up by cells through a non-specific endocytosis mechanism. However, the micelles without cRGD groups did not exhibit any significant binding to the two cell lines. The cellular uptake efficiency of the non-cRGD micelles is almost one order lower than that of the cRGD targeting moieties-containing micelles. The spectral analysis result correlates very well with the trend of the result of the confocal fluorescence microscopy. Thus, it can be concluded that cRGD functional group-containing micelles exhibited high affinity αvβ3 integrin specific binding in vitro, indicating the micelles are new fluorescence probes for targeted imaging of tumor cells possessing αvβ3 integrin.Factors affecting cellular internalization: temperature and Ca2+ ion-a confirmation of the cellular uptake is through the endocytosis
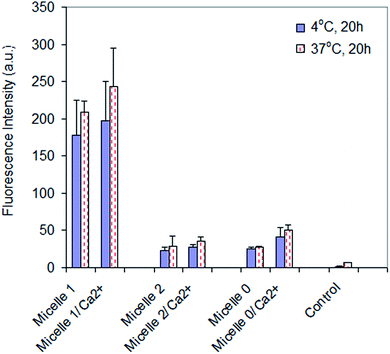
Cellular internalization of micelle 1, micelle 2, and micelle 0 with U87MG cells was studied under varying conditions in order to determine if the modality of cellular uptake was through endocytosis. It has been reported that several basic criteria must be fulfilled if a given material is internalized by endocytosis. The process is energy dependent, thus the extent of internalization is reduced at low temperature47,48 but enhanced by calcium ion.49,50 In the present studies, U87MG cells incubated with the micelles and internalized a smaller amount of micelles at 4 °C than 37 °C (Fig. 5). In addition, the internalization of the micelles in the presence of calcium ions (3 mM) was higher than that without the addition of calcium ions. These data indicated that the cellular internalization of the micelles was through endocytosis, including the receptor-mediated endocytosis for micelle 1 and the non-specific endocytosis for micelles 2 and 0.Experimental
Materials
P1 (Mn (GPC) = 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800, Mw/Mn = 1.43, Mn (NMR) = 12
800, Mw/Mn = 1.43, Mn (NMR) = 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100; the molecular weights of RED, PCL, and PEG are 1072, 4200, and 6800 respectively) with α,ω-diamino terminal groups was prepared according to our previous publication.42 4-Maleimidopropionic acid N-hydroxysuccinimide ester (compound 1) shown in Scheme 1 was prepared according to procedures described in literature.44 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), acetic anhydride, and N,N′-dimethylaminopyridine were purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. Poly(ethylene oxide-b-ε-caprolactone) (P4, Mw/Mn = 1.30, PEO molecular weight is 2000, PCL molecular weight is 5000) was purchased from Polymer Source (Montreal, Canada). Cyclo(Arg-Gly-Asp-D-Phe-Cys) (cRGDfc, catalogue number PCI 3686-PI) was purchased from Peptide International Inc (Louisville, KT). Dialysis membranes (regenerate cellulose, Mw cut off 12
100; the molecular weights of RED, PCL, and PEG are 1072, 4200, and 6800 respectively) with α,ω-diamino terminal groups was prepared according to our previous publication.42 4-Maleimidopropionic acid N-hydroxysuccinimide ester (compound 1) shown in Scheme 1 was prepared according to procedures described in literature.44 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), acetic anhydride, and N,N′-dimethylaminopyridine were purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. Poly(ethylene oxide-b-ε-caprolactone) (P4, Mw/Mn = 1.30, PEO molecular weight is 2000, PCL molecular weight is 5000) was purchased from Polymer Source (Montreal, Canada). Cyclo(Arg-Gly-Asp-D-Phe-Cys) (cRGDfc, catalogue number PCI 3686-PI) was purchased from Peptide International Inc (Louisville, KT). Dialysis membranes (regenerate cellulose, Mw cut off 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) were purchased from Pierce (Rockford, IL). Dulbecco's modified Eagle's medium (DMEM) and nuclei staining dye Hoechst 33342 were acquired from Invitrogen (Carlsbad, CA). Human glioblastoma tumor U87MG and cervical cancer HeLa cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA).
