Open Access Article
Open Access ArticleEvolutionary approaches in protein engineering towards biomaterial construction
Brindha J.
a,
Balamurali M. M.
*a and
Kaushik Chanda
 *b
*b
aDepartment of Chemistry, School of Advanced Science, Vellore Institute of Technology, Chennai Campus, Vandalur-Kelambakkam Road, Chennai-600 127, Tamil Nadu, India. E-mail: mmbala@gmail.com
bDepartment of Chemistry, School of Advanced Science, Vellore Institute of Technology, Vellore-632014, Tamil Nadu, India. E-mail: chandakaushik1@gmail.com
First published on 29th October 2019
Abstract
The tailoring of proteins for specific applications by evolutionary methods is a highly active area of research. Rational design and directed evolution are the two main strategies to reengineer proteins or create chimeric structures. Rational engineering is often limited by insufficient knowledge about proteins' structure–function relationships; directed evolution overcomes this restriction but poses challenges in the screening of candidates. A combination of these protein engineering approaches will allow us to create protein variants with a wide range of desired properties. Herein, we focus on the application of these approaches towards the generation of protein biomaterials that are known for biodegradability, biocompatibility and biofunctionality, from combinations of natural, synthetic, or engineered proteins and protein domains. Potential applications depend on the enhancement of biofunctional, mechanical, or other desired properties. Examples include scaffolds for tissue engineering, thermostable enzymes for industrial biocatalysis, and other therapeutic applications.
1. Introduction
Today, strategies for improving proteins' functionalities include immensely varying combinations of rational design along with random mutagenesis via directed evolution followed by screening. To move forward in this process of enhancing proteins' desirable properties, protein engineers ought to compare the two main protein engineering strategies – rational design and directed evolution, combine the effective methodologies, and successfully create protein variants with ease by employing various protein engineering methods. Proteins or enzymes obtained by directed evolution have found a firm place in industrial and therapeutic applications, but are still striving for improvement in other biomedical fields of application. To date, the demand for an efficient method to achieve improved biofunctionalities or modified thermodynamic properties persists. Rational design and directed evolution are the two approaches being employed to achieve this.Engineered proteins with their enhanced biofunctionality/other properties and ability to form hydrogels or other forms of biomaterials are becoming vital these days, for their potential applications in biomedical fields such as drug delivery, synthetic extracellular matrix materials for tissue engineering,1–4 etc. The history of biomaterials dates far back into ancient civilization. According to the first Consensus Conference of the European Society for Biomaterials (ESB) in 1976, a biomaterial was defined as ‘an inert material used in medical applications, to connect with biological systems’, which was later changed as ‘a material that can combine biological systems to check, heal, improve or restore any part or function of the human body’.5 The subtle modification in the definition indicates that biomaterials have been evolving with the needs and challenges in healthcare. The capability to repair or replace damaged tissues or organs has enhanced the life expectancy of several individuals. Among the earliest were the use of waxes, glues and tissues, by the Indians and Chinese in restoring the missing parts of the body, and also the artificial body parts that were seen in Egyptian mummies.6 The requirement for basic research on implantable materials was initiated when physicians started using implants in medical field.7 Early biomedical implants were not successful because the critical concepts associated with infection, biomaterials, and the biological responses to those biomaterials were not fully understood. Until Joseph Lister's sterile surgical method8 was established in the 1860s, different metallic implants like pins, wires made up of iron, gold, silver etc. and tissue replacements failed due to contamination after implanting. Moreover, dislodgement, contamination with infectious agents at the association of implant and tissue, rupture and migration occurs with time when foreign implants are used. The development of biomaterials has evolved since then, from titanium that was reported to be biocompatible with bone.9 The calcium and phosphate ion immobilized titanium finds application as bone tissue implant by stimulation of the responses of pre-osteoblast cells.10 Currently, biomaterials find a vast range of biomedical applications including prosthetic implants,11,12 drug delivery systems,13,14 tissue engineering15,16 and regenerative medicine.17,18
Biomaterials, as a multidimensional field has grown steadily since its outset with a constant influx of new concepts and advancements. Biomaterials have evolved since ancient times for the betterment of humans. The main goal during 1960s to 1970s, in the design of first generation biomaterials was to keep a balance between mechanical and physical properties along with minimal toxicity to host tissue.19 The limitations within contemporary medicine led to the development of tissue engineering and regenerative medicine. With the progress in molecular biology concepts, recombinant DNA technology and genetic engineering, new path has been opened up for biomaterial design that can coordinate with the normal human physiology and integrate with human body. Various combinations of biomaterials with distinct properties come together as novel materials for their use in controlled drug delivery, gene therapy or therapeutics, tissue engineering applications, etc. Several generations of biomaterials have evolved with time and advancements in various branches of science. New smart biomaterials20,21 are being created by integrating various technical concepts and biological constituents with the aim of expanding its applications in human healthcare.
This review aims to highlight the two important protein engineering approaches – rational design and directed evolution along with selective examples that can pave way for their constitution into biomaterials. Here, the readers can also find a brief account on various protein based biomaterials used in biomedical applications. On the whole, this review will lead the readers to ponder about the possibilities of using protein engineering strategies and combinations of engineered proteins/protein domains, towards the creation of novel protein based biomaterials for use in biomedical field.
2. Protein engineering and biomaterial construction
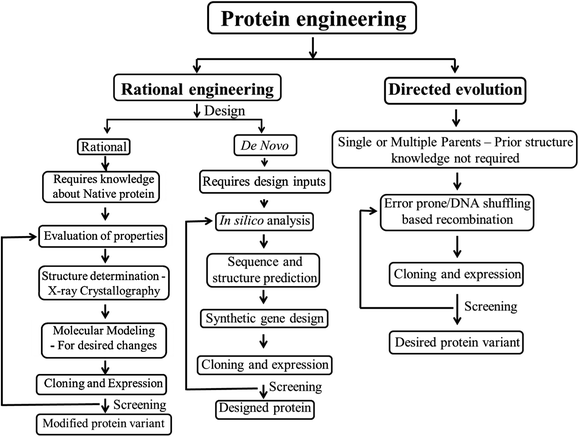
Protein engineering employs recombinant DNA technology to modify amino acid sequences, for designing new enzymes or proteins with modified or enhanced properties. Earlier protein engineering was known to depend on techniques like X-ray crystallography, chemical synthesis of DNA, computational modeling of protein structure and folding, etc. A combined approach involving protein crystal structure and chemical synthesis of genes has been the most practiced one for creating proteins with desired functions.22 The potential of protein engineering had been identified by then towards engineering biocatalysts with enhanced efficiency.23 In recent years, protein engineering methods and applications have become extensive and significant due to the advancements in recombinant DNA technology and high-throughput screening techniques. Protein engineering can be broadly classified under the following categories – rational design, de novo design and directed evolution. The first protein design was done rationally based on manual analysis of the existing protein sequences, amino acid charges and the desired structural geometry.24 Gutte et al. pioneered the protein design by rationally designing a 70 amino acid peptide analog of ribonuclease S with enzyme activity via solid phase synthesis.25 Rational design is effective only when the structure and/or folding mechanism of the proteins of interest are well-established. The limitation of these data about protein structure and mechanism of protein folding led to the use of other evolutionary methods like directed evolution. This method of directed evolution involve “random mutagenesis and selection” to achieve desired properties of the protein,26 but then the limitation roots in screening of the randomly evolved proteins. The manual analyses of protein designs was then replaced by a fully automated computational algorithm based de novo protein design which can screen a library of amino acid sequences for compatibility with the desired target design.27 Later, in the beginning of 21st century, the potential of de novo enzyme engineering was established.28 This method includes computational rational design, from the scratch and not based on another similar parent protein associated with a substrate/reaction mechanism. The advantages of rational and directed evolution have been combined together, as localized/region specific random mutations to design proteins, often known as semi-rational protein engineering.29,30 An outline of the protein engineering strategies is given as a flowchart in Fig. 1.Research on biomaterials, a multidisciplinary field is consistently expanding with substantial introduction of new ideas and approaches, particularly to aid human healthcare. Use of biomaterials dates back to a few decades and the demand for new reliable biomaterials has led to developments in this field. Natural proteins offer biofunctionality by mimicking the extracellular matrices in human body, thereby enabling biological interaction with the cells/tissues with lesser immunogenic responses as compared to other synthetic biomaterials. Proteins also offer biodegradability to the biomaterials. In addition, proteins can also be engineered for controlled degradation, where the scaffold material degradation can be synchronized for tissue formation over a period of time. There is a shift from synthetic and natural biomaterials to hybrid biomaterials that have synergistic effect and improved characteristics suitable for use in drug delivery systems, implants, tissue engineering scaffolds, biosensors and other biomedical applications. Herein, we discuss the applications of various protein engineering strategies towards the generation of biomaterials and categorize them based on the source proteins used for their construction. The biomaterials constructed with a combination of well characterized natural proteins or peptide/protein domains with known structure/function along with/without synthetic/ceramic materials are known as rationally designed protein biomaterials. Whereas, those biomaterials constructed with random combinations of randomly evolved proteins/protein domains/ensembles with no prior structural data are known to be directed evolution based protein biomaterials.
