Open Access Article
Open Access ArticleRetracted Article: Long noncoding RNA DLEU1 promotes cell proliferation and migration of Wilms tumor through the miR-300/HOXC8 axis†
Wen'an Ge and
Shengxing Wang *
*
Department of Urology, Xi'an Children's Hospital, Xi'an 710003, China. E-mail: shengxing_wang1@126.com; Fax: +86-29-87692009; Tel: +86-29-87692210
First published on 4th December 2019
Abstract
Wilms tumor (nephroblastoma) is the most common primary renal tumor occurring in children. Long noncoding RNA (lncRNA) deleted in lymphocytic leukemia 1 (DLEU1) is an identified cancer-associated lncRNA that plays an important role in various cancers. However, the role of DLEU1 in Wilms tumor remains unclear. In the present study, we examined the expression and role of DLEU1 in Wilms tumor. We demonstrated that DLEU1 expression was upregulated in Wilms tumor tissues and cell lines. Knockdown of DLEU1 significantly inhibited the proliferation, migration and invasion of GHINK-1 cells. Furthermore, DLEU1 directly sponged miR-300 and regulated the expression level of miR-300 in GHINK-1 cells. Inhibition of miR-300 reversed the inhibitory effects of DLEU1 downregulation on cell proliferation, migration and invasion. Homeobox C8 (HOXC8) was found to be a target gene of miR-300 and mediated the role of miR-300 in GHINK-1 cells. In conclusion, these findings indicated that DLEU1 executed an oncogenic role in Wilms tumor via regulating the miR-300/HOXC8 axis, indicating that DLEU1 might be a therapeutic target for the treatment of Wilms tumor.
1. Introduction
Wilms tumor, also known as nephroblastoma, is the most common primary renal tumor occurring in children, accounting for 5% of pediatric malignant tumors.1,2 It is an embryonic tumor composed of stromal, blastemal, and epithelial cells, and is histologically similar to renal embryogenesis.3 According to the diagnosis and staging assessment of Wilms tumor cases, the treatments include surgical resection, radiotherapy, chemotherapy, or multimodal therapies.4 Although the overall 5-year survival rate exceeds 90%, a more successful treatment may reduce the late adverse effects, such as secondary malignancies and cardiac dysfunction.5 Therefore, better understanding of the molecular pathogenesis of Wilms tumor is conducive to improve the treatment and outcomes of patient with Wilms tumor.Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides with no or limited protein-coding ability.6 LncRNAs can interact with DNAs, RNAs and proteins to regulate gene expression at epigenetic, transcriptional and post-transcriptional levels.7,8 Increasing amounts of evidence have demonstrated that lncRNAs are closely associated with the occurrence and progression of cancers.9,10 Recent studies have indicated that lncRNAs may be therapeutic target in the cancer treatments.11 However, the bio-functions and molecular mechanisms of lncRNAs in Wilms tumor remain limited. Zhu et al.12 reported that LINC00473 functions as an oncogene in Wilms tumor via microRNA (miR)-195/IκB kinase (IKK). LncRNA LINP1 enhances cell metastasis and proliferation of Wilms tumor via inducing the Wnt/β-catenin signaling pathway.13
LncRNA deleted in lymphocytic leukemia 1 (DLEU1) is an identified oncogenic lncRNA in various cancers, such as lung cancer, colorectal cancer, cervical cancer, endometrial cancer, and ovarian carcinoma.14–18 DLEU1 expression is upregulated in colorectal cancer tissues, the higher expression of DLEU1 indicates lower survival rate and poorer prognosis.15 DLEU1 promotes tumorigenesis and progression of non-small cell lung cancer (NSCLC) and might be a promising therapeutic target for NSCLC.14 These findings suggest that DLEU1 play a crucial role in the tumorigenesis. However, the role of DLEU1 in Wilms tumor remains unclear.
In the present study, we found that DLEU1 expression was increased in Wilms tumor tissues and exerted an oncogenic role. Therefore, we speculated that DLEU1 might be a therapeutic target in the Wilms tumor treatment.