000) were purchased from Pierce (Rockford, IL). Dulbecco's modified Eagle's medium (DMEM) and nuclei staining dye Hoechst 33342 were acquired from Invitrogen (Carlsbad, CA). Human glioblastoma tumor U87MG and cervical cancer HeLa cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA).
General methods
1H NMR spectra were measured using a Varian liquid-state NMR operated at 400 MHz. Molecular weights of polymers were determined using a Waters 1515 gel permeation chromatography (GPC) coupled with a RI detector, in reference with a series of polystyrene standards with tetrahydrofuran (THF) as the eluent. Shimadzu RF-5301 spectrofluorophotometer (Shimadzu Scientific Instruments, Columbia, MD) was used for fluorescence measurements. Dynamic light scattering (DLS) measurements for micelle diameters were performed using a Malvern Nano-ZS instrument (Worcestershire, UK) equipped with a 4 mW He–Ne laser (633 nm) with an output at a scattering angle of 173°. The solution was passed through a 0.45 μm Nylon micro-filter (VWR, Batavia, IL) to remove dust before the DLS measurements.Synthesis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, Mw/Mn = 1.27. Mn (NMR) = 12
500, Mw/Mn = 1.27. Mn (NMR) = 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400.
400.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700, Mw/Mn = 1.22. Mn (NMR) = 12
700, Mw/Mn = 1.22. Mn (NMR) = 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200.
200.Micelle preparation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) for three days with water exchange approximately 12 hours each time. The solution was then filtered through a 200 nm micro-filter to remove possible large particles. Concentration of the fluorophore RED in the micelles was determined using UV-vis spectrophotometer. The concentration of RED in micelle 1 was determined to be 65 μM. The corresponding polymer concentration was calculated to be 1.5 mg mL−1.
000) for three days with water exchange approximately 12 hours each time. The solution was then filtered through a 200 nm micro-filter to remove possible large particles. Concentration of the fluorophore RED in the micelles was determined using UV-vis spectrophotometer. The concentration of RED in micelle 1 was determined to be 65 μM. The corresponding polymer concentration was calculated to be 1.5 mg mL−1.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) for three days with water exchange approximately 12 hours each time. The solution was then filtered through a 200 nm micro-filter to remove possible large particles. Concentration of RED in micelles 2 was determined to be 100 μM. The corresponding polymer concentration was calculated to be 2.4 mg mL−1. The solution was further diluted 1.5 fold using HEPES buffer to get the RED-fluorophore concentration of 65 μM for use.
000) for three days with water exchange approximately 12 hours each time. The solution was then filtered through a 200 nm micro-filter to remove possible large particles. Concentration of RED in micelles 2 was determined to be 100 μM. The corresponding polymer concentration was calculated to be 2.4 mg mL−1. The solution was further diluted 1.5 fold using HEPES buffer to get the RED-fluorophore concentration of 65 μM for use.Cell culture and imaging
U87MG and HeLa cells were cultured in the DMEM medium supplemented with 10% fetal bovine serum (FBS), 5% penicillin, 2 mM L-glutamine (Sigma-Aldrich, St. Louis, MO), and incubated in a 5% CO2 atmosphere at 37 °C. The cells were then seeded onto 96-well plates at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and incubated for 1 day.
000 cells per well and incubated for 1 day.
