Open Access Article
Open Access ArticleFour rare structurally characterized hetero-pentanuclear [Zn4Ln] bis(salamo)-type complexes: syntheses, crystal structures and spectroscopic properties†
Lu-Mei Pu*a,
Lan Wangb,
Xiao-Yan Lib,
Yin-Xia Sunb,
Quan-Peng Kangb,
Hai-Tao Longa,
Wei-Bing Xu a and
Wen-Kui Dong
a and
Wen-Kui Dong *b
*b
aCollege of Science, Gansu Agricultural University, Lanzhou, Gansu 730070, P. R. China. E-mail: pulm@gsau.edu.cn
bCollege of Chemical and Biological Engineering, Lanzhou Jiaotong University, Lanzhou, Gansu 730070, P. R. China. E-mail: dongwk@126.com
First published on 15th November 2019
Abstract
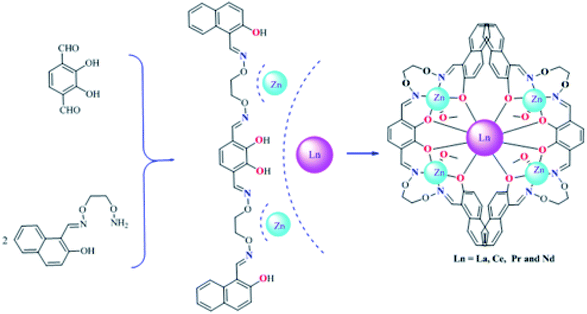
Four new hetero-pentanuclear 3d–4f complexes [Zn4(L)2La(NO3)2(OEt)(H2O)] (1), [Zn4(L)2Ce(NO3)2(OMe)(MeOH)] (2), [Zn4(L)2Pr(NO3)2(OEt)(EtOH)] (3) and [Zn4(L)2Nd(NO3)2(OMe)(MeOH)] (4) were synthesized by the reactions of a newly synthesized octadentate bis(salamo)-based tetraoxime ligand (H4L) with Zn(OAc)2·2H2O and Ln(NO3)3·6H2O (Ln = La, Ce, Pr and Nd), respectively, and characterized via elemental analyses, FT-IR, UV-Vis spectroscopy and single crystal X-ray crystallography. The X-ray crystallographic investigation revealed that all ZnII ions were located in N2O3 coordination spheres, and possessed a trigonal bipyramid coordination environment. The LnIII ion lay in an O8 coordination sphere, and adopted a distorted square antiprismatic coordination environment. Furthermore, supramolecular interactions and fluorescence properties were investigated.
1 Introduction
Salen-type ligands and their analogues are very versatile chelating ligands in inorganic and organometallic chemistry.1 Their complexes have considerable intrinsic value due to their wide applications in electrochemistry,2 building supramolecular structures,3 catalysis fields,4 magnetism,5 biological fields6 and so forth.In recent years, a preferable class of salen-type compounds (salamo: (R–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–O–(CH2)n–O–N
N–O–(CH2)n–O–N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–R)) has been reported,7 and the large electronegativity of O atoms is expected to lead to different and novel structures and properties of the resulting complexes. The 3d–4f complexes have attracted much attention due to the visible and near-infrared luminescence produced by lanthanide f–f transitions.8 Luminescence applications of lanthanides are a consequence of their narrow emission bands, large Stokes shifts, negligible environmental influences and relatively long luminescence lifetimes. However, 3d–4f complexes with salamo-like ligands have been rarely reported.9
CH–R)) has been reported,7 and the large electronegativity of O atoms is expected to lead to different and novel structures and properties of the resulting complexes. The 3d–4f complexes have attracted much attention due to the visible and near-infrared luminescence produced by lanthanide f–f transitions.8 Luminescence applications of lanthanides are a consequence of their narrow emission bands, large Stokes shifts, negligible environmental influences and relatively long luminescence lifetimes. However, 3d–4f complexes with salamo-like ligands have been rarely reported.9
Herein, a series of rare heteropentanuclear [Zn4Ln] (Ln = La, Ce, Pr and Nd) complexes containing octadentate bis(salamo)-based tetraoxime ligand H4L were synthesized and structurally characterized. Meanwhile, the luminescence properties of complexes 1–4 were studied.