2. Results
2.1 DLEU1 is highly expressed in Wilms tumor tissues and cell lines
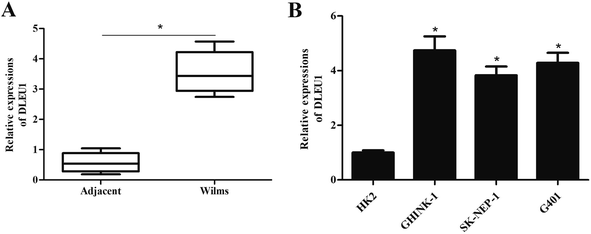
To reveal the role of DLEU1 in Wilms tumor, qRT-PCR analysis was performed to determine the expression level of DLEU1 in Wilms tumor samples and adjacent non-tumor tissues. As revealed in Fig. 1A, DLEU1 was upregulated in Wilms tumor samples compared with non-tumor tissues (p < 0.05). Then we tested the DLEU1 expression in Wilms tumor cell lines including GHINK-1, SK-NEP-1 and G401 cells. The results showed that DLEU1 was highly expressed in Wilms tumor cell lines (Fig. 1B).2.2 Knockdown of DLEU1 inhibits the proliferation, migration and invasion of Wilms tumor cells
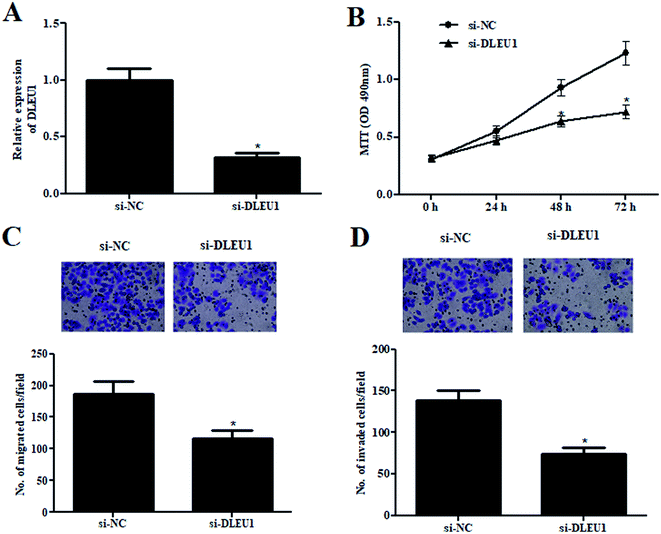
To further investigate the roles of DLEU1 on Wilms tumor, GHINK-1 cells were transfected with si-DLEU1 or si-NC. The knockdown of DLEU1 was confirmed by qRT-PCR analysis (Fig. 2A). The results from MTT assay indicated that GHINK-1 cells with lower DLEU1 exhibited reduced proliferation as compared to the GHINK-1 cells transfected with si-NC (Fig. 2B). Additionally, cell migration and invasion were respectively decreased by si-DLEU1 (Fig. 2C and D). Furthermore, we found that DLEU1 had no significant effect on cell proliferation and migration of normal kidney HK2 cells (ESI Fig. 1†).2.3 DLEU1 targets miR-300 in Wilms tumor cells
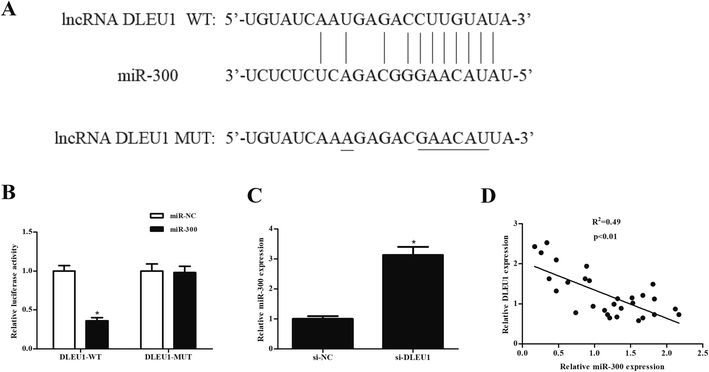
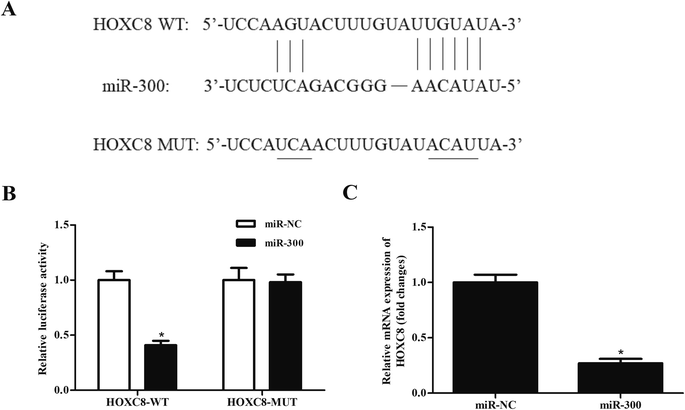
We next analyzed the molecular mechanism of DLEU1 using the online software to predict the target miRNA of DLEU1. As shown in Fig. 3A, putative complementary sequences between DLEU1 and miR-300 were found. Luciferase reporter assay proved that co-transfection with psiCHECK2-DLEU1 and miR-300 mimics significantly reduced the luciferase activity (Fig. 3B). In addition, miR-300 expression was significantly increased by si-DLEU1 in GHINK-1 cells (Fig. 3C). Furthermore, correlation analysis showed that there was a negative correlation between DLEU1 expression and miR-300 expression in Wilms tumor tissues (Fig. 3D).2.4 MiR-300 regulates the effects of DLEU1 on cell proliferation, migration and invasion
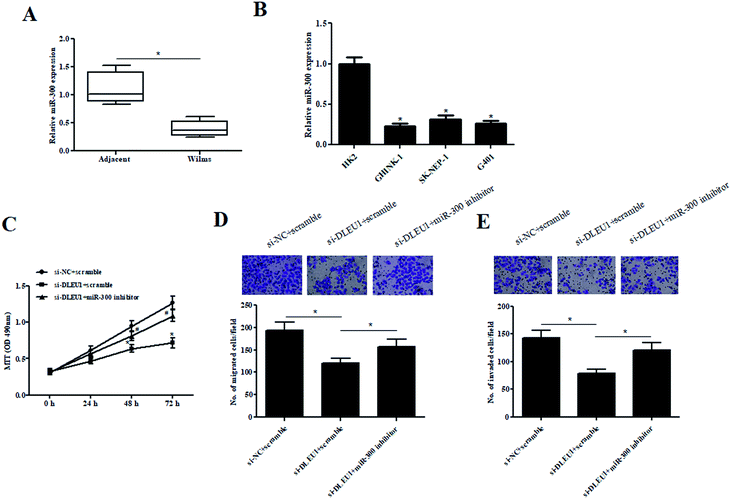