The micelles were diluted in the DMEM growth medium to give final RED concentrations of 1.3, 2.2, and 4.4 μM and polymer concentrations of 0.032, 0.055, and 0.11 mg mL−1 for cell staining. The media in the wells were replaced with 100 μL of the pre-prepared medium containing micelles. The plates were then returned to the incubator and maintained in 5% CO2 at 37 °C for 20 hours. Upon removal of the micelles-containing media, the cells were washed once with 100 μL of phosphate buffer (PBS) solution (pH: 7.2). Hoechst 33342 with a concentration of 10 μM in 100 μL of fresh DMEM medium was then added into the wells and incubated for 30 minutes for nuclei staining. Live cells were imaged with a Nikon Eclipse TE2000E confocal microscope (Melville, NY). Note, the confocal microscope is based on single-photon excitation mechanism, therefore, it is also called single-photon confocal fluorescence microscope. Blue emission of Hoechst 33342 was generated by a 402 nm laser and the emission was collected using a filter set at 450/35 nm. Red emission of RED was generated using a 488 nm laser and collected using a filter set at 605/75 nm. Negligible background fluorescence of cells was detected under the settings used.
Protocol for measurements of cellular uptakes of micelles
HeLa and U87 cell lines were plated at a concentration of 4 × 105 cells per mL into 48-well plates (300 μL in each well). After letting the cells grow for 20 hours, micelles were added to their specific predetermined wells. Final concentrations of RED were controlled to be 1.3, 2.2 and 4.4 μM, similar to the concentrations used for bioimaging. After 20 hours internalization, cell culture media were aspirated and the cells were gently washed once with PBS buffer. 200 μL of CelLytic M (Sigma-Aldrich, Catalog Number C2978) lysis buffer was then added to each well and the plate was put on the shaker for 20–30 minutes. 500 μL of dimethyl sulfoxide (DMSO) was then added to each well. The contents from each well were then loaded in a cuvette with 1300 μL of distilled water to result a total volume of 2 mL for fluorescence measurement using spectofluorophotometer. 488 nm was used as an excitation wavelength and 650 nm emission was collected.Cytotoxicity study
The assay was performed by an in vitro MTT based toxicology assay kit (Sigma-Aldrich) which is based on the intracellular reduction of a tetrazolium (MTT) dye to a formazan product measured spectrophotometrically and is used for high-throughput screening.45,46 Cells incubated with micelles for 20 hours in the 96-well plate were washed with PBS buffer and then incubated in fresh DMEM medium (100 μL) and 10 μL of MTT solution (5 mg mL−1) in 5% CO2 at 37 °C for another 3 h. 60 μL of the culture medium was taken out and 50 μL of DMSO was added to each well to dissolve the internalized purple formazan crystals by gentle pipetting up and down. The absorbance was measured at a wavelength of 540 nm using SpectraMax 190 from Molecular Devices (Downingtown, PA). Each experiment was conducted twice in triplicate. The result was expressed as a percentage of the absorbance of the blank control.Determination of two-photon cross-sections
Two-photon excitation spectra were measured using a two-photon-induced fluorescence technique17,18 using a mode-locked Ti:Sapphire laser excitation source (Coherent; Mira 900; Bloomfield, CT). The laser provides a pulse of ∼120 fs pulse width at a pulse repetition frequency of 76 MHz in the wavelength range of 710–1000 nm. The pumping wavelengths were determined by a monochromator-CCD system. Fluorescein in pH 11 water, which has been well characterized in literature,17,18 was used as a reference (r). The two-photon absorption cross section of a sample compound (s) can be calculated at each wavelength according to eqn (1)
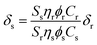 | (1) |
Two-photon fluorescence microscope