2 Experimental
2.1. Materials and methods
1,2-Dimethoxybenzene, 1,2-dibromoethane, TMEDA, n-butyllithium, boron tribromide and 2-hydroxy-1-naphthaldehyde (99%) were purchased from Alfa Aesar and used without further purification. Other reagents and solvents were analytical grade reagents from Tianjin Chemical Reagent Factory.Elemental analyses for carbon, hydrogen and nitrogen were obtained using a GmbH VariuoEL V3.00 automatic elemental analysis instrument (Berlin, Germany). LaIII, CeIII, PrIII and NdIII were gained using an IRIS ER/S-WP-1 ICP atomic emission spectrometer (Berlin, Germany). Melting points were obtained via a microscopic melting point apparatus made by Beijing Taike Instrument Company Limited. IR spectra (4000–400 cm−1) were determined via a Vertex 70 FT-IR spectrophotometer (Bruker, Billerica, MA, USA), with samples prepared as KBr pellets. UV-Vis absorption spectra were determined using a Shimadzu UV-3900 spectrometer (Shimadzu, Japan). 1H NMR spectra were determined via German Bruker AVANCE DRX-400/600 spectroscopy. X-ray single crystal structure determinations for complexes 1, 2, 3 and 4 were carried out on a Bruker APEX-II CCD diffractometer. Fluorescence spectra were recorded on an F-7000 FL spectrophotometer. Near infrared (NIR) spectra were determined through PTI QM4 spectrofluorometer with a PTI QM4 Near infrared InGaAs detector.
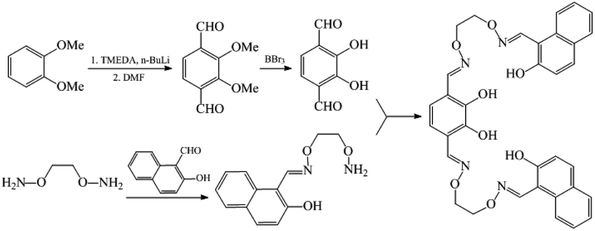
2.2. Synthesis of the H4L
The reaction steps of the ligand (H4L) can be seen from Scheme 1. 1,2-Bis(aminooxy)ethane, 2,3-dihydroxybenzene-1,4-dicarbaldehyde and 2-[O-(1-ethyloxyamide)]oxime-2-naphthol were prepared according to analogous methods reported earlier.10An ethanol solution (10 mL) of 2,3-dihydroxybenzene-1,4-dicarbaldehyde (166.2 mg, 1.0 mmol) was added to an ethanol solution (20 mL) of 2-[O-(1-ethyloxyamide)]oxime-2-naphthol (492.6 mg, 2 mmol). The mixed solution was stirred at 55 °C for 8 h, cooling to room temperature, the precipitate was filtered and washed with n-hexane to obtain a yellow powder. Yield: 87%. mp.: 198–200 °C. Anal. calc. for C34H30N4O8: C, 65.59; H, 4.86; N, 9.00%. Found: C, 65.65; H, 4.94; N, 8.92%. 1H NMR (400 MHz, CDCl3) δ 10.82 (s, 2H), 9.70 (s, 2H), 9.17 (s, 2H), 8.25 (d, J = 2.5 Hz, 2H), 7.96 (d, J = 8.7 Hz, 2H), 7.80–7.72 (m, 4H), 7.50 (t, J = 7.7 Hz, 2H), 7.35 (t, J = 7.5 Hz, 2H), 7.20 (d, J = 9.0 Hz, 2H), 6.75 (s, 2H), 4.56 (s, 8H).
2.3. General procedure for the preparation of complexes 1–4
The synthesis methods of complexes 2–4 are similar to that of complex 1 (Scheme 2). An ethanol solution (3 mL) of Zn(OAc)2·2H2O (13.155 mg, 0.065 mmol) was added to a chloroform solution (5 mL) of H4L (18.675 mg, 0.03 mmol) under constant magnetic stirring, and an ethanol solution (3 mL) of La(NO3)3·6H2O (4.33 mg, 0.015 mmol) was then added. The mixed solution was stirred for 15 minutes at room temperature and then filtered off, and the filtrate was transferred to a cillin bottle. Sealed the opening of the bottle with tinfoil and let it stand for two weeks, some block-like crystals suitable for X-ray diffraction were formed.Complex 1, yellow block-like crystals. Yield: 52%. Elemental analysis: anal. calc. for [Zn4(L)2La(NO3)2(OEt)(H2O)] (C70H59LaZn4N10O24) (%): C, 46.08; H, 3.26; N, 7.68; Zn, 14.33; La, 7.61. Found (%): C, 46.19; H, 3.38; N, 7.53; Zn, 14.41; La, 7.48.
Complex 2, yellow block-like crystals. Yield: 62%. Elemental analysis: anal. calc. for [Zn4(L)2Ce(NO3)2(OMe)(MeOH)] (C70H59CeZn4N10O24) (%): C, 46.05; H, 3.26; N, 7.67; Zn, 14.32; Ce, 7.67. Found (%): C, 46.12; H, 3.37; N, 7.56; Zn, 14.39; Ce, 7.81.