To better understand the role of miR-300 in Wilms tumor, the expression of miR-300 was examined. As illustrated in Fig. 4A and B, the expressions of miR-300 were markedly decreased in Wilms tumor tissues and cell lines. Furthermore, we observed that the decreased cell proliferation caused by si-DLEU1 was mitigated by transfection with miR-300 inhibitor (Fig. 4C). Moreover, miR-300 inhibitor also attenuated si-DLEU1-caused decrease in cell migration and invasion (Fig. 4D and E). Furthermore, we found that miR-300 mimics reserved the promotion effects of over-DLEU1 on GHINK-1 cell proliferation, migration and invasion (ESI Fig. 2A–C†).2.5 MiR-300 targets HOXC8 in Wilms tumor cells
It has been well-known that miRNAs usually bind to the 3′UTR of target mRNA and thereby regulating gene expression.19,20 Then we used bioinformatic prediction software to explore the potential target gene of miR-300. We identified that miR-300 was able to bind to the 3′UTR of HOXC8 (Fig. 5A). To confirm the interaction between miR-300 and HOXC8, a luciferase assay was performed and demonstrated that miR-300 dramatically reduced the luciferase activity of the psiCHECK2-HOXC8 (Fig. 5B). Moreover, miR-300 mimics significantly suppressed the HOXC8 expression in GHINK-1 cells (Fig. 5C).2.6 HOXC8 overexpression reverses the effects of miR-300 mimics on cell proliferation, migration and invasion
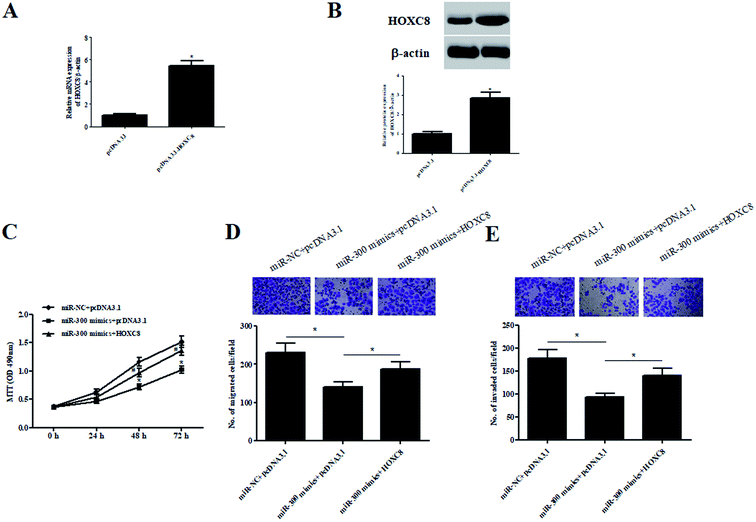
To validate whether HOXC8 mediated the role of miR-300, HOXC8-overexpressing plasmid pcDNA3.1-HOXC8 was transfected into GHINK-1 cells. The results from qRT-PCR and western blot showed that HOXC8 expression was markedly elevated after transfection with pcDNA3.1-HOXC8, compared with transfection with pcDNA3.1 vector (Fig. 6A and B). Moreover, the inhibitory effects of miR-300 mimics on cell proliferation, migration and invasion were mitigated by HOXC8 overexpression (Fig. 6C–E).3. Discussion
Increasing evidences have indicated that many lncRNAs are involved in the tumorigenesis of various types of cancers. However, the role of lncRNAs in Wilms tumor remains largely unknown. In the present study, we first evaluated the function of DLEU1 in Wilms tumor. The results showed that DLEU1 expression was upregulated in Wilms tumor tissues and acted as an oncogene. Further investigations proved that DLEU1 sponged miR-300 in GHINK-1 cells and regulated the expression of HOXC8, indicating that DLEU1 exerted its oncogenic role via regulating miR-300/HOXC8 axis.Previous studies have demonstrated that DLEU1 promotes the progression of many cancers. DLEU1 is highly expressed in endometrial carcinoma and contributes to tumorigenesis and development of endometrial carcinoma both in vitro and in vivo.21 DLEU1 expression is closely associated with advanced