Two-photon fluorescence microscopy images of micelle 1-labeled U87MG and HeLa cells were obtained with a multiphoton Ultima IV In Vivo Laser Scanning Microscope (Prairie Technologies, Middleton, WI) by exciting the probes with a titanium-sapphire laser (Mai Tai, Spectra Physics, Irvine, CA) set at wavelength 780–900 nm and output power 1230 mW, which corresponded to approximately 10 mW average power in the focal plane. To obtain images at 585–629 nm range, internal PMTs were used to collect the signals in an 8 bit unsigned 512 × 512 pixels at 400 Hz scan speed. Fluorescence signals were acquired using the Ultima IV In Vivo Laser Scanning Microscope with a 63× water immersion lens.Conclusions
For the first time, micelles with BTD-conjugated two-photon absorbing red emitter was grafted with cRGD peptide as a typical targeting group for targeted imaging αvβ3 integrin rich human glioblastoma U87MG cells. αvβ3 integrin deficient human cervical cancer HeLa cells were used as counterparts. Significant cellular uptake of the cRGD-containing micelles by U87MG cells, in contrast to the minimum cellular uptake of the same micelles by HeLa cells, were observed under both the single-photon confocal fluorescence microscope and two-photon fluorescence microscope and quantitatively measured by using spectrofluorophotometer. This study demonstrated that the cRGD targeting groups played a critical role to enhance the delivery efficiency of fluorophore labeled micelles to αvβ3 integrin rich cells, indicating the potential application of the micellar probe for αvβ3 integrin rich tumor cells detection. This study may also provide insights for understanding the cellular internalization of micelles with cells. Further investigation using the 2PA fluorophore-conjugated micelles for in vivo imaging and drug delivery is in progress.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support was provided by the Microscale Life Sciences Center, a NIH Center of Excellence in Genomic Sciences at Arizona State University (Grant 5P50 HG002360, Dr Deirdre Meldrum, PI, Director), National Natural Science Foundation of China (21574061, 21601037 and 21604036), Guangdong Industry Polytechnic (KJ2019-003), and Guangdong Province Engineering Technology Centre for Molecular Probe and Biomedicine Imagining. Authors also would like to thank Dr Yongzhong Li and Dr Weiwen Zhang for discussions.Notes and references
- H. Gao, H. Feng, Y. Bai, Z. Li, L. Chen, L. Jin, J. Wang, E. F. Georges, G. Liu, J. Li and M. Wang, J. Biomed. Nanotechnol., 2019, 15, 1764–1770 CrossRef PubMed
.
- B. Kulkarni and M. Jayakannan, ACS Biomater. Sci. Eng., 2017, 3, 2185–2197 CrossRef CAS
.
- K. Shen, D. Li, J. Guan, J. Ding, Z. Wang, J. Gu, T. Liu and X. Chen, Nanomedicine: Nanotechnology, 2017, 13, 1279–1288 CrossRef CAS
.
- Q. Wang, P. Zhang, Z. Li, X. Feng, C. Lv, H. Zhang, H. Xiao, J. Ding and X. Chen, Theranostics, 2019, 9, 1426–1452 CrossRef CAS
.
- H. Xiao, L. Yan, E. Dempsey, W. Song, R. Qi, W. Li, Y. Huang, X. Jing, D. Zhou, J. Ding and X. Chen, Prog. Polym. Sci., 2018, 87, 70–106 CrossRef CAS
.
- Q. Wang, X. Zhang, Y. Sun, L. Wang, L. Ding, W.-H. Zhu, W. Di and Y.-R. Duan, Biomaterials, 2019, 212, 73–86 CrossRef CAS
.
- L. Jie, D. Lang, X. Kang, Z. Yang, Y. Du and X. Ying, J. Nanosci. Nanotechnol., 2019, 19, 5456–5462 CrossRef
.
- H. T. T. Duong, T. Thambi, Y. Yin, J. E. Lee, Y. K. Seo, J. H. Jeong and D. S. Lee, ACS Appl. Mater. Interfaces, 2019, 11, 13058–13068 CrossRef CAS
.
- R. Savić, L. Luo, A. Eisenberg and D. Maysinger, Science, 2003, 300, 615–618 CrossRef
.
- A. Sami, J. Public Health Biol. Sci., 2012, 01, 121–126 Search PubMed
.
- N. Tang, G. J. Du, N. Wang, C. C. Liu, H. Y. Hang and W. Liang, J. Natl. Cancer Inst., 2007, 99, 1004–1015 CrossRef CAS
.
- C. K. Huang, C. L. Lo, H. H. Chen and G. H. Hsiue, Adv. Funct. Mater., 2007, 17, 2291–2297 CrossRef CAS
.
- J. Liaw, T. Aoyagi, K. Kataoka, Y. Sakurai and T. Okano, Pharm. Res., 1999, 16, 213–220 CrossRef CAS PubMed
.