Complex 3, yellow block-like crystals. Yield: 69%. Elemental analysis: anal. calc. for [Zn4(L)2Pr(NO3)2(OEt)(EtOH)] (C72H63PrZn4N10O24) (%): C, 46.62; H, 3.42; N, 7.55; Zn, 14.10; Pr, 7.60. Found (%): C, 46.79; H, 3.48; N, 7.50; Zn, 14.15; Pr, 7.68.
Complex 4, yellow block-like crystals. Yield: 64%. Elemental analysis: anal. calc. for [Zn4(L)2Nd(NO3)2(OMe)(MeOH)] (C70H59NdZn4N10O24) (%): C, 45.94; H, 3.25; N, 7.65; Zn, 14.29; Nd, 7.88. Found (%): C, 46.09; H, 3.38; N, 7.53; Zn, 14.38; Nd, 7.96.
2.4. X-ray crystallographic analysis
Crystal data for complexes 1–4 were collected on a Bruker APEX-II CCD area detector with Mo Kα radiation (λ = 0.71073 Å) at 296(2), 173(2), 173(2) and 173(2) K. respectively. Reflection data were corrected for LP factors semi-empirical absorption were using SADABS. The single crystal structures were solved by the direct methods (SHELXS-2016).11a All hydrogen atoms were included at the calculated positions, and their positions were refined by a riding model. All non-hydrogen atoms were refined anisotropically using a full-matrix least-squares procedure on F2 with SHELXL-2016.11b Crystallographic data and the structure refinements for complexes 1–4 are presented in Table 3.3 Results and discussion
3.1. IR spectra
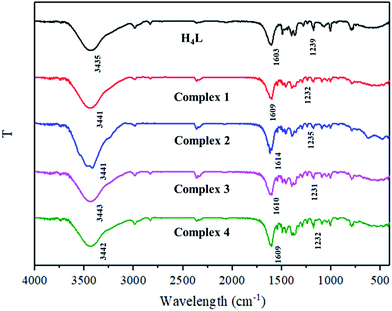
IR spectra of H4L and its corresponding complexes 1–4 displayed various bands in the 4000–400 cm−1 region (Fig. 1).In the infrared spectrum of H4L, a typical C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching band appeared at ca. 1603 cm−1, and C
N stretching band appeared at ca. 1603 cm−1, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching bands of complexes 1–4 were observed at 1609–1614 cm−1, indicating that H4L has coordinated with ZnII ions.12 In addition, the free ligand H4L exhibited a typical Ar–O stretching frequency at ca. 1239 cm−1, while the Ar–O stretching frequencies in complexes 1–4 were observed at ca. 1232, 1235, 1231 and 1232 cm−1, which are shifted to lower frequencies, indicating that the Zn–O or Ln–O bond is formed between the oxygen atoms of phenolic group and the metal ions.13 Meanwhile, the hydroxyl stretching band of H4L was observed at ca. 3435 cm−1 that belongs to the phenolic O–H groups. These absorption bands in complexes 1–4 were observed at ca. 3441–3443 cm−1, indicating the existence of coordinated water, methanol or ethanol molecules.14