tumor-node-metastasis stage, vascular metastasis and poor overall survival in hepatocellular carcinoma patients, indicating that DLEU1 promotes hepatocellular carcinoma progression.22 DLEU1 is overexpressed in osteosarcoma tissue specimens, moreover, downregulation of DLEU1 in osteosarcoma cells inhibits the cell proliferation, migration and invasion, indicating that DLEU1 aggravates osteosarcoma carcinogenesis.23 Our results proved that DLEU1 expression was significantly upregulated in Wilms tumor tissues. In vitro investigations revealed that downregulation of DLEU1 inhibited the proliferation, migration and invasion of Wilms tumor cells. However, according to a previous study, DLEU1 functions as a tumor suppressor gene and confer chemoimmunotherapy resistance in children and adolescents with Burkitt lymphoma (BL).24 These findings suggest that the role of DLEU1 in cancers may depend on the cancer type.
It has been suggested that DLEU1 acts as endogenous miRNA sponges by binding to target miRNAs. Chen et al.23 reported that DLEU1 acts as an oncogene in osteosarcoma by directly sponging miR-671-5p. DLEU1 promotes cervical cancer cell proliferation and invasion by interacting with miR-381.16 Therefore, we used bioinformatic software to predict the target miRNA of DLEU1, and we found putative complementary sequences between DLEU1 and miR-300. DLEU1 directly bound to miR-300 and regulated the miR-300 expression in GHINK-1 cells. Recent studies have denoted that miR-300 is associated with tumor development. For instance, Liu et al.25 showed that miR-300 levels in the serum of osteosarcoma patients are higher than healthy controls and correlates with clinical stage, distant metastasis and poor survival, indicating that serum miR-300 may be an independent prognostic marker for osteosarcoma. MiR-300 promotes proliferation and epithelial–mesenchymal transition (EMT)-mediated migration and invasion in colorectal cancer cells.26 Additionally, miR-300 expression is downregulated in laryngeal squamous cell carcinoma (LSCC) tissues. Overexpression of miR-300 inhibits cell proliferation and invasion in Hep-2 cells.27 Our results proved that miR-300 overexpression reduced cell proliferation, migration and invasion of GHINK-1 cells. Inhibition of miR-300 reversed the effects of DLEU1 knockdown on cell proliferation, migration and invasion. These findings suggested that miR-300 exhibited tumor suppressive activity, which contributed to the oncogenic activity of DLEU1.