2.1 Evolution of proteins by rational engineering – towards biomaterial construction
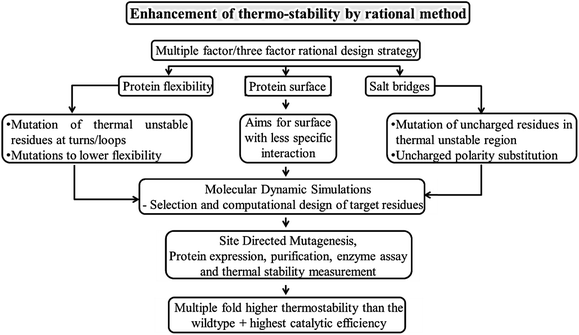
Rational design is one of the traditional strategies to engineer proteins with enhanced or desired properties.31 The process of rational design starts with choosing a suitable protein scaffold, identifying the important residues for modification, and screening of mutants followed by their characterization. In rational approach it is necessary to choose the protein scaffold based on prior knowledge of that protein's structure or by the structure of a homologous protein. The residues or regions to be changed in the protein scaffold are decided according to the function of the protein on hand as well as the anticipated function after rational designing. These modifications involve mutation, deletion, or insertion of one or several of the amino acids in the native protein sequence. This is carried out through rational engineering by site directed mutagenesis where amplification of variants occurs with mutated oligonucleotides by methods like megaprimer-based PCR, inverse PCR and overlap extension PCR. Gene truncation or deletions is also one another strategy to identify specific function of a domain or region. Alteration of functions by rational approaches32 can be achieved through single-point mutation, exchange of elements in secondary structure, exchange of whole domains, or generation of fusion proteins.33 For instance, the amino acid residues responsible for thermostability are identified by comparing the sequences of stable or less stable proteins.34 Multiple factor strategy has been designed rationally by the integration of three factors that affect the structure, including salt bridges, protein flexibility and protein surface as illustrated in Fig. 2. It was reported that mutants with a combination of multiple factors of thermostable sites were found to have doubled thermostability and also enhanced catalytic efficiency. These kinds of rational mutations could be applied towards construction of biomaterials as well. The significant strategies employed for the rationally engineered biomaterials and its applications are discussed below.• Rational designing of protein biomaterials include natural proteins because of their known bio functionality, biodegradability and biocompatibility, which are usually more preferred for biomedical applications. Natural proteins including collagen, silk/silk fibroin, keratin, gelatin, fibrinogen, fibrin, fibronectin, elastin, actin, myosin, resilin, titin, soy, zein, wheat gluten, casein, serum albumin, etc. are employed for biomaterial construction. Here we limit our discussion to protein biomaterials from natural proteins like collagen, silk, engineered/modular proteins and their combinations with synthetic polymers, ceramics or other proteins/peptides along with their biomedical applications. The Table 1 gives a list of significant biomedical applications along with the base material used.
| Biomedical Applications | Base materials | References |
|---|---|---|
| Wound healing | • Collagen | 39 |
| • Chitosan nano-particles | 40 and 41 | |
| • Silk protein | 42 | |
| Biological adhesives/sealants | • Collagen, gelatin based | 43 |
| • Fibrin based | 44 | |
| • Chitin and chitosan membranes | 39 | |
| • Polyurethane and PEG based sealants | 45 and 46 | |
| Biodegradable sutures | • Silk sutures | 47 and 48 |
| • Metal sutures | 49 | |
| • Polymer sutures | 50 | |
| Carriers in drug delivery systems | • Albumin | 51 |
| • Plant proteins | 52 | |
| • Chitosan and its nano composites | 53 | |
| • Polymeric biomaterial and lipid based nanoparticles | 54 | |
| • Nano bioceramics | 55 | |
| Biosensor materials | • Resilin like polypeptides (rec1-Resilin) | 56 |
| • Conducting polymers-polypyrrole, polythiophene, polyaniline | 57 and 58 |
• Two or more materials are rationally combined for their distinct composition, structure and other physical properties in rational designing of hybrid or semi synthetic biomaterials. It usually follows the approach of combining the biomaterials in the form of natural/synthetic biopolymers and/or bioceramics. The natural polymers are known to have weak mechanical properties and these can be enhanced by combination of inorganic hydroxyapatite or polymers like polycaprolactone.
• By means of genetic engineering, properties of proteins can be rationally tuned by incorporating functional sequences, unnatural amino acids, and specific peptide domains with unique properties forming combinatorial design and also formation of stimuli-induced systems. Also, as isolation of protein in large scale from natural sources is not affordable, genetically engineered proteins are developed with homogeneity. Modular proteins with a combination of silk-like-proteins and elastin-like-proteins by recombinant technology forms silk-elastin-like-proteins, a stimuli-responsive system, which are found responsive for parameters like pH, temperature, redox, ionic strength, electric fields and enzymatic stimuli, that can be used for controlled delivery of therapeutics.35 These rationally designed recombinant protein polymers possess tunable structure–function property,36 enhanced mechanical and elastic property, biocompatibility, degradability and target specificity.
• Rationally designed modular protein engineered biomaterials are another form of hybrid materials, that involves selection of desired peptide domains with desired functionality, designing of structural orientation of those peptide domains, selection of their amino acid sequences and encoding gene sequences, followed by cloning and polypeptide purification using a host organism37 as shown in Fig. 3. These protein engineered materials with known structure and function, ensures both tunability and biofunctionality required for a particular application, which cannot be customized in many native or synthetic materials.38
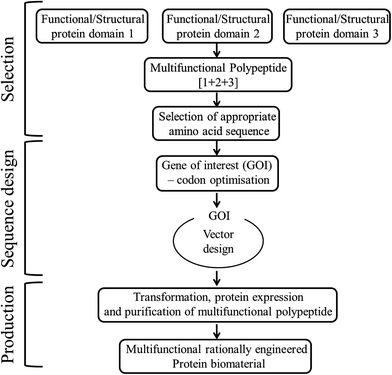 | ||
| Fig. 3 Steps involved in the rational design of multifunctional modular protein engineered biomaterial construction. | ||
Wound healing. Several wound dressings with specific properties are being developed, to meet the healing requirements of the different kinds of wounds. More recently, nanofibrous matrices were developed using a combination of non-mulberry silk protein sericin, chitosan and polyvinyl alcohol, mimicking the structure of extracellular matrices.42 These matrices have proved to promote the growth of keratinocytes in vitro and wound healing in vivo with the formation of epithelial cells of skin along with angiogenesis. For the treatment of chronic wounds where the normal healing process has failed, biomaterials with mesenchymal stem cells have been used in the recent years. Soy protein was integrated with chitin to form a moisture retaining biomaterial59 that unites with the human mesenchymal stem cells derived from adipose tissue, resulting in the regeneration of wound tissues by enhanced anti-inflammatory effect due to indoleamine 2,3-dioxygenase production. Collagen60 and chitosan39 based blends have also been largely exploited for wound dressing applications due to their promising biocompatibility. Also, gelatin films grafted with carboxy methylated guar gum incorporated with a natural antimicrobial compound curcumin serves as an excellent wound healing biomaterial.61 Wound healing by collagen, chitosan or silk proteins sericin/fibroin have also been enhanced by incorporating anti-inflammatories, antiseptics, antibiotics, nanoparticles and other bioactive agents.62,63 Use of plasma modified collagen is yet another new approach towards wound healing64 mainly in diabetic and immuno-compromised patients. Plasma treatment along with collagen component was reported to enhance the rate of angiogenesis and the wound healing process.