N stretching bands of complexes 1–4 were observed at 1609–1614 cm−1, indicating that H4L has coordinated with ZnII ions.12 In addition, the free ligand H4L exhibited a typical Ar–O stretching frequency at ca. 1239 cm−1, while the Ar–O stretching frequencies in complexes 1–4 were observed at ca. 1232, 1235, 1231 and 1232 cm−1, which are shifted to lower frequencies, indicating that the Zn–O or Ln–O bond is formed between the oxygen atoms of phenolic group and the metal ions.13 Meanwhile, the hydroxyl stretching band of H4L was observed at ca. 3435 cm−1 that belongs to the phenolic O–H groups. These absorption bands in complexes 1–4 were observed at ca. 3441–3443 cm−1, indicating the existence of coordinated water, methanol or ethanol molecules.14
3.2. UV-Vis spectra
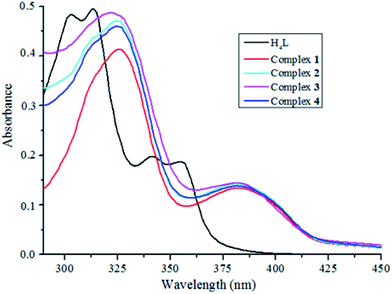
The UV-Vis absorption spectra of H4L and its complexes 1–4 in CHCl3/CH3CH2OH solution (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) are shown in Fig. 2. The absorption spectrum of H4L (1.0 × 10−5 M) showed four relatively strong absorption peaks at ca. 302, 313, 341 and 355 nm, the former two peaks can be assigned to the π–π* transitions of the naphthalene rings. The later two absorption peaks can be assigned to the intra-ligand π–π* transition of the oxime group.15 Compared with the absorption peaks of the free ligand H4L, the first absorption peaks were observed at 326, 325, 321 and 325 nm in complexes 1–4, respectively. These peaks are bathochromically shifted, indicating coordination of the (L)4− moieties with metal(II/III) ions. Meanwhile, the new peaks emerged at ca. 382 nm in complexes 1–4, respectively, which belong to the n–π* charge transfer transitions from the lone-pair electrons of the N atoms of C
1) are shown in Fig. 2. The absorption spectrum of H4L (1.0 × 10−5 M) showed four relatively strong absorption peaks at ca. 302, 313, 341 and 355 nm, the former two peaks can be assigned to the π–π* transitions of the naphthalene rings. The later two absorption peaks can be assigned to the intra-ligand π–π* transition of the oxime group.15 Compared with the absorption peaks of the free ligand H4L, the first absorption peaks were observed at 326, 325, 321 and 325 nm in complexes 1–4, respectively. These peaks are bathochromically shifted, indicating coordination of the (L)4− moieties with metal(II/III) ions. Meanwhile, the new peaks emerged at ca. 382 nm in complexes 1–4, respectively, which belong to the n–π* charge transfer transitions from the lone-pair electrons of the N atoms of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N groups to benzene rings.16
N groups to benzene rings.16
3.3. Crystal structure descriptions
X-ray crystallographic analysis revealed the crystal structures of complexes 1–4. Selected bond lengths and angles are given in Table 1| Complex | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Empirical formula | C70H59LaZn4N10O24 | C70H59CeZn4N10O24 | C72H63PrZn4N10O24 | C70H59NdZn4N10O24 |
| Formula weight | 1824.66 | 1824.86 | 1854.71 | 1829.99 |
| T (K) | 296(2) | 173(2) | 173(2) | 173(2) |