- S. M. Moghimi, A. C. Hunter, J. C. Murray and A. Szewczyk, Science, 2004, 303, 626–627 CrossRef CAS
.
- G. S. He, L. S. Tan, Q. Zheng and P. N. Prasad, Chem. Rev., 2008, 108, 1245–1330 CrossRef CAS PubMed
.
- X. L. Yue, A. R. Morales, G. W. Githaiga, A. W. Woodward, S. Tang, J. Sawada, M. Komatsu, X. Liu and K. D. Belfield, Org. Biomol. Chem., 2015, 13, 10716–10725 RSC
.
- P. T. C. So, C. Y. Dong, B. R. Masters and K. M. Berland, Annu. Rev. Biomed. Eng., 2000, 2, 399–429 CrossRef CAS
.
- C. Xu and W. W. Webb, J. Opt. Soc. Am. B, 1996, 13, 481–491 CrossRef CAS
.
- W. R. Zipfel, R. M. Williams and W. W. Webb, Nat. Biotechnol., 2003, 21, 1368–1376 CrossRef PubMed
.
- M. Albota, D. Beljonne, J. L. Bredas, J. E. Ehrlich, J. Y. Fu, A. A. Heikal, S. E. Hess, T. Kogej, M. D. Levin, S. R. Marder, D. McCord-Maughon, J. W. Perry, H. Rockel, M. Rumi, C. Subramaniam, W. W. Webb, X. L. Wu and C. Xu, Science, 1998, 281, 1653–1656 CrossRef CAS
.
- M. Rumi, J. E. Ehrlich, A. A. Heikal, J. W. Perry, S. Barlow, Z. Y. Hu, D. McCord-Maughon, T. C. Parker, H. Rockel, S. Thayumanavan, S. R. Marder, D. Beljonne and J. L. Bredas, J. Am. Chem. Soc., 2000, 122, 9500–9510 CrossRef CAS
.
- L. Ventelon, S. Charier, L. Moreaux, J. Mertz and M. Blanchard-Desce, Angew. Chem., Int. Ed., 2001, 40, 2098–2101 CrossRef CAS
.
- J. Yoo, S. K. Yang, M. Y. Jeong, H. C. Ahn, S. J. Jeon and B. R. Cho, Org. Lett., 2003, 5, 645–648 CrossRef CAS
.
- H. Y. Woo, B. Liu, B. Kohler, D. Korystov, A. Mikhailovsky and G. C. Bazan, J. Am. Chem. Soc., 2005, 127, 14721–14729 CrossRef CAS
.
- H. Y. Woo, J. W. Hong, B. Liu, A. Mikhailovsky, D. Korystov and G. C. Bazan, J. Am. Chem. Soc., 2005, 127, 820–821 CrossRef CAS
.
- Y. Tian, C. Y. Chen, Y. J. Cheng, A. C. Young, N. M. Tucker and A. K. Y. Jen, Adv. Funct. Mater., 2007, 17, 1691–1697 CrossRef CAS
.
- W. Qin, P. F. Zhang, H. Li, J. W. Y. Lam, Y. J. Cai, R. T. K. Kwok, J. Qian, W. Zheng and B. Z. Tang, Chem. Sci., 2018, 9, 2705–2710 RSC
.
- S. Kato, T. Matsumoto, M. Shigeiwa, H. Gorohmaru, S. Maeda, T. Ishi-i and S. Mataka, Chem.–Eur. J., 2006, 12, 2303–2317 CrossRef CAS
.
- R. F. Liu, J. Tang, Y. X. Xu and Z. F. Dai, ACS Nano, 2019, 13, 5124–5132 CrossRef CAS
.
- B. Kim, X. Yue, B. Sui, X. Zhang, Y. Xiao, M. V. Bondar, J. Sawada, M. Komatsu and K. D. Belfield, Eur. J. Org. Chem., 2015, 25, 5563–5571 CrossRef
.