Biological adhesives/sealants. Biological or tissue adhesives/sealants have several potential medical applications that include topical wound closure, replacement of sutures or staples in internal surgical procedures, adhesion of dental/corneal onlays or inlays and to prevent post-surgical adhesions. Tissue adhesive made up of albumin or gelatin is the commonly available protein based adhesives with less toxicity and high mechanical strength. Gelatin is a hydrolyzed form of collagen which is a fibrous insoluble protein found in bone, skin and connective tissues. Highly porous alkaline treated gelatin adhesives exhibits good cell adhesion as well as angiogenesis.65 Similarly, a novel tissue adhesive named “BCD” was developed from, bovine serum albumin (BSA), citrate acid (CA) and dopamine66 which exhibited 10 times better adhesion stress than the commercially existing fibrin glue within a time span of 30 minutes. Membranes made up of chitin and chitosan polysaccharides also serve as biological adhesives.67,68
Biodegradable Sutures. Suture materials are the mostly used biomaterials during a surgery to help the wound healing process by sealing the wounded area. The suture material should ideally be biocompatibile with appropriate physical strength. They can be either absorbable or non-absorbable sutures. Biodegradable/absorbable sutures are more preferred as it is not necessary to remove the suture material. Silk has been used as medical suture material since a long time, but the silk fibroin present in it is reported to undergo a very slow degradation in human body by proteolysis.69 Recent findings have proved that incorporation of an antiseptic, 4-hexylresorcinol, in silk sutures exhibited anti-microbial as well as biodegradable properties comparable to a commercial bioresorbable suture, polyglactin 910 sutures. It also enhances the expression of metalloproteinases (MMP-2, -3, and -9) in macrophages that are responsible for the degradation of a wide range of proteins. Apart from these, biodegradable metal49 and biopolymeric sutures50 are also exploited for soft and hard tissue sutures. It is interesting to note the development of a regenerative biomedical device where an in situ photochemical deposition technology is used to deposit silver on an absorbable co-polymer, poly(lactic-co-glycolic acid) sutures, functionalized with silk sericin.48
Carriers in drug delivery systems. Drug delivery systems are used to overcome the critical issues of drug insolubility, instability in normal physiological environments, poor uptake/selectivity for target cells/tissues and other side effects. Among the protein based drug delivery systems, the ones prepared from albumin of serum or chicken eggs is the mainly explored protein for developing nanostructures of protein based drug delivery systems.51,70 Albumin possesses the advantages of natural origin, ability to solubilize wide range of binding sites that can be exploited for drug delivery. Naturally occurring plant proteins52 such as, zein from maize, gliadin from wheat and corn, legumin from leguminous seeds and lectins binding selectively to carbohydrate residues, are formulated into nanoparticles and used in drug delivery. These have been reported to exhibit biocompatibility, increased solubility, prolonged drug release, reduction in toxicity and side effects of the drugs. In addition to all these drug delivery systems, chitosan,53 polymer54 and inorganic bioceramic55 based drug delivery systems, usually in the form of nanoparticles or carriers also exists to deliver antibiotics/specific drugs to the surgical site or tumors for cancer treatment.71
Biosensor materials. Conducting polymers have drawn keen interest of researchers to explore their use in biomedical applications like drug delivery systems, biomedical implants, tissue engineering and biosensors. These conducting polymers possess optical and electrical properties like those of semiconductors and metals, and are also easy to synthesize72–74 by electrochemical or chemical methods.75,76 Synthetic nerve conduits have been fabricated from the blends of polypyrrole and poly(D,L-)lactic acid77 which has the potential to be used in nerve tissue regeneration. Polyaniline and its derivatives have also been studied for its biomedical applications including, neural probes, biosensors, controlled drug delivery, and tissue engineering applications.57,58 Recently, silicon nanowires have been used to detect electrical signals from viable cells, tissues and organs, which have the potential to be used as biosensors that monitor the ionic species traversing the cell membrane with the change in potential of the electric field.78 Further research and development of these materials are being investigated for clinical applications.
| Functional Domains | Domain Functionality | References |
|---|---|---|
| Peptides: | Cell adhesion: | |
| RGD, REDV, LDV peptides | • Promotes attachment of cells by interaction with integrin receptors and selective to cell lines | 89, 79 and 90 |
| YIGSR, CDPGYIGSR, IKVAV, RNIAEIIKDI, YFQRYLI, PDSGR- laminin derived peptides | • Laminin derived peptides selectively for nerve cell adhesion | 91–96 |
| KQAGDV- peptide from fibronectin | • Elicits certain specific α5β1 integrin mediated cell behavior | 97 |
| Growth factors: | Growth factor activity: | |
| Platelet-derived growth factor (PDGF) – homo- (PDGF-AA, PDGF-BB, PDGF-CC, and PDGF-DD) and heterodimeric (PDGF-AB) polypeptide dimers | • Wound healing of hard and soft tissues, re-epithelialization, central nervous system (CNS) development and also angiogenesis in some tissues | 80, 85 and 98 |
| Vascular Endothelium Growth factor (VEGF) – VEGF-A, -B, -C, -D, and -E – VEGF-A | • For nerve repair and treatment of neurodegenerative conditions by stimulating neurogenesis and neuroprotection | 84, 85 and 99 |
| • Promotes vasculogenesis and angiogenesis by stimulation of endothelial cell migration and proliferation. | ||
| Transforming Growth factor (TGF)-β: TGF-β1 – predominant isoform bone morphogenetic proteins (BMPs)-TGF subfamily(Platelets – major source of TGF-β) | • Wound healing, angiogenesis, reepithelialization, and connective tissue generation | 85 and 98 |
| • ECM synthesis and reinforcement, bone formation, healing and osteoblast deposition on the collagen matrix of bone | ||
| Structural domain: | Thermo-responsive and mechanical properties like native elastin | 86 |
| Elastin like polypeptides (VPGXG)n | ||
| X = any amino acid except proline | 100 | |
| X = lysine, allows crosslinking via primary amine side chains | ||
| X = cysteine, allows surface immobilization and crosslining | 101 | |
| Resilin like polypeptides (Dros16, (GGRPSDSYGAPGGGN)n) from Drosophila melanogaster (An16,(AQTPSSQYGAP)n) from Anopheles gambiae | Mechanical properties including high elasticity, high resilience and heat stability | 87 |
The laminin derived peptides such as YIGSR, CDPGYIGSR, IKVAV, etc., were reported to adhere nerve cells selectively79 and a fibronectin derived peptide KQAGDV shows specific α5β1 integrin mediated cell behavior. The stromal cells and growth factors from patient's blood, including platelet rich plasma and platelet rich fibrin preparations, have been tested and are in clinical use for wound healing and tissue regeneration.40,41 These preparations can be made personalized to the specific needs of the patients where the growth factors are released from these platelets and intracellular signaling pathways are activated for specific tissue formation/repair. The platelet growth factors including, homo- (PDGF-AA, PDGF-BB, PDGF-CC, and PDGF-DD) and heterodimeric (PDGF-AB) polypeptide dimers, have been reported to aid in wound healing by re-epithelialization.80 Further, human platelet lysates are used as supplements for stem cell therapy in order to retain its biological properties, reduce the culture time and promote regeneration of target tissue by growth and differentiation of cells.81,82 Whereas, Vascular endothelial growth factor A (VEGF-A), a member of cysteine knot family, is well known for its vital roles in blood vessel growth. In mammals, it also includes VEGF-B, VEGF-C, VEGF-D and placental growth factor (PlGF, or PGF), as inducer of blood vessel development.83 These have also been reported to be involved in neuron generation.84 Transforming growth factor (TGF)-β are mostly found in platelets, aiding in inflammation, re-epithelialisation, matrix formation and remodeling.85 Apart from these growth factors, there are structural protein domains like, elastin like polypeptides86 and resilin like polypeptides,87 which have been crosslinked by different methods88 to enhance the mechanical, elastic or resilience properties forming hydrogels or other biomaterials.
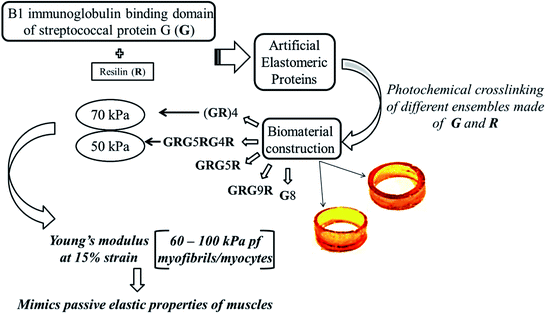
Another significant rational design from protein domains includes, a hybrid of mechanically strong GB1(B1 immunoglobulin binding domain of streptococcal protein G) protein domain102 and highly elastic resilin protein domain which were integrated by recombinant techniques to obtain artificial elastomeric proteins.103 The tandem repeats of GB1 and resilin were then photochemically crosslinked104 in different random combinations, (GR)4, GRG5RG4R, GRG5R, GRG9R, G8, that resulted in biomaterials that were hydrated and mimicked elastic properties of muscles as illustrated in Fig. 4. Each of the randomly combined ensembles were reported to possess varied mechanical properties, with two of the hydrogels formed by ensembles (GR)4 and GRG5RG4R possessing equivalent mechanical properties, to that of the muscle proteins in humans, and could potentially be used as scaffolds/matrices for tissue engineering of muscles. Rational design of protein biomaterials can thus be constructed without much complications, with the only limitation being the need for prior knowledge about structural data of the parent proteins.