| Wavelength (Å) | 0.71073 | 0.71073 | 0.71073 | 0.71073 |
| Crystal system | Monoclinic | Monoclinic | Monoclinic | Monoclinic |
| Space group | C2/c | C2/c | C2/c | C2/c |
| a (Å) | 23.8927(11) | 16.9226(7) | 16.7809(7) | 16.9226(7) |
| b (Å) | 15.5212(7) | 23.3361(10) | 23.4499(7) | 23.3361(10) |
| c (Å) | 45.813(2) | 24.4208(13) | 24.1484(11) | 24.4208(13) |
| α (°) | 90 | 90 | 90 | 90 |
| β (°) | 97.7300(10) | 109.4290(10) | 108.736(5) | 109.4290(10) |
| γ (°) | 90 | 90 | 90 | 90 |
| V (Å3) | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 835.2(13) 835.2(13) |
9094.8(7) | 8999.1(7) | 9094.8(7) |
| Z | 8 | 4 | 4 | 4 |
| Dcalc. (g cm−3) | 1.440 | 1.333 | 1.369 | 1.336 |
| Absorption coefficient (mm−1) | 1.694 | 1.599 | 1.653 | 1.669 |
| F (000) | 7344 | 3672 | 3744 | 3684 |
| Crystal size (mm) | 0.270 × 0.250 × 0.220 | 0.220 × 0.190 × 0.160 | 0.220 × 0.200 × 0.180 | 0.220 × 0.190 × 0.160 |
| θ Range (°) | 2.044–25.010 | 2.485–25.008 | 1.548–26.000 | 2.485–25.008 |
| Index ranges | −25 ≤ h ≤ 28 | −20 ≤ h ≤ 20 | −20 ≤ h ≤ 20 | −20 ≤ h ≤ 17 |
| −18 ≤ k ≤ 18 | −22 ≤ k ≤ 27 | −27 ≤ k ≤ 28 | −27 ≤ k ≤ 27 | |
| −51 ≤ l ≤ 54 | −29 ≤ l ≤ 29 | −29 ≤ l ≤ 29 | −29 ≤ l ≤ 28 | |
| Reflections collected/unique | 59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 539/14 539/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 818 [Rint = 0.0347] 818 [Rint = 0.0347] |
32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 276/7999 [Rint = 0.0371] 276/7999 [Rint = 0.0371] |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 902/8832 [Rint = 0.0179] 902/8832 [Rint = 0.0179] |
32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 970/8002 [Rint = 0.0391] 970/8002 [Rint = 0.0391] |
| Completeness to θ | 99.8% (θ = 25.010) | 99.8% (θ = 25.008) | 99.7% (θ = 25.242) | 99.8% (θ = 25.008) |
| Data/restraints/parameters | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 818/0/1009 818/0/1009 |
7999/0/493 | 8832/5/496 | 8002/6/487 |
| GOF | 0.963 | 1.049 | 1.043 | 1.030 |
| Final R1, wR2 indices | 0.0399, 0.1006 | 0.0363, 0.1003 | 0.0315, 0.0971 | 0.0377, 0.0974 |
| R1, wR2 indices (all data) | 0.0454, 0.1044 | 0.0448, 0.1059 | 0.0412, 0.1007 | 0.0465, 0.1020 |
| Largest diff. peak and hole (e Å−3) | 1.492 and −1.009 | 1.563 and −0.981 | 1.048 and −0.805 | 1.814 and −0.783 |
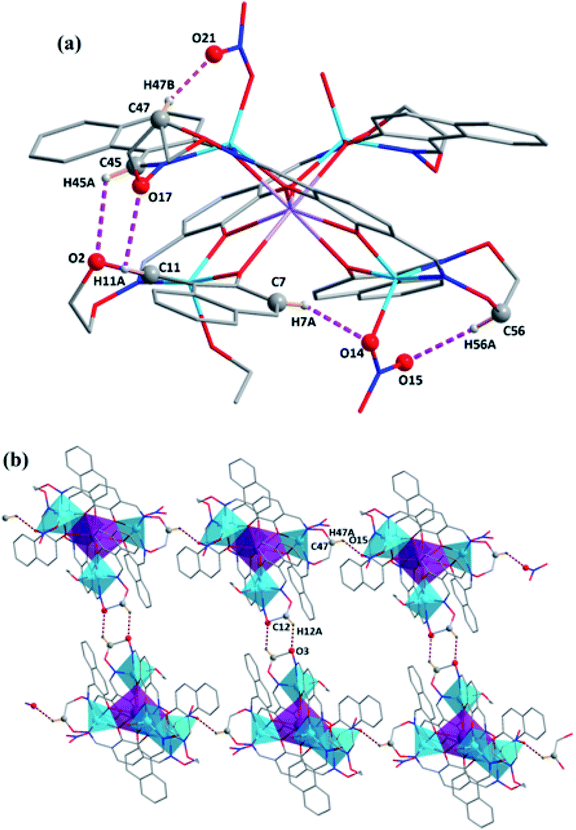
The intramolecular and intermolecular hydrogen bonds for complex 3 are presented in Table 2. Each molecule formed five intramolecular hydrogen bonds (C23–H23A⋯O12 and C8–H8A⋯O10) as shown in Fig. 4.19 Meanwhile, a self-assembled infinite 2D supramolecular structure was formed by C12–H12A⋯O3 and C47–H47A⋯O15 hydrogen bond interactions20 (Fig. 4).
| Complex 1 | |||||
|---|---|---|---|---|---|
| Bond | Lengths | Bond | Lengths | Bond | Lengths |