- S. Kim, H. E. Pudavar and P. N. Prasad, Chem. Commun., 2006, 19, 2071–2073 RSC
.
- G. Lemercier, M. Four and S. Chevreux, Coord. Chem. Rev., 2018, 368, 1–12 CrossRef CAS
.
- Y. Gao, G. Feng, T. Jiang, C. Goh, L. Ng, B. Liu, B. Li, L. Yang, J. Hua and H. Tian, Adv. Funct. Mater., 2015, 25, 2857–2866 CrossRef CAS
.
- J. L. Geng, K. Li, D. Ding, X. H. Zhang, W. Qin, J. Z. Liu, B. Z. Tang and B. Liu, Small, 2012, 8, 3655–3663 CrossRef CAS
.
- A. R. Morales, C. O. Yanez, Y. Zhang, X. Wang, S. Biswas, T. Urakami, M. Komatsu and K. D. Belfield, Biomaterials, 2012, 33, 8477–8485 CrossRef CAS
.
- L. Hu, H. Wang, X. Xu, M. Zhang, X. Tian, J. Wu, H. Zhou, J. Yang and Y. Tian, Sens. Actuators, B, 2017, 241, 1082–1089 CrossRef CAS
.
- Z. Zhen, W. Tang, H. Chen, X. Lin, T. Todd, G. Wang, T. Cowger, X. Chen and J. Xie, ACS Nano, 2013, 7, 4830–4837 CrossRef CAS
.
- W. Su, H. Wang, S. Wang, Z. Liao, S. Kang, Y. Peng, L. Han and J. Chang, Int. J. Pharm., 2012, 426, 170–181 CrossRef CAS PubMed
.
- D. Deng, L. Qu, J. Zhang, Y. Ma and Y. Gu, ACS Appl. Mater. Interfaces, 2013, 5, 10858–10865 CrossRef CAS
.
- C. Wang, C. Bao, S. Liang, H. Fu, K. Wang, M. Deng, Q. Liao and D. Cui, Nanoscale Res. Lett., 2014, 9, 264 CrossRef
.
- Y. Miura, T. Takenaka, K. Toh, S. R. Wu, H. Nishihara, M. R. Kano, Y. Ino, T. Nomoto, Y. Matsumoto, H. Koyama, H. Cabral, N. Nishiyama and K. Kataoka, ACS Nano, 2013, 7, 8583–8592 CrossRef CAS
.
- Y. Q. Tian, W. C. Wu, C. Y. Chen, T. Strovas, Y. Z. Li, Y. G. Jin, F. Y. Su, D. R. Meldrum and A. K. Y. Jen, J. Mater. Chem., 2010, 20, 1728–1736 RSC
.
- O. Engebraaten, M. Trikha, S. Juell, S. Garman-Vik and O. Fodstad, Anticancer Res., 2009, 29, 131–137 CAS
.
- K. Thibaudeau, R. Leger, X. C. Huang, M. Robitaille, O. Quraishi, C. Soucy, N. Bousquet-Gagnon, P. van Wyk, V. Paradis, J. P. Castaigne and D. Bridon, Bioconjugate Chem., 2005, 16, 1000–1008 CrossRef CAS
.
- J. Carmichael, W. G. Degraff, A. F. Gazdar, J. D. Minna and J. B. Mitchell, Cancer Res., 1987, 47, 936–942 CAS
.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS
.
- R. M. Steinman, I. S. Mellman, W. A. Muller and Z. A. Cohn, J. Cell Biol., 1983, 96, 1–27 CrossRef CAS
.
- L. B. Luo, J. Tam, D. Maysinger and A. Eisenberg, Bioconjugate Chem., 2002, 13, 1259–1265 CrossRef CAS
.
- D. Beutner, T. Voets, E. Neher and T. Moser, Neuron, 2001, 29, 681–690 CrossRef CAS
.
- A. Yuan, C. H. Siu and C. P. Chia, Cell Calcium, 2001, 29, 229–238 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06694b |
| This journal is © The Royal Society of Chemistry 2019 |