To analyze the molecular mechanism of miR-300, we found that HOXC8 might be a target gene of miR-300. MiR-300 significantly suppressed the expression of HOXC8 in GHINK-1 cells. Interestingly, HOXC8, a member of HOX family, has been proven to play a crucial role in cancer development. HOXC8 represents an interesting tumor suppressor candidate in breast cancer since it can regulate self-renewal, differentiation and transformation of breast cancer stem cells.28 HOXC8 is involved in the tumorigenesis of osteosarcoma with predictive potential and oncogenic effects.29 Significantly increased expression of HOXC8 has been observed in cervical cancer tissues and cell lines. Downregulation of HOXC8 inhibits the proliferation rate of cervical cancer cells, which suggest that HOXC8 may serve as a therapeutic target for cervical cancer.30 Liu et al. reported that HOXC8 was upregulated in NSCLC specimens, and overexpression of HOXC8 significantly promoted the proliferation, anchorage-independent growth and migration of NSCLC cells.31 These results indicated that targeting HOXC8 may be new strategy for the therapeutic intervention. Our study proved that HOXC8 was a target gene of miR-300 and mediated the effects of miR-300 on GHINK-1 cells, suggesting an oncogenic role of HOXC8 in Wilms tumor.
In summary, we demonstrated that DLEU1 expression was upregulated in Wilms tumor tissues and cell lines. Knockdown of DLEU1 inhibited the proliferation, migration and invasion of Wilms tumor cells. Furthermore, DLEU1 executed its oncogenic role in Wilms tumor via regulating miR-300/HOXC8 axis.
4. Materials and methods
4.1 Clinical tissues and cell culture
Thirty pairs of Wilms tumor tissues and adjacent normal tissues were collected from patients who has Wilms tumor underwent surgery at Xi'an Children's Hospital (Xi'an, China) from July 2016 to June 2017. All samples were removed and immediately frozen in liquid nitrogen and stored at −80 °C until use. All experiments were performed in compliance with the Ethics Committee of Xi'an Children's Hospital and approved by the Ethics Committee of Xi'an Children's Hospital. Informed consent was obtained for any experimentation with human subjects.A human immortalized normal kidney cell line (HK2; Shanghai Model Cell Bank, Shanghai, China) and Wilms' Tumor cell lines including GHINK-1 (RIKEN, Wako, Japan), SK-NEP-1 and G401 (Shanghai Model Cell Bank) were cultured in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (Invitrogen). All cells were maintained in an incubator with 5% CO2 at 37 °C.
4.2 Cell transfection
Small interfering RNA (siRNA) targeting DLEU1 (si-DLEU1) and nonspecific negative control siRNA (si-NC) were synthesized (Biotend, Shanghai, China). MiR-300 mimics, miR-300 inhibitor and non-targeting scramble controls were purchased from Genechem (Shanghai, China). Homeobox C8 (HOXC8) coding sequence was inserted into pcDNA3.1 vector to construct HOXC8-overexpressing plasmid pcDNA3.1-HOXC8. Cell transfection was performed using Lipofectamine 3000 Reagent (Invitrogen) according to the manufacturer's protocols.4.3 Cell proliferation assay
Cell proliferation was evaluated by MTT assay. The cells (5 × 103 cell per well) with different treatments were cultured for 0, 24, 48, or 72 h, 20 μl MTT were added into each well and continued to culture for another 4 h. After that, 150 μl DMSO was added to the cells, and then the plates were placed on the oscillator for 10 min to dissolve the crystal. The optical density (OD) values were detected and recorded with a microplate reader (Bio-Tek, Winooski, VT, USA) at 490 nm wavelength.4.4 Cell migration and invasion assays
GHINK-1 cells (5 × 104 cells) in serum-free culture medium were placed in the upper chamber of Transwell chambers (Corning Inc., Corning, NY, USA) for the transwell assay. For the determination of cell invasive ability, the chambers were coated with Matrigel (Sigma). The lower chamber was added with normal medium containing 10% FBS. After 24 h incubation, the cells remained on the upper side of the filters were removed with a cotton swab. Cells moved to the lower side of the filters were fixed using methanol and stained with 0.05% crystal violet. Five random pictures were taken, and the cell numbers were counted under microscope.4.5 qRT-PCR