Scaffold biomaterials for tissue regeneration. As collagen fibers have strength, flexibility, and the ability to be oriented and woven, they are used as scaffolds in skin repair,105,106 engineering vascular grafts,107 bone repair108 and also as carriers in drug delivery systems109 and wound healing.110 Though collagen finds various applications it lacks mechanical strength and control of degradation rate, hence collagen are combined with other synthetic or natural biomaterials to overcome these limitations. In recent days, microporous electro spun scaffolds with nano fiber mesh composed of collagen type I and synthetic polymer, polycaprolactone, are used for skin repair and regeneration.111 Silk is also a structured fibrous protein known for its mechanical strength and self-assembling ability that can form fibers.112 These silk fibers are formed by insects like silkworm and spiders for its cocoon and nest construction.113 Silk fibroin offers significant mechanical strength, degradability and biocompatibility, forming highly loaded grafts that fits for musculoskeletal tissue engineering.114,115 Keratins formed by α-helical coiled coil dimers, are structural proteins present in the epidermis appendages like nails, wool, hair and in the cytoskeletons of epithelial cells.116,117 These are biocompatible and are cell adhesive due to the presence of cell binding domain (LDV or Leu-Asp-Val), which makes it suitable for preparation of scaffolds for tissue engineering.118,119 Nerve regeneration using keratin hydrogel scaffolds have also been developed and patented.120 Further, primary evidence is available for the application of keratin biomaterial in providing protective effect or treatment, for the cells in hypoxia or low oxygen levels, which also helps in inducing cardiac regeneration after myocardial infarction.121 The bioresorbable calcium phosphate ceramics are found to be a remarkable scaffold material for the regeneration of bone due to its ability to stimulate osteogenesis.122 Based on the calcium and phosphate ratio, crystalline phase and crystallinity of calcium phosphate ceramics, release of calcium and phosphate ions and mineralization of bone occurs.123,124 This unique property separates them from other types of ceramics used in bones like alumina or zirconia which are biologically inert. In this regard, alumina and zirconia are used together as composites125,126 or with bioactive agents or polymers. Calcium phosphate based composites with natural polymers like apatite-collagen, apatite-cellulose, octa calcium phosphate-collagen127,128 have been reported to mimic the mechanical properties of bones, for their use in bone tissue engineering. Several biological molecules like bone morphogenetic proteins, laminin, heparin have been incorporated into the apatite layer,129,130 allowing further addition of active agents and extending the application of these composites in drug delivery systems.131 Electro spun nano-fibre scaffolds or films from a combination of well-known proteins like collagen, silk fibroin and a synthetic polymer polycaprolactone not only adds to mechanical strength, but have also been proven as appropriate scaffold for tissue engineering urethral reconstruction.132 Biocompatibility and conductivity for cell proliferation were proved to be feasible with the rationally designed composite films of collagen protein with nano fibres of a conducting polymer, polyaniline.133 Polyanilines have also been studied for its use as biosensor materials, due to its electrical conductivity and biocompatibility when combined with natural proteins like collagen.57,58 For further reading about tissue engineering applications and their respective base biomaterials, selective references listed in the Table 3 can be referred.
| Tissue Engineering Applications | Source of scaffold materials | References |
|---|---|---|
| Bone repair | Collagen calcium phosphate, hydroxyapatite ceramics | 3, 79 and 138 |
| Neural tissue engineering | Type I collagen, keratin, hyaluronic acid derivatives, polyglycolic acid (neurotube), polycaprolactone (neuroLac) | 139–141 |
| Skin repair/regeneration | Collagen and chitosan, polycaprolactone (PCL), and poly-ethylene-glycol (PEG), gelatin/poly (vinyl alcohol) (Gel/PVA) | 142 and 143 |
| Vascular grafts | Poly(L-lactic acid) (PLLA) and segmented polyurethane (PHD) with surface modified heparin, polycaprolactone and collagen | 144 and 145 |
| Cartilage tissue | Decellularized cartilage, devitalized cartilage | 146 and 147 |
| Tendon/ligament tissue engineering | Silk, collagen | 148 and 149 |
Thermo-responsive biomaterial. Elastin-like polypeptides (ELPs) are chosen as a representative to explain the rational design of protein with temperature induced responses for biomedical application. ELPs are artificial proteins obtained from the natural protein elastin, major component of vascular wall, with repeating VPGXG amino acid sequences. These proteins exhibit an inverse phase transition at a specific temperature at which they are in soluble aqueous form, precipitates when raised to a gelatinous aggregate called a coacervate.134,135 Urry et al.136 reported that the temperature for the transition of polypeptide depends on mean residue hydrophobicity. Based on these results from Urry et al., incorporation of valine, glycine and alanine in a 5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio, respectively, at the guest position X of the residue VPG-X-G, a ten pentapeptide sequence was genetically engineered. It was reported137 that the liquid like property of these ELPs below the transition temperature, makes it suitable for mixing cells and cell seeding, while the gel coacervate formed above the transition temperature, exhibits three times increase in complex shear modulus and comparable mechanical properties to that of hyaluronan and collagen, the extracellular matrix proteins. In addition these ELPs also maintains histological appearance, cell viability and chondrocyte phenotype in cell culture studies, proving its potential use as scaffolds for cartilaginous tissue repair.
2 ratio, respectively, at the guest position X of the residue VPG-X-G, a ten pentapeptide sequence was genetically engineered. It was reported137 that the liquid like property of these ELPs below the transition temperature, makes it suitable for mixing cells and cell seeding, while the gel coacervate formed above the transition temperature, exhibits three times increase in complex shear modulus and comparable mechanical properties to that of hyaluronan and collagen, the extracellular matrix proteins. In addition these ELPs also maintains histological appearance, cell viability and chondrocyte phenotype in cell culture studies, proving its potential use as scaffolds for cartilaginous tissue repair.
2.2 Protein engineering strategies via directed evolution
Directed evolution guides the scientists to evolve RNA, proteins, metabolic pathways, genetic circuits and also whole cells in a repetitive manner to obtain properties that are not conferred by nature. Directed evolution of proteins is usually carried out to obtain proteins with new or enhanced properties. It enables us to exploit a protein or enzyme that can confer desired properties for their applications in pharmaceutical or chemical industries. The strategy of protein design typically involves four main steps,(i) Identification of appropriate parent sequences
(ii) Mutation of parent sequences by one of the different methodologies to generate mutant libraries
(iii) Screening of the library for enhanced or desirable function/property
(iv) Repeating the mutational process to obtain variants with improved/desired properties
Diverse libraries are generally created by mutagenesis or recombination, though many other methodologies are being created from time to time. In the same way, high throughput screening techniques are being developed to reduce the experimental loads in finding variants with desired properties.150–152 Despite incomplete knowledge about protein folding, structure, expression and the mechanism of action of enzymes, directed evolution can be employed to enhance the inherent properties of enzymes.153 It includes enzyme activity, selectivity, substrate scope or stability for the application in preparation of pharmacological intermediates or fine industrial chemicals.154 The most common methods of gene mutagenesis includes, error prone PCR, saturation mutagenesis and DNA shuffling. Few of the notable strategies and examples are discussed as follows.
• Directed evolution approaches like error prone PCR,155 gene shuffling,156 site saturation mutagenesis along with appropriate screening steps, enhancement of thermostability in enzymes like laccases, cellobiohydrolases, α-amylase157 has been made possible. Directed evolution have always had the problem of screening of large number of resulting variants. Cornvik and his group had made efforts to make screening simpler by developing a new screening method for thermostability of engineered proteins, known as the HotCoFi method.158 This method is based on colony filtration (CoFi) blot and incubation of the expressed colonies at wide range of temperatures, where the proteins unfold and aggregate, the filter membrane retains the aggregates and allows the soluble proteins to pass on to a nitrocellulose membrane. It is followed by detection with affinity reagents against the target or with the help of tags fused to targets, where high signal corresponds to soluble proteins and the loss in signal corresponds to aggregated or unfolded proteins.
• Computational modeling has been combined with directed evolution approaches, to design a transaminase with desirable activity for amination of prositagliptin ketone. Synthesis of an antidiabetic drug, sitagliptin159 has been made possible by the directed evolution of a transaminase, as biocatalyst for chiral amine synthesis. In silico methods were integrated with directed evolution, more like a semi-rational approach in cases where well annotated structural data are missing for the protein of interest.
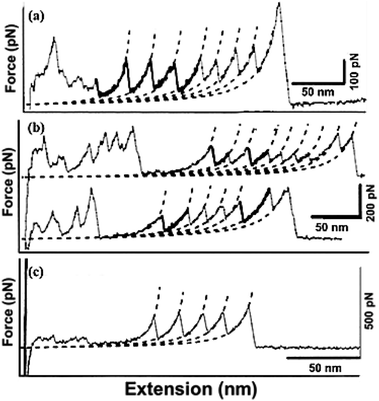
• Attempts using directed evolution based recombination or shuffling of secondary structures of the two homologous proteins, immunoglobulin domain 27 of titin (I27) and immunoglobulin domain 32 of titin (I32) to develop new mechanical proteins160 is illustrated in Fig. 5. Single molecule atomic force microscopy studies were carried out with tandem repeats161 of a well characterized protein GB1 (B1 immunoglobulin binding domain of streptococcal protein G) with contour length of 18 nm162,163 and the hybrids (i.e. [GB1-hybrid]4), to probe their unfolding events as shown as force–extension curves in Fig. 6.160 Six out of 13 daughter protein hybrids were found to be mechanically stable with contour length of 28 nm as depicted in Fig. 6(a), similar to that of the wild type parent proteins I27 and I32.164 Whereas, three protein hybrids were categorized to possess dual personality (Fig. 6(b)), with two kinds of mechanical stabilities with unfolding events similar to parent proteins as well as unfolding at forces of undetectable limits. Also, four of the hybrids were found to be mechanically labile Fig. 6(c), with no unfolding events of Ig domain of contour length 28 nm. This study enlightens the importance of adjacent residues of secondary structures as well as the non-local bindings, in protein folding. It was suggested that computational methods like SCHEMA165,166 could be used for increasing the success rate while designing proteins by using DNA shuffling based recombination approaches. SCHEMA,166 involves prediction of fragments of homologous proteins that can be recombined without disturbing the structural integrity of proteins. This computational tool finds compact polypeptides with large number of intra-block interactions that corresponds to fragments that can be swapped while preserving the structure. The resulting fragments or schemata could be then recombined by laboratory based recombination methods.