| a Symmetry transformations used to generate equivalent atoms: #13/2 − x, −1/2 + y, 3/2 − z; #2−x + 1, y, −z + 1/2; #3−x, y, −z + 1/2; #4−x + 1, y, −z + 1/2. | |||||
| Zn1–O19 | 2.001(3) | Zn1–O20 | 2.085(3) | Zn1–O22 | 2.054(6) |
| Zn2–O1 | 2.051(2) | Zn2–O4 | 1.957(2) | Zn1–O22#1 | 2.019(14) |
| Zn2–O24 | 2.014(3) | Zn3–O5 | 1.975(2) | Zn3–O6 | 1.983(3) |
| Zn3–O7 | 2.081(2) | Zn4–O12 | 1.999(3) | Zn4–O13 | 2.057(3) |
| Zn4–O14 | 2.059(3) | Zn1–N3 | 2.149(3) | Zn1–N4 | 2.048(3) |
| Zn2–N2 | 2.011(3) | Zn2–N6 | 2.108(3) | Zn3–N1 | 2.127(3) |
| Zn3–N5 | 2.019(3) | Zn4–N7 | 2.038(4) | Zn4–N8 | 2.120(3) |
| La1–O5 | 2.491(2) | La1–O12 | 2.497(2) | La1–O19 | 2.500(2) |
| La1–O20 | 2.496(3) | La1–O4 | 2.505(2) | La1–O7 | 2.514(2) |
| La1–O1 | 2.534(3) | La1–O13 | 2.537(3) | ||
| Complex 1 | |||||
|---|---|---|---|---|---|
| Bond | Angles | Bond | Angles | Bond | Angles |
| O19–Zn1–O22 | 109.0(2) | N4–Zn1–O22 | 123.2(2) | O19–Zn1–O20 | 81.62(10) |
| O19–Zn1–O22#1 | 134.0(5) | O19–Zn1–N4 | 124.08(12) | O22#1–Zn1–N4 | 101.9(4) |
| O22#1–Zn1–O20 | 105.3(4) | N4–Zn1–O20 | 83.62(11) | O22–Zn1–O20 | 85.79(17) |
| O19–Zn1–N3 | 86.56(12) | O22#1–Zn1–N3 | 89.3(4) | N4–Zn1–N3 | 95.80(14) |
| O22–Zn1–N3 | 106.58(18) | O20–Zn1–N3 | 165.20(13) | O4–Zn2–N2 | 129.50(12) |
| O4–Zn2–O24 | 109.62(13) | N2–Zn2–O24 | 120.27(14) | O4–Zn2–O1 | 83.74(10) |
| N2–Zn2–O1 | 85.55(11) | O24–Zn2–O1 | 93.95(12) | O4–Zn2–N6 | 89.59(11) |
| N2–Zn2–N6 | 95.24(12) | O24–Zn2–N6 | 92.61(13) | O1–Zn2–N6 | 171.90(12) |
| O5–Zn3–O6 | 113.02(12) | O5–Zn3–N5 | 128.10(12) | O6–Zn3–N5 | 117.44(14) |
| O5–Zn3–O7 | 83.24(10) | O6–Zn3–O7 | 90.74(12) | N5–Zn3–O7 | 84.77(11) |
| O5–Zn3–N1 | 87.25(11) | O6–Zn3–N1 | 97.52(14) | N5–Zn3–N1 | 97.33(13) |
| O7–Zn3–N1 | 169.29(12) | O12–Zn4–N7 | 124.57(13) | O12–Zn4–O13 | 82.80(10) |
| N7–Zn4–O13 | 84.94(12) | O12–Zn4–O14 | 103.94(11) | N7–Zn4–O14 | 130.82(13) |
| O13–Zn4–O14 | 94.37(12) | O12–Zn4–N8 | 87.66(12) | N7–Zn4–N8 | 96.06(14) |
| O13–Zn4–N8 | 169.00(13) | O14–Zn4–N8 | 93.24(14) | ||
| O5–La1–O20 | 91.87(9) | O5–La1–O12 | 129.19(8) | O20–La1–O12 | 126.29(8) |
| O5–La1–O19 | 148.96(9) | O20–La1–O19 | 64.64(8) | O12–La1–O19 | 63.26(8) |
| O5–La1–O4 | 62.34(8) | O20–La1–O4 | 78.16(8) | O12–La1–O4 | 146.66(8) |
| O19–La1–O4 | 126.01(8) | O5–La1–O7 | 65.16(8) | O20–La1–O7 | 89.17(9) |
| O12–La1–O7 | 81.51(8) | O19–La1–O7 | 93.10(8) | O4–La1–O7 | 125.23(8) |
| O5–La1–O1 | 124.73(8) | O20–La1–O1 | 90.06(9) | O12–La1–O1 | 91.02(8) |
| O19–La1–O1 | 77.66(8) | O4–La1–O1 | 64.15(8) | O7–La1–O1 | 170.10(8) |
| O5–La1–O13 | 77.97(8) | O20–La1–O13 | 169.01(8) | O12–La1–O13 | 64.41(8) |
| O19–La1–O13 | 126.35(8) | O4–La1–O13 | 93.23(9) | O7–La1–O13 | 90.31(9) |
| O1–La1–O13 | 92.31(9) | ||||
| Complex 2 | |||||