TRIzol reagent (Invitrogen) was used for the extraction of the total RNA from tissue samples or culture cells. Then the total RNA (500 ng) was transcribed to cDNA using the PrimeScriptRT Kit (TaKaRa, Dalian, China). The relative expression levels of DLEU1 and HOXC8 were evaluated by qRT-PCR using STBR Premix Ex Taq (TakaRa) according to the instructions. The relative expression levels of miR-300 were measured using All-in-One miRNA qRT-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA). The primer sequences were listed as following: DLEU1 F 5′-TCA GAA TGC CGA CTC TAT GCT-3′; R 5′-GGT GAG GAC AGA GTT AAA CGC-3′; HOXC8 F 5′-GGC CAT AGA GAT TAG GGG TTC-3′; R 5′-GGG GCT CAT GAC CTA AGC TA-3′; GAPDH F 5′-AAT GGG CAG CCG TTA GGA AA-3′, R 5′-TGA AGG GGT CAT TGA TGG CA-3′; MiR-300 F 5′-TAT ACA AGG GCA GAC TCT CTC T-3′; R 5′-GTG CAG GTT CCG AGG T-3′; U6 F 5′-CTC GCT TCG GCA GCA CAT ATA CT-3′, R 5′-ACG CTT CAC GAA TTT GCG TGT C-3′.4.6 Western blot
GHINK-1 cells were harvested and lysed with RIPA lysis buffer containing 1% v/v PMSF (Beyotime, Haimen, China). A total of 40 μg proteins were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and then the separated proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Thermo Fisher Scientific, Waltham, MA, USA). The membranes were then blocked in 5% w/v non-fat milk solution at room temperature for 1 h, followed by an incubation with a specific primary antibody against HOXC8 (PA5-27979; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000; Invitrogen) or β-actin (PA5-85490; 1
3000; Invitrogen) or β-actin (PA5-85490; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000; Invitrogen) overnight at 4 °C. Then the membranes were incubated with HRP-conjugated secondary antibody (A32740; 1
5000; Invitrogen) overnight at 4 °C. Then the membranes were incubated with HRP-conjugated secondary antibody (A32740; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2500; Invitrogen) at room temperature for 1 h. Immune complexes were visualized using the enhanced chemiluminescence (ECL) system (Thermo).
2500; Invitrogen) at room temperature for 1 h. Immune complexes were visualized using the enhanced chemiluminescence (ECL) system (Thermo).
4.7 Target prediction
LncRNATargets (http://starbase.sysu.edu.cn) was used to predict target miRNAs of DLEU1. TargetScan (http://www.targetscan.org) was applied for the prediction of the target genes of miR-300.4.8 Luciferase reporter assay
According to the predicted results from bioinformatics software, the wide-type sequences of DLEU1 (DLEU1-WT) and the HOXC8 3′UTRs containing the predicted miR-300 binding sites were respectively inserted into the psiCHECK vector (Promega Corporation, Madison, WI, USA) to generate psiCHECK-DLEU1-WT and psiCHECK-HOXC8-WT plasmids. Meanwhile, the mutant type of DLEU1 (DLEU1-MUT) or HOXC8 (HOXC8-MUT) was inserted into psiCHECK vector to construct psiCHECK-DLEU1-MUT and psiCHECK-HOXC8-MUT plasmids. GHINK-1 cells (2.5 × 105 cells per well) were cultured in a 24-well plate 24 h prior to transfection. The psiCHECK-DLEU1-WT/psiCHECK-DLEU1-MUT or psiCHECK-HOXC8-WT/psiCHECK-HOXC8-MUT and miR-300 mimics/control mimics were used for co-transfection of GHINK-1 cells using Lipofectamine 2000 (Invitrogen). After 48 h, luciferase activities were determined with a dual-luciferase assay kit (Promega) following the manufacturer's protocols.4.9 Statistical analysis
All experiments were repeated for three times, and the data were presented as mean ± standard deviation. The analyses of the data were performed by SPSS 19.0 software (IBM, Chicago, IL, USA) using unpaired two-tailed Student's t-test or one-way analysis of variance (ANOVA). A value of p < 0.05 was considered as statistically significant.Conflicts of interest
There are no conflicts to declare.References
- D. M. Green, G. J. D'Angio, J. B. Beckwith, N. E. Breslow, P. E. Grundy, M. L. Ritchey and P. R. M. Thomas, Wilms tumor, Ca-Cancer J. Clin., 1995, 46, 46–63 CrossRef PubMed.