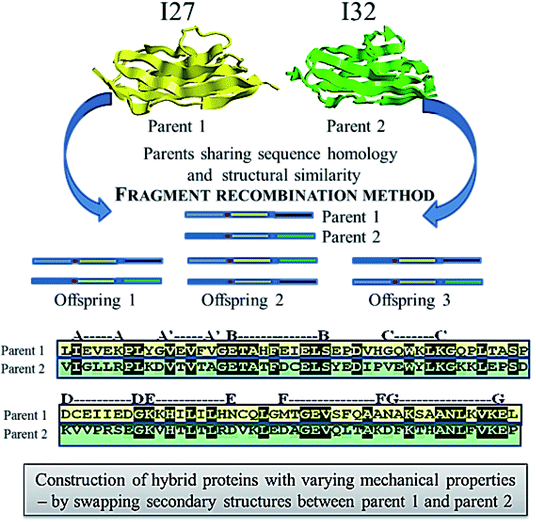 | ||
| Fig. 5 Schematics showing the method of fragment recombination used to construct hybrid proteins from the two distantly related or non-related parent proteins 1 and 2. | ||
In case of the contemporary directed evolution, several variants are generated from parental sequences, screened for preferred properties, and then the best variants are selected to act as parents for the next round of mutation/evolution, discarding other variants. When machine learning technique is applied in evolving a protein of interest, the sequences and screening data of all the variants can be exploited to train a set of models including linear, kernel, neural network, and ensemble methods. The model exhibiting highest accuracy is then employed for screening of the variants in a round of in silico evolution, in which models simulate the variant sequences for fitness and rank them accordingly. This results in a restricted library of variants with the highest simulated fitness which are then generated and screened experimentally.168 The best variant from these restricted libraries are then chosen as parent sequence for the next round of evolution with mutations at new positions, followed by identification of further improvised variants. Thereby the in silico screening using sequence–fitness relationship in machine learning investigates the entire combinatorial space of mutations at multiple positions. More recently, the advantages of in silico screening by means of machine learning technique was examined and proved to be rapid,169 using the dataset collected by Wu and coworkers,170 who reported the effects of mutations at four positions in human GB1 binding protein on antibody binding. Protein engineering applications of machine learning have identified favorable mutations171 and best possible combinations of protein fragments172 for enhanced enzyme activity and protein stability.173
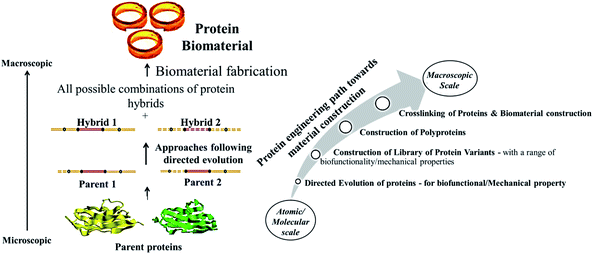
3. Future scope
The strategy of directed evolution has been found to be simpler than the traditional rational method to successfully design/engineer novel proteins with desired properties, despite of insufficient knowledge about existing parent proteins. Though several methodologies do exist in directed evolution approach, easy, rapid, sensitive and affordable screening techniques are yet to be established for identifying the variants with enhanced desirable traits. Proteins or enzymes obtained by directed evolution have found a firm place in industrial and therapeutic applications, and are still striving for improvement in other biomedical fields of applications. In the same way proteins are to be mainly engineered considering their biofunctional and mechanical properties by rational design or directed evolution in creating a library of desirable variants, and crosslink them to form macroscopic potential biomaterials as illustrated in Fig. 6. This can be anticipated to broaden the toolbox of engineered proteins for their application as biomaterials. Protein biomaterials are known for their biocompatibility, inherent biofunctionality and biodegradability, making them more suitable as scaffold materials for tissue engineering. Combinations of synthetic polymers, ceramics and also various functional/structural peptide domains along with the natural/engineered proteins as base biomaterials are to be explored more to make them fit for suitable biomedical applications. Also, it is necessary to prevent non-specific protein adsorption on the surface of these protein biomaterials that elicits several biological responses. Though polymer coatings are available to protect the biomaterials from non-specific adsorption, further research is required to develop more stable, bioinert and biospecific polymer coatings that lasts for a longer time on biomaterial surfaces under physiological conditions.Conflicts of interest
We have declared no conflict of interest.Acknowledgements
The authors are thankful to Vellore Institute of Technology, India for their support and encouragement in preparing this review. We also thank DST SERB for their financial support (DST Project id: YSS/2014/00296) to perform experiments for preliminary investigations.References
- R. V. Ulijn, N. Bibi, V. Jayawarna, P. D. Thornton, S. J. Todd, R. J. Mart, A. M. Smith and J. E. Gough, Mater. Today, 2007, 10, 40–48 CrossRef CAS.
- L. T. Saldin, M. C. Cramer, S. S. Velankar, L. J. White and S. F. Badylak, Acta Biomater., 2017, 49, 1–15 CrossRef CAS.
- N. E. Fedorovich, J. Alblas, J. R. de Wijn, W. E. Hennink, A. J. Verbout and W. J. Dhert, Tissue Eng., 2007, 13, 1905–1925 CrossRef CAS.
- M. C. Cushing and K. S. Anseth, Science, 2007, 316, 1133–1134 CrossRef CAS.
- F. J. O'brien, Mater. Today, 2011, 14, 88–95 CrossRef.
- D. F. Williams and J. Cunningham, Materials in clinical dentistry, Oxford University Press, USA, 1979 Search PubMed.
- C. W. Hall, J. Biomed. Mater. Res., 1971, 5, 1–4 CrossRef.
- J. Lister, Br. Med. J., 1967, 2, 9–12 CrossRef CAS.
- P. I. Branemark, J. Prosthet. Dent., 1983, 50, 399–410 CrossRef CAS.
- R. Toita, K. Tsuru and K. Ishikawa, Mater. Sci. Eng. C, 2016, 68, 291–298 CrossRef.
- Q. Chen and G. A. Thouas, Mater. Sci. Eng. R Rep., 2015, 87, 1–57 CrossRef.
- B. Altmann, R.-J. Kohal, T. Steinberg, P. Tomakidi, M. Bächle-Haas, A. Wennerberg and W. Att, Tissue Eng., Part C, 2013, 19, 850–863 CrossRef CAS.
- W. She, N. Li, K. Luo, C. Guo, G. Wang, Y. Geng and Z. Gu, Biomaterials, 2013, 34, 2252–2264 CrossRef CAS.
- Y. Zhang, H. F. Chan and K. W. Leong, Adv. Drug Deliv. Rev., 2013, 65, 104–120 CrossRef CAS.
- J. Lam, E. C. Clark, E. L. Fong, E. J. Lee, S. Lu, Y. Tabata and A. G. Mikos, Biomaterials, 2016, 83, 332–346 CrossRef CAS.
- T. H. Qazi, D. J. Mooney, M. Pumberger, S. Geissler and G. N. Duda, Biomaterials, 2015, 53, 502–521 CrossRef CAS.
- L. Germain, P. De Berdt, J. Vanacker, J. Leprince, A. Diogenes, D. Jacobs, G. Vandermeulen, C. Bouzin, V. Preat, C. Dupont-Gillain and A. des Rieux, Regen. Med., 2015, 10, 153–167 CrossRef CAS.
- P. Whiting, J. Kerby, P. Coffey, L. da Cruz and R. McKernan, Phil. Trans. R. Soc. Lond. Ser. B Biol. Sci., 2015, 370, 20140375 CrossRef.
- L. L. Hench, Science, 1980, 208, 826–831 CrossRef CAS.
- J.-A. Yang, J. Yeom, B. W. Hwang, A. S. Hoffman and S. K. Hahn, Prog. Polym. Sci., 2014, 39, 1973–1986 CrossRef CAS.
- X. Xu, Z. Xu, X. Yang, Y. He and R. Lin, Int. J. Biol. Macromol., 2017, 95, 294–298 CrossRef CAS.