|---|---|---|---|---|---|
| Bond | Lengths | Bond | Lengths | Bond | Lengths |
| O12–Zn1 | 2.020(3) | O11–Zn1 | 2.043(2) | O10–Zn2 | 2.068(2) |
| O9–Zn2 | 2.026(3) | O4–Zn2 | 1.985(2) | O3–Zn1 | 1.972(2) |
| N4–Zn2 | 2.015(3) | N3–Zn2 | 2.137(3) | N2–Zn1 | 2.110(3) |
| N1–Zn1 | 2.019(3) | ||||
| Ce1–O3 | 2.475(2) | Ce1–O10#2 | 2.475(2) | Ce1–O11#2 | 2.511(2) |
| Ce1–O3#2 | 2.475(2) | Ce1–O4#2 | 2.479(2) | Ce1–O11 | 2.511(2) |
| Ce1–O10 | 2.475(2) | Ce1–O4 | 2.479(2) | ||
| Complex 2 | |||||
|---|---|---|---|---|---|
| Bond | Angles | Bond | Angles | Bond | Angles |
| O3–Zn1–O12 | 113.98(12) | O3–Zn1–N1 | 127.13(12) | O12–Zn1–N1 | 117.85(14) |
| O3–Zn1–O11 | 81.90(10) | O12–Zn1–O11 | 93.03(11) | N1–Zn1–O11 | 85.67(12) |
| O3–Zn1–N2 | 88.94(11) | O12–Zn1–N2 | 95.64(12) | N1–Zn1–N2 | 95.67(13) |
| O11–Zn1–N2 | 169.37(12) | O4–Zn2–N4 | 122.97(12) | O4–Zn2–O9 | 109.03(11) |
| N4–Zn2–O9 | 127.60(13) | O4–Zn2–O10 | 82.84(9) | N4–Zn2–O10 | 84.60(11) |
| O9–Zn2–O10 | 96.52(12) | O4–Zn2–N3 | 87.24(11) | N4–Zn2–N3 | 95.14(13) |
| O9–Zn2–N3 | 93.18(14) | O10–Zn2–N3 | 167.93(12) | ||
| O3–Ce1–O3#2 | 126.22(11) | O3#2–Ce1–O10 | 92.34(8) | O3#2–Ce1–O10#2 | 127.67(8) |
| O3–Ce1–O10 | 127.67(8) | O3–Ce1–O10#2 | 92.34(8) | O10–Ce1–O10#2 | 87.78(12) |
| O3–Ce1–O4#2 | 146.35(8) | O4#2–Ce1–O4 | 130.09(11) | O3–Ce1–O11 | 63.72(8) |
| O3#2–Ce1–O4#2 | 63.21(8) | O3–Ce1–O11#2 | 80.04(8) | O3#2–Ce1–O11 | 80.03(8) |
| O10–Ce1–O4#2 | 78.78(8) | O3#2–Ce1–O11#2 | 63.72(8) | O10–Ce1–O11 | 168.47(8) |
| O10#2–Ce1–O4#2 | 65.57(8) | O10–Ce1–O11#2 | 90.07(8) | O10#2–Ce1–O11 | 90.07(8) |
| O3–Ce1–O4 | 63.21(8) | O10#2–Ce1–O11#2 | 168.48(8) | O4#2–Ce1–O11 | 90.02(8) |
| O3#2–Ce1–O4 | 146.35(8) | O4#2–Ce1–O11#2 | 125.03(8) | O4–Ce1–O11 | 125.03(8) |
| O10–Ce1–O4 | 65.57(8) | O4–Ce1–O11#2 | 90.03(8) | O11#2–Ce1–O11 | 94.20(11) |
| O10#2–Ce1–O4 | 78.78(8) | ||||
| Complex 3 | |||||
|---|---|---|---|---|---|
| Bond | Lengths | Bond | Lengths | Bond | Lengths |
| N1–Zn1 | 2.015(2) | N2–Zn1 | 2.143(3) | N3–Zn2 | 2.110(3) |
| N4–Zn2 | 2.018(3) | O1–Zn1 | 2.0699(19) | O5–Zn2 | 1.9785(19) |
| O8–Zn2 | 2.0372(19) | O12–Zn2 | 2.015(2) | ||
| O1–Pr1 | 2.4646(19) | O8–Pr1 | 2.5097(19) | O5#3–Pr1 | 2.4471(19) |
| O4–Pr1 | 2.4605(19) | O1#3–Pr1 | 2.4646(19) | O8#3–Pr1 | 2.5097(19) |
| O5–Pr1 | 2.4471(19) | O4#3–Pr1 | 2.4605(19) | ||
| Complex 3 | |||||
|---|---|---|---|---|---|
| Bond | Angles | Bond | Angles | Bond | Angles |
| O4–Zn1–N1 | 122.13(9) | O4–Zn1–O9 | 110.45(9) | N1–Zn1–O9 | 127.21(10) |
| O4–Zn1–O1 | 82.56(8) | N1–Zn1–O1 | 85.70(9) | O9–Zn1–O1 | 97.27(9) |
| O4–Zn1–N2 | 86.69(9) | N1–Zn1–N2 | 94.43(10) | O9–Zn1–N2 | 92.74(10) |
| O1–Zn1–N2 | 167.33(9) | O5–Zn2–O12 | 113.73(10) | O5–Zn2–N4 | 126.50(9) |
| O12–Zn2–N4 | 118.79(11) | O5–Zn2–O8 | 81.60(8) | O12–Zn2–O8 | 93.65(9) |