- A. D. Friedman, Wilms tumor, Pediatr. Rev., 2013, 34, 328–330 CrossRef ; discussion 330.
- N. E. Breslow, J. B. Beckwith, E. J. Perlman and A. E. Reeve, Age distributions, birth weights, nephrogenic rests, and heterogeneity in the pathogenesis of Wilms tumor, Pediatr. Blood Cancer, 2010, 47, 260–267 CrossRef PubMed.
- J. S. Dome, G. Norbert, J. I. Geller, C. V. Fernandez, E. A. Mullen, S. Filippo, H. E. Marry and P. J. Kathy, Advances in Wilms Tumor Treatment and Biology: Progress Through International Collaboration, J. Clin. Oncol., 2011, 33, 2999–3007 CrossRef PubMed.
- K. P. Davenport, F. C. Blanco and A. D. Sandler, Pediatric malignancies: neuroblastoma, Wilm's tumor, hepatoblastoma, rhabdomyosarcoma, and sacroccygeal teratoma, Surg. Clin. N. Am., 2012, 92, 745–767 CrossRef PubMed.
- M. Wang, L. Zhou, F. Yu, Y. Zhang, P. Li and K. Wang, The functional roles of exosomal long non-coding RNAs in cancer, Cell. Mol. Life Sci., 2019, 76, 2059–2076 CrossRef CAS PubMed.
- I. M. Dykes and C. Emanueli, Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA, Genom. Proteom. Bioinform., 2017, 15, 177–186 CrossRef PubMed.
- X. Chen, Y. Sun, R. Cai, G. Wang, X. Shu and W. Pang, Long noncoding RNA: multiple players in gene expression, BMB Rep., 2018, 51, 280–289 CrossRef CAS PubMed.
- H. Liu, J. Luo, S. Luan, C. He and Z. Li, Long non-coding RNAs involved in cancer metabolic reprogramming, Cell. Mol. Life Sci., 2019, 76, 495–504 CrossRef CAS PubMed.
- J. R. Prensner and A. M. Chinnaiyan, The emergence of lncRNAs in cancer biology, Cancer Discovery, 2011, 1, 391 CrossRef CAS PubMed.
- L. Chen, E. E. Dzakah and G. Shan, Targetable long non-coding RNAs in cancer treatments, Cancer Lett., 2018, 418, 119–124 CrossRef CAS PubMed.
- S. Zhu, W. Fu, L. Zhang, K. Fu, J. Hu, W. Jia and G. Liu, LINC00473 antagonizes the tumour suppressor miR-195 to mediate the pathogenesis of Wilms tumour via IKKα, Cell Proliferation, 2018, 51, 381–392 Search PubMed.
- K. R. Zhu, Q. F. Sun and Y. Q. Zhang, Long non-coding RNA LINP1 induces tumorigenesis of Wilms' tumor by affecting Wnt/beta-catenin signaling pathway, Eur. Rev. Med. Pharmacol. Sci., 2019, 23, 5691–5698 Search PubMed.
- S. Zhang, Y. Guan, X. Liu, M. Ju and Q. Zhang, Long non-coding RNA DLEU1 exerts an oncogenic function in non-small cell lung cancer, Biomed. Pharmacother., 2019, 109, 985–990 CrossRef CAS PubMed.
- T. Liu, Z. Han, H. Li, Y. Zhu, Z. Sun and A. Zhu, LncRNA DLEU1 contributes to colorectal cancer progression via activation of KPNA3, Mol. Cancer, 2018, 17, 118, DOI:10.1186/s12943-018-0873-2.
- C. Liu, X. Tian, J. Zhang and L. Jiang, Long Non-coding RNA DLEU1 Promotes Proliferation and Invasion by Interacting With miR-381 and Enhancing HOXA13 Expression in Cervical Cancer, Front. Genet., 2018, 9, 629–651 CrossRef CAS PubMed.
- W. Shao, Y. Li, F. Chen, H. Jia, J. Jia and Y. Fu, Long non-coding RNA DLEU1 contributes to the development of endometrial cancer by sponging miR-490 to regulate SP1 expression, Pharmazie, 2018, 73, 379–385 CAS.