- K. M. Ulmer, Science, 1983, 219, 666–671 CrossRef CAS.
- M. N. Gupta, Eur. J. Biochem., 1992, 203, 25–32 CrossRef CAS.
- J. S. Richardson and D. C. Richardson, Trends Biochem. Sci., 1989, 14, 304–309 CrossRef CAS.
- B. Gutte, J. Biol. Chem., 1975, 250, 889–904 CAS.
- F. Arnold, FASEB J., 1993, 7, 744–749 CrossRef CAS.
- B. I. Dahiyat and S. L. Mayo, Science, 1997, 278, 82–87 CrossRef CAS.
- M. V. Golynskiy and B. Seelig, Trends Biotechnol., 2010, 28, 340–345 CrossRef CAS.
- S. Lutz, Curr. Opin. Biotechnol., 2010, 21, 734–743 CrossRef CAS.
- R. A. Chica, N. Doucet and J. N. Pelletier, Curr. Opin. Biotechnol., 2005, 16, 378–384 CrossRef CAS.
- K. Steiner and H. Schwab, Comput. Struct. Biotechnol. J., 2012, 2, e201209010 CrossRef.
- I. V. Korendovych, in Protein Eng., Springer, 2018, pp. 15–23 Search PubMed.
- M. Sobti and B. C. Mabbutt, in Protein Nanotechnology, Springer, 2013, pp. 233–250 Search PubMed.
- M. Lehmann and M. Wyss, Curr. Opin. Biotechnol., 2001, 12, 371–375 CrossRef CAS.
- W. Huang, A. Rollett and D. L. Kaplan, Expert Opin. Drug Deliv., 2015, 12, 779–791 CrossRef CAS.
- X. X. Xia, Q. Xu, X. Hu, G. Qin and D. L. Kaplan, Biomacromolecules, 2011, 12, 3844–3850 CrossRef CAS.
- R. L. DiMarco and S. C. Heilshorn, Adv. Mater., 2012, 24, 3923–3940 CrossRef CAS.
- N. H. Romano, D. Sengupta, C. Chung and S. C. Heilshorn, Biochim. Biophys. Acta, 2011, 1810, 339–349 CrossRef CAS.
- D. Archana, P. K. Dutta and J. Dutta, in Chitin and Chitosan for Regenerative Medicine, Springer, 2016, pp. 193–227 Search PubMed.
- E. Anitua, M. Sanchez, G. Orive and I. Andia, Biomaterials, 2007, 28, 4551–4560 CrossRef CAS.
- G. Intini, Biomaterials, 2009, 30, 4956–4966 CrossRef CAS.
- S. Sapru, S. Das, M. Mandal, A. K. Ghosh and S. C. Kundu, Acta Biomater., 2018, 78, 137–150 CrossRef CAS.
- C. M. Elvin, T. Vuocolo, A. G. Brownlee, L. Sando, M. G. Huson, N. E. Liyou, P. R. Stockwell, R. E. Lyons, M. Kim, G. A. Edwards, G. Johnson, G. A. McFarland, J. A. Ramshaw and J. A. Werkmeister, Biomaterials, 2010, 31, 8323–8331 CrossRef CAS.
- S. P. Mandell and N. S. Gibran, Expert Opin. Biol. Ther., 2014, 14, 821–830 CrossRef.
- G. Lee, C. K. Lee and M. Bynevelt, Spine, 2010, 35, E1522–1524 CrossRef.
- T. W. Gilbert, S. F. Badylak, J. Gusenoff, E. J. Beckman, D. M. Clower, P. Daly and J. P. Rubin, Plast. Reconstr. Surg., 2008, 122, 95–102 CrossRef CAS PubMed.
- Y. Y. Jo, H. Kweon, D. W. Kim, M. K. Kim, S. G. Kim, J. Y. Kim, W. S. Chae, S. P. Hong, Y. H. Park, S. Y. Lee and J. Y. Choi, Sci. Rep., 2017, 7, 42441 CrossRef CAS.
- A. L. Gallo, M. Pollini and F. Paladini, J. Mater. Sci.: Mater. Med., 2018, 29, 133 CrossRef.
- J. M. Seitz, M. Durisin, J. Goldman and J. W. Drelich, Adv. Healthcare Mater., 2015, 4, 1915–1936 CrossRef CAS PubMed.
- B. Joseph, A. George, S. Gopi, N. Kalarikkal and S. Thomas, Int. J. Pharm., 2017, 524, 454–466 CrossRef CAS PubMed.
- M. Karimi, S. Bahrami, S. B. Ravari, P. S. Zangabad, H. Mirshekari, M. Bozorgomid, S. Shahreza, M. Sori and M. R. Hamblin, Expert Opin. Drug Delivery, 2016, 13, 1609–1623 CrossRef CAS PubMed.
- H. Malekzad, H. Mirshekari, P. Sahandi Zangabad, S. Moosavi Basri, F. Baniasadi, M. Sharifi Aghdam, M. Karimi and M. R. Hamblin, Crit. Rev. Biotechnol., 2018, 38, 47–67 CrossRef CAS.
- A. Ali and S. Ahmed, Int. J. Biol. Macromol., 2018, 109, 273–286 CrossRef CAS.
- F. Dilnawaz, Curr. Med. Chem., 2017, 24, 2423–2438 CrossRef CAS.
- S. Kumar Ts and K. Madhumathi, Ther. Deliv., 2016, 7, 573–588 CrossRef PubMed.
- M. Y. Truong, N. K. Dutta, N. R. Choudhury, M. Kim, C. M. Elvin, A. J. Hill, B. Thierry and K. Vasilev, Biomaterials, 2010, 31, 4434–4446 CrossRef CAS.
- L. Ghasemi-Mobarakeh, M. P. Prabhakaran, M. Morshed, M. H. Nasr-Esfahani, H. Baharvand, S. Kiani, S. S. Al-Deyab and S. Ramakrishna, J. Tissue Eng. Regenerat. Med., 2011, 5, e17–35 CrossRef CAS.
- P. Humpolicek, V. Kasparkova, P. Saha and J. Stejskal, Synth. Met., 2012, 162, 722–727 CrossRef CAS.
- K. Las Heras, S. Santos-Vizcaino, A. Etxabide, J. Uranga, E. Santos-Vizcaino, P. Guerrero, J. Pedraz, M. Igartua, K. De La Caba and R. Hernandez, FarmaJournal, 2019, 4, 65 Search PubMed.
- J. Chu, P. Shi, W. Yan, J. Fu, Z. Yang, C. He, X. Deng and H. Liu, Nanoscale, 2018, 10, 9547–9560 RSC.
- P. J. Manna, T. Mitra, N. Pramanik, V. Kavitha, A. Gnanamani and P. Kundu, Int. J. Biol. Macromol., 2015, 75, 437–446 CrossRef CAS.
- J. Liu, L. Yan, W. Yang, Y. Lan, Q. Zhu, H. Xu, C. Zheng and R. Guo, Bioactive Materials, 2019, 4, 151–159 CrossRef PubMed.
- G. Tao, R. Cai, Y. Wang, L. Liu, H. Zuo, P. Zhao, A. Umar, C. Mao, Q. Xia and H. He, Mater. Des., 2019, 180, 107940 CrossRef CAS.
- L. O'Neill, P. Dobbyn, M. Kulkarni and A. Pandit, Clinical Plasma Medicine, 2018, 12, 23–32 CrossRef.
- K. Yoshizawa, R. Mizuta and T. Taguchi, Biomaterials, 2015, 63, 14–23 CrossRef CAS PubMed.
- W. Zhu, Y. Peck, J. Iqbal and D.-A. Wang, Biomaterials, 2017, 147, 99–115 CrossRef CAS.
- A. Anitha, N. Deepa, K. Chennazhi, S. Nair, H. Tamura and R. Jayakumar, Carbohydr. Polym., 2011, 83, 66–73 CrossRef CAS.
- K. Azuma, M. Nishihara, H. Shimizu, Y. Itoh, O. Takashima, T. Osaki, N. Itoh, T. Imagawa, Y. Murahata, T. Tsuka, H. Izawa, S. Ifuku, S. Minami, H. Saimoto, Y. Okamoto and M. Morimoto, Biomaterials, 2015, 42, 20–29 CrossRef CAS.
- Y. Cao and B. Wang, Int. J. Mol. Sci., 2009, 10, 1514–1524 CrossRef CAS PubMed.
- R. C. Oppenheim, in Polymeric nanoparticles and microspheres, CRC Press, 2018, pp. 1–27 Search PubMed.
- J. Fan, S. Wang, W. Sun, S. Guo, Y. Kang, J. Du and X. Peng, AIChE J., 2018, 64, 835–859 CrossRef CAS.
- N. K. Guimard, N. Gomez and C. E. Schmidt, Prog. Polym. Sci., 2007, 32, 876–921 CrossRef CAS.
- B. Guo, L. Glavas and A.-C. Albertsson, Prog. Polym. Sci., 2013, 38, 1263–1286 CrossRef CAS.