| N4–Zn2–O8 | 85.60(9) | O5–Zn2–N3 | 89.31(9) | O12–Zn2–N3 | 94.94(10) |
| N4–Zn2–N3 | 95.59(10) | O8–Zn2–N3 | 169.44(9) | ||
| O5#3–Pr1–O5 | 125.71(9) | O5#3–Pr1–O1#3 | 128.04(6) | O1–Pr1–O8#3 | 88.83(7) |
| O5#3–Pr1–O4 | 146.19(6) | O5–Pr1–O1#3 | 91.97(6) | O1#3–Pr1–O8#3 | 167.92(6) |
| O5–Pr1–O4 | 63.29(6) | O4–Pr1–O1#3 | 79.09(6) | O5#3–Pr1–O8 | 80.03(6) |
| O5#3–Pr1–O4#3 | 63.29(6) | O4#3–Pr1–O1#3 | 65.99(6) | O5–Pr1–O8 | 63.92(6) |
| O5–Pr1–O4#3 | 146.19(6) | O1–Pr1–O1#3 | 89.04(9) | O4–Pr1–O8 | 125.13(6) |
| O4–Pr1–O4#3 | 130.59(9) | O5#3–Pr1–O8#3 | 63.92(6) | O4#3–Pr1–O8 | 89.19(6) |
| O5#3–Pr1–O1 | 91.97(6) | O5–Pr1–O8#3 | 80.03(6) | O1–Pr1–O8 | 167.93(6) |
| O5–Pr1–O1 | 128.04(6) | O4–Pr1–O8#3 | 89.19(6) | O1#3–Pr1–O8 | 88.83(7) |
| O4–Pr1–O1 | 65.99(6) | O4#3–Pr1–O8#3 | 125.13(6) | O8#3–Pr1–O8 | 95.65(9) |
| O4#3–Pr1–O1 | 79.10(6) | ||||
| Complex 4 | |||||
|---|---|---|---|---|---|
| Bond | Lengths | Bond | Lengths | Bond | Lengths |
| N1–Zn1 | 2.020(3) | N2–Zn1 | 2.110(3) | N3–Zn2 | 2.133(3) |
| N4–Zn2 | 2.017(3) | O3–Zn1 | 1.968(3) | O4–Zn2 | 1.985(2) |
| O9–Zn2 | 2.023(3) | O10–Zn2 | 2.061(3) | O11–Zn1 | 2.046(3) |
| O12–Zn1 | 2.017(3) | ||||
| Nd1–O4 | 2.444(2) | Nd1–O3 | 2.450(2) | Nd1–O11#4 | 2.481(3) |
| Nd1–O4#4 | 2.444(2) | Nd1–O10 | 2.452(2) | Nd1–O11 | 2.481(3) |
| Nd1–O3#4 | 2.450(2) | Nd1–O10#4 | 2.452(2) | ||
| Complex 4 | |||||
|---|---|---|---|---|---|
| Bond | Angles | Bond | Angles | Bond | Angles |
| O3–Zn1–O12 | 113.61(13) | O3–Zn1–N1 | 127.19(13) | O12–Zn1–N1 | 118.26(15) |
| O3–Zn1–O11 | 81.28(10) | O12–Zn1–O11 | 93.77(12) | N1–Zn1–O11 | 86.07(12) |
| O3–Zn1–N2 | 89.29(12) | O12–Zn1–N2 | 95.15(13) | N1–Zn1–N2 | 95.29(14) |
| O11–Zn1–N2 | 169.03(13) | O4–Zn2–N4 | 122.85(13) | O4–Zn2–O9 | 109.21(11) |
| N4–Zn2–O9 | 127.58(13) | O4–Zn2–O10 | 82.07(10) | N4–Zn2–O10 | 85.26(12) |
| O9–Zn2–O10 | 96.81(13) | O4–Zn2–N3 | 87.26(12) | N4–Zn2–N3 | 94.59(14) |
| O9–Zn2–N3 | 93.47(14) | O10–Zn2–N3 | 167.13(12) | ||
| O4–Nd1–O4#4 | 129.52(12) | O4–Nd1–O10#4 | 78.43(9) | O10–Nd1–O11#4 | 90.19(9) |
| O4–Nd1–O3#4 | 145.82(8) | O4#4–Nd1–O10#4 | 65.72(8) | O10#4–Nd1–O11#4 | 167.33(8) |
| O4#4–Nd1–O3#4 | 63.98(8) | O3#4–Nd1–O10#4 | 128.56(8) | O4–Nd1–O11 | 126.22(8) |
| O4–Nd1–O3 | 63.98(8) | O3–Nd1–O10#4 | 91.74(8) | O4#4–Nd1–O11 | 89.50(8) |
| O4#4–Nd1–O3 | 145.82(8) | O10–Nd1–O10#4 | 88.34(12) | O3#4–Nd1–O11 | 79.37(8) |
| O3#4–Nd1–O3 | 125.79(12) | O4–Nd1–O11#4 | 89.50(8) | O3–Nd1–O11 | 64.06(9) |
| O4–Nd1–O10 | 65.72(8) | O4#4–Nd1–O11#4 | 126.21(8) | O10–Nd1–O11 | 167.33(8) |
| O4#4–Nd1–O10 | 78.43(9) | O3#4–Nd1–O11#4 | 64.06(8) | O10#4–Nd1–O11 | 90.20(9) |
| O3#4–Nd1–O10 | 91.73(8) | O3–Nd1–O11#4 | 79.37(8) | O11#4–Nd1–O11 | 93.93(12) |
| O3–Nd1–O10 | 128.56(8) | ||||