- L. L. Wang, K. X. Sun, D. D. Wu, Y. L. Xiu, X. Chen, S. Chen, Z. H. Zong, X. B. Sang, Y. Liu and Y. Zhao, DLEU1 contributes to ovarian carcinoma tumourigenesis and development by interacting with miR-490-3p and altering CDK1 expression, J. Cell. Mol. Med., 2017, 21, 3055–3065 CrossRef CAS PubMed.
- B. Liu, J. Li and M. J. Cairns, Identifying miRNAs, targets and functions, Briefings Bioinf., 2014, 15, 1–19 CrossRef CAS PubMed.
- S. P. Kabekkodu, V. Shukla, V. K. Varghese, J. D. Souza, S. Chakrabarty and K. Satyamoorthy, Clustered miRNAs and their role in biological functions and diseases, Biol. Rev., 2018, 93, 1955–1986 CrossRef PubMed.
- Y. Du, L. Wang, S. Chen, Y. Liu and Y. Zhao, lncRNA DLEU1 contributes to tumorigenesis and development of endometrial carcinoma by targeting mTOR, Mol. Carcinog., 2018, 57, 1191–1200 CrossRef CAS PubMed.
- W. Zhang, S. Liu, K. Liu and Y. Liu, Long non-coding RNA deleted in lymphocytic leukaemia 1 promotes hepatocellular carcinoma progression by sponging miR-133a to regulate IGF-1R expression, J. Cell. Mol. Med., 2019, 23, 5154–5164 CrossRef CAS PubMed.
- X. Chen, C. Zhang and X. Wang, Long noncoding RNA DLEU1 aggravates osteosarcoma carcinogenesis via regulating the miR-671-5p/DDX5 axis, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 3322–3328 CrossRef CAS.
- S. Lee, W. Luo, T. Shah, C. Yin, T. O'Connell, T. H. Chung, S. L. Perkins, R. R. Miles, J. Ayello, E. Morris, L. Harrison, C. van de Ven and M. S. Cairo, The effects of DLEU1 gene expression in Burkitt lymphoma (BL): potential mechanism of chemoimmunotherapy resistance in BL, Oncotarget, 2017, 8, 27839–27853 Search PubMed.
- J. D. Liu, Q. Xin, C. S. Tao, P. F. Sun, P. Xu, B. Wu, L. Qu and S. Z. Li, Serum miR-300 as a diagnostic and prognostic biomarker in osteosarcoma, Oncol. Lett., 2016, 12, 3912–3918 CrossRef CAS PubMed.
- L. Wang and P. Yu, miR-300 promotes proliferation and EMT-mediated colorectal cancer migration and invasion by targeting p53, Oncol. Rep., 2016, 36, 3225–3232 CrossRef CAS PubMed.
- W. Ge, C. Han, J. Wang and Y. Zhang, MiR-300 suppresses laryngeal squamous cell carcinoma proliferation and metastasis by targeting ROS1, Am. J. Transl. Res., 2016, 8, 3903–3911 CAS.
- M. Shah, R. Cardenas, B. Wang, J. Persson, N. P. Mongan, A. Grabowska and C. Allegrucci, HOXC8 regulates self-renewal, differentiation and transformation of breast cancer stem cells, Mol. Cancer, 2017, 16, 38–77 CrossRef PubMed.
- L. Cheng, X. Wei, K. Zhao, F. Wu, W. Lu, S. Tong and G. Yu, The predictive potential and oncogenic effects of HOXC8 expression on osteosarcoma, Tumor Biol., 2016, 37, 14961–14967 CrossRef CAS PubMed.
- Y. Huang, L. Chen and A. Guo, Upregulated expression of HOXC8 is associated with poor prognosis of cervical cancer, Oncol. Lett., 2018, 15, 7291–7296 Search PubMed.
- H. Liu, M. Zhang, S. Xu, J. Zhang, J. Zou, C. Yang, Y. Zhang, C. Gong, Y. Kai and Y. Li, HOXC8 promotes proliferation and migration through transcriptional up-regulation of TGFβ1 in non-small cell lung cancer, Oncogenesis, 2018, 7, 1–15 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra07215b |
| This journal is © The Royal Society of Chemistry 2019 |