- A. J. Heeger, Angew. Chem., Int. Ed., 2001, 40, 2591–2611 CrossRef CAS.
- A. Diaz, K. K. Kanazawa and G. P. Gardini, J. Chem. Soc., Chem. Commun., 1979, 635–636 RSC.
- K. Kaneto, K. Yoshino and Y. Inuishi, Solid State Commun., 1983, 46, 389–391 CrossRef CAS.
- H. Xu, J. M. Holzwarth, Y. Yan, P. Xu, H. Zheng, Y. Yin, S. Li and P. X. Ma, Biomaterials, 2014, 35, 225–235 CrossRef CAS PubMed.
- X. Duan, R. Gao, P. Xie, T. Cohen-Karni, Q. Qing, H. S. Choe, B. Tian, X. Jiang and C. M. Lieber, Nat. Nanotechnol., 2012, 7, 174–179 CrossRef CAS.
- M. Fittkau, P. Zilla, D. Bezuidenhout, M. Lutolf, P. Human, J. A. Hubbell and N. Davies, Biomaterials, 2005, 26, 167–174 CrossRef CAS.
- J. Andrae, R. Gallini and C. Betsholtz, Genes Dev., 2008, 22, 1276–1312 CrossRef CAS.
- E. Anitua, R. Prado and G. Orive, Trends Biotechnol., 2013, 31, 364–374 CrossRef CAS.
- R. Crespo-Diaz, A. Behfar, G. W. Butler, D. J. Padley, M. G. Sarr, J. Bartunek, A. B. Dietz and A. Terzic, Cell Transplant., 2011, 20, 797–811 Search PubMed.
- T. Tammela, T. V. Petrova and K. Alitalo, Trends Cell Biol., 2005, 15, 434–441 CrossRef CAS.
- F. Mackenzie and C. Ruhrberg, Development, 2012, 139, 1371–1380 CrossRef CAS.
- S. Barrientos, O. Stojadinovic, M. S. Golinko, H. Brem and M. Tomic-Canic, Wound Repair Regen., 2008, 16, 585–601 CrossRef.
- S. Roberts, M. Dzuricky and A. Chilkoti, FEBS Lett., 2015, 589, 2477–2486 CrossRef CAS.
- R. S.-C. Su, Y. Kim and J. C. Liu, Acta Biomater., 2014, 10, 1601–1611 CrossRef CAS.
- J. Brindha, K. Chanda and M. Balamurali, Mater. Sci. Eng. C, 2019, 95, 312 CrossRef CAS.
- S. L. Bellis, Biomaterials, 2011, 32, 4205–4210 CrossRef CAS.
- U. Hersel, C. Dahmen and H. Kessler, Biomaterials, 2003, 24, 4385–4415 CrossRef CAS.
- J. P. Ranieri, R. Bellamkonda, E. J. Bekos, J. A. Gardella Jr, H. J. Mathieu, L. Ruiz and P. Aebischer, Int. J. Dev. Neurosci., 1994, 12, 725–735 CrossRef CAS.
- J. P. Ranieri, R. Bellamkonda, E. J. Bekos, T. G. Vargo, J. A. Gardella Jr and P. Aebischer, J. Biomed. Mater. Res., 1995, 29, 779–785 CrossRef CAS PubMed.
- L. Aucoin, C. Griffith, G. Pleizier, Y. Deslandes and H. Sheardown, J. Biomater. Sci., Polym. Ed., 2002, 13, 447–462 CrossRef CAS PubMed.
- M. Huber, P. Heiduschka, S. Kienle, C. Pavlidis, J. Mack, T. Walk, G. Jung and S. Thanos, J. Biomed. Mater. Res., 1998, 41, 278–288 CrossRef CAS.
- J. C. Schense, J. Bloch, P. Aebischer and J. A. Hubbell, Nat. Biotechnol., 2000, 18, 415 CrossRef CAS.
- Y. W. Tong and M. S. Shoichet, J. Biomed. Mater. Res., 1998, 42, 85–95 CrossRef CAS.
- Y. Maeshima, U. L. Yerramalla, M. Dhanabal, K. A. Holthaus, S. Barbashov, S. Kharbanda, C. Reimer, M. Manfredi, W. M. Dickerson and R. Kalluri, J. Biol. Chem., 2001, 276, 31959–31968 CrossRef CAS.
- S. Graham, A. Leonidou, M. Lester, M. Heliotis, A. Mantalaris and E. Tsiridis, Expert Opin. Investig. Drugs, 2009, 18, 1633–1654 CrossRef CAS.
- C. Ruiz de Almodovar, D. Lambrechts, M. Mazzone and P. Carmeliet, Physiol. Rev., 2009, 89, 607–648 CrossRef CAS.
- H. Wang, L. Cai, A. Paul, A. Enejder and S. C. Heilshorn, Biomacromolecules, 2014, 15, 3421–3428 CrossRef CAS PubMed.
- Y. Yuan and P. Koria, J. Biomed. Mater. Res. A, 2016, 104, 697–706 CrossRef CAS.
- Y. Cao and H. Li, Nat. Mater., 2007, 6, 109–114 CrossRef CAS.
- S. Lv, D. M. Dudek, Y. Cao, M. M. Balamurali, J. Gosline and H. Li, Nature, 2010, 465, 69–73 CrossRef CAS PubMed.
- D. A. Fancy and T. Kodadek, Proc. Natl. Acad. Sci. U.S.A., 1999, 96, 6020–6024 CrossRef CAS.
- C. Helary, I. Bataille, A. Abed, C. Illoul, A. Anglo, L. Louedec, D. Letourneur, A. Meddahi-Pelle and M. M. Giraud-Guille, Biomaterials, 2010, 31, 481–490 CrossRef CAS PubMed.
- F. Wehrhan, E. Nkenke, I. Melnychenko, K. Amann, K. A. Schlegel, C. Goerlach, W. H. Zimmermann and S. Schultze-Mosgau, Dermatol. Surg., 2010, 36, 919–930 CrossRef CAS.
- I. S. Park, S. H. Kim, Y. H. Kim, I. H. Kim and S. H. Kim, J. Biomater. Sci. Polym. Ed., 2009, 20, 1645–1660 CrossRef CAS.
- F. G. Lyons, A. A. Al-Munajjed, S. M. Kieran, M. E. Toner, C. M. Murphy, G. P. Duffy and F. J. O'Brien, Biomaterials, 2010, 31, 9232–9243 CrossRef CAS PubMed.
- G. Kleinmann, S. Larson, B. Hunter, S. Stevens, N. Mamalis and R. J. Olson, Ophthalmologica, 2006, 221, 51–56 CrossRef.
- M. Maeda, K. Kadota, M. Kajihara, A. Sano and K. Fujioka, J. Control. Release, 2001, 77, 261–272 CrossRef CAS.
- P. P. Bonvallet and S. L. Bellis, Microsc. Microanal., 2016, 22, 3 CrossRef.
- M. Heim, L. Römer and T. Scheibel, Chem. Soc. Rev., 2010, 39, 156–164 RSC.
- F. G. Omenetto and D. L. Kaplan, Science, 2010, 329, 528–531 CrossRef CAS.
- T. Nau and A. Teuschl, World J. Orthop., 2015, 6, 127–136 CrossRef.
- A. Teuschl, P. Heimel, S. Nurnberger, M. van Griensven, H. Redl and T. Nau, Am. J. Sports Med., 2016, 44, 1547–1557 CrossRef.
- P. Hill, H. Brantley and M. Van Dyke, Biomaterials, 2010, 31, 585–593 CrossRef CAS.
- E. B. Lane and W. H. McLean, J. Pathol., 2004, 204, 355–366 CrossRef CAS.
- S. Reichl, Biomaterials, 2009, 30, 6854–6866 CrossRef CAS.
- A. Tachibana, S. Kaneko, T. Tanabe and K. Yamauchi, Biomaterials, 2005, 26, 297–302 CrossRef CAS.
- M. E. Van Dyke, US Pat., 8,968,764, 2015.
- B. Lund, L. Burleson, M. James, S. Burris, P. Link and M. Brinkley, Keratin biomaterials attenuate hypoxia-R mediated cell toxicity, Washington, 2015 Search PubMed.
- I. Denry and L. T. Kuhn, Dent. Mater., 2016, 32, 43–53 CrossRef CAS.
- S. V. Dorozhkin, Materials, 2013, 6, 3840–3942 CrossRef CAS.
- A. F. Khan, M. Saleem, A. Afzal, A. Ali, A. Khan and A. R. Khan, Mater. Sci. Eng. C, 2014, 35, 245–252 CrossRef CAS.
- H. C. Ko, J. S. Han, M. Bachle, J. H. Jang, S. W. Shin and D. J. Kim, Dent. Mater., 2007, 23, 1349–1355 CrossRef CAS.
- G. Schierano, F. Mussano, M. G. Faga, G. Menicucci, C. Manzella, C. Sabione, T. Genova, M. M. von Degerfeld, B. Peirone, A. Cassenti, P. Cassoni and S. Carossa, BioMed Res. Int., 2015, 2015, 157360 Search PubMed.
- A. Yoshida, T. Miyazaki, M. Ashizuka and E. Ishida, J. Biomater. Appl., 2006, 21, 179–194 CrossRef CAS.
- T. Kawai, K. Matsui, S. Iibuchi, T. Anada, Y. Honda, K. Sasaki, S. Kamakura, O. Suzuki and S. Echigo, Clin. Implant Dent. Relat. Res., 2011, 13, 112–123 CrossRef.
- T. Kodama, T. Goto, T. Miyazaki and T. Takahashi, Int. J. Oral Maxillofac. Implant., 2008, 23, 1013–1019 Search PubMed.
- A. Oyane, M. Uchida and A. Ito, J. Biomed. Mater. Res. A, 2005, 72, 168–174 CrossRef.
- T. Miyazaki, K. Ishikawa, Y. Shirosaki and C. Ohtsuki, Biol. Pharm. Bull., 2013, 36, 1670–1675 CrossRef CAS PubMed.
- G. Wei, C. Li, Q. Fu, Y. Xu and H. Li, Int. Urol. Nephrol., 2015, 47, 95–99 CrossRef CAS PubMed.
- H.-S. Kim, H. L. Hobbs, L. Wang, M. J. Rutten and C. C. Wamser, Synth. Met., 2009, 159, 1313–1318 CrossRef CAS.
- D. W. Urry, J. Protein Chem., 1988, 7, 1–34 CrossRef CAS PubMed.
- D. W. Urry, Prog. Biophys. Mol. Biol., 1992, 57, 23–57 CrossRef CAS.
- D. W. Urry, C. H. Luan, T. M. Parker, D. C. Gowda, K. U. Prasad, M. C. Reid and A. Safavy, J. Am. Chem. Soc., 1991, 113, 4346–4348 CrossRef CAS.
- H. Betre, L. A. Setton, D. E. Meyer and A. Chilkoti, Biomacromolecules, 2002, 3, 910–916 CrossRef CAS.
- B. Fei, H. Xu, Y. Cao, S. Ma, H. Guo, T. Song, D. Qiao and Y. Cao, J. Ind. Microbiol. Biotechnol., 2013, 40, 457–464 CrossRef CAS.
- R. Boni, A. Ali, A. Shavandi and A. N. Clarkson, J. Biomed. Sci., 2018, 25, 90 CrossRef CAS.
- F. Mohamadi, S. Ebrahimi-Barough, M. Reza Nourani, M. Ali Derakhshan, V. Goodarzi, M. Sadegh Nazockdast, M. Farokhi, R. Tajerian, R. Faridi Majidi and J. Ai, J. Biomed. Mater. Res. A, 2017, 105, 1960–1972 CrossRef CAS.
- P. Sensharma, G. Madhumathi, R. D. Jayant and A. K. Jaiswal, Mater. Sci. Eng. C, 2017, 77, 1302–1315 CrossRef CAS.
- H. Mahnama, S. Dadbin, M. Frounchi and S. Rajabi, Artif. Cells, Nanomed., Biotechnol., 2017, 45, 928–935 CrossRef CAS.
- A. A. Chaudhari, K. Vig, D. R. Baganizi, R. Sahu, S. Dixit, V. Dennis, S. R. Singh and S. R. Pillai, Int. J. Mol. Sci., 2016, 17 CAS.
- S. J. Lee, J. Liu, S. H. Oh, S. Soker, A. Atala and J. J. Yoo, Biomaterials, 2008, 29, 2891–2898 CrossRef CAS.
- P. C. Caracciolo, M. I. Rial-Hermida, F. Montini-Ballarin, G. A. Abraham, A. Concheiro and C. Alvarez-Lorenzo, Mater. Sci. Eng. C, 2017, 75, 1115–1127 CrossRef CAS PubMed.
- E. A. Kiyotake, E. C. Beck and M. S. Detamore, Ann. N. Y. Acad. Sci., 2016, 1383, 139–159 CrossRef CAS PubMed.
- J. M. Aamodt and D. W. Grainger, Biomaterials, 2016, 86, 68–82 CrossRef CAS PubMed.
- S. Yunoki, H. Hatayama, M. Ebisawa, E. Kondo and K. Yasuda, J. Biomed. Mater. Res. A, 2015, 103, 3054–3065 CrossRef CAS PubMed.
- D. S. Musson, D. Naot, A. Chhana, B. G. Matthews, J. D. McIntosh, S. T. Lin, A. J. Choi, K. E. Callon, P. R. Dunbar, S. Lesage, B. Coleman and J. Cornish, Tissue Eng., Part A, 2015, 21, 1539–1551 CrossRef CAS.
- Y. Yu and S. Lutz, Trends Biotechnol., 2011, 29, 18–25 CrossRef CAS.
- R. D. Gupta and D. S. Tawfik, Nat. Methods, 2008, 5, 939 CrossRef CAS.
- S. A. Benner, S. O. Sassi and E. A. Gaucher, Adv. Enzymol. Relat. Areas Mol. Biol., 2007, 75, 1–132 CAS.
- A. S. Bommarius, J. K. Blum and M. J. Abrahamson, Curr. Opin. Chem. Biol., 2011, 15, 194–200 CrossRef CAS.
- M. T. Reetz, P. Soni, L. Fernandez, Y. Gumulya and J. D. Carballeira, Chem. Commun., 2010, 46, 8657–8658 RSC.
- I. Wu and F. H. Arnold, Biotechnol. Bioeng., 2013, 110, 1874–1883 CrossRef CAS.
- I. Pardo, A. I. Vicente, D. M. Mate, M. Alcalde and S. Camarero, Biotechnol. Bioeng., 2012, 109, 2978–2986 CrossRef CAS.
- C. Wang, R. Huang, B. He and Q. Du, BMC Bioinf., 2012, 13, 263 CrossRef CAS PubMed.
- I. Asial, Y. X. Cheng, H. Engman, M. Dollhopf, B. Wu, P. Nordlund and T. Cornvik, Nat. Commun., 2013, 4, 2901 CrossRef PubMed.
- C. K. Savile, J. M. Janey, E. C. Mundorff, J. C. Moore, S. Tam, W. R. Jarvis, J. C. Colbeck, A. Krebber, F. J. Fleitz and J. Brands, Science, 2010, 329, 305–309 CrossRef CAS.
- M. Balamurali, D. Sharma, A. Chang, D. Khor, R. Chu and H. Li, Protein Sci., 2008, 17, 1815–1826 CrossRef CAS PubMed.
- P. J. Bujalowski and A. F. Oberhauser, Methods, 2013, 60, 151–160 CrossRef CAS PubMed.
- Y. Cao, C. Lam, M. Wang and H. Li, Angew. Chem., 2006, 118, 658–661 CrossRef.
- Y. Cao and H. Li, Nat. Mater., 2007, 6, 109 CrossRef CAS.
- M. Rief, M. Gautel, F. Oesterhelt, J. M. Fernandez and H. E. Gaub, Science, 1997, 276, 1109–1112 CrossRef CAS.
- C. A. Voigt, C. Martinez, Z.-G. Wang, S. L. Mayo and F. H. Arnold, Nat. Struct. Mol. Biol., 2002, 9, 553 CAS.
- M. M. Meyer, J. J. Silberg, C. A. Voigt, J. B. Endelman, S. L. Mayo, Z. G. Wang and F. H. Arnold, Protein Sci., 2003, 12, 1686–1693 CrossRef CAS.
- K. T. Butler, D. W. Davies, H. Cartwright, O. Isayev and A. Walsh, Nature, 2018, 559, 547 CrossRef CAS.
- T. M. Jacobs, H. Yumerefendi, B. Kuhlman and A. Leaver-Fay, Nucleic Acids Res., 2014, 43, e34 CrossRef PubMed.
- Z. Wu, S. J. Kan, R. D. Lewis, B. J. Wittmann and F. H. Arnold, Proc. Natl. Acad. Sci. U.S.A., 2019, 116, 8852–8858 CrossRef CAS PubMed.
- N. C. Wu, L. Dai, C. A. Olson, J. O. Lloyd-Smith and R. Sun, Elife, 2016, 5, e16965 CrossRef.
- R. J. Fox, S. C. Davis, E. C. Mundorff, L. M. Newman, V. Gavrilovic, S. K. Ma, L. M. Chung, C. Ching, S. Tam and S. Muley, Nat. Biotechnol., 2007, 25, 338 CrossRef CAS.
- P. A. Romero, A. Krause and F. H. Arnold, Proc. Natl. Acad. Sci. U.S.A., 2013, 110, E193–E201 CrossRef CAS.
- K. K. Yang, Z. Wu and F. H. Arnold, arXiv preprint arXiv:1811.10775, 2018.
| This journal is © The Royal Society of Chemistry 2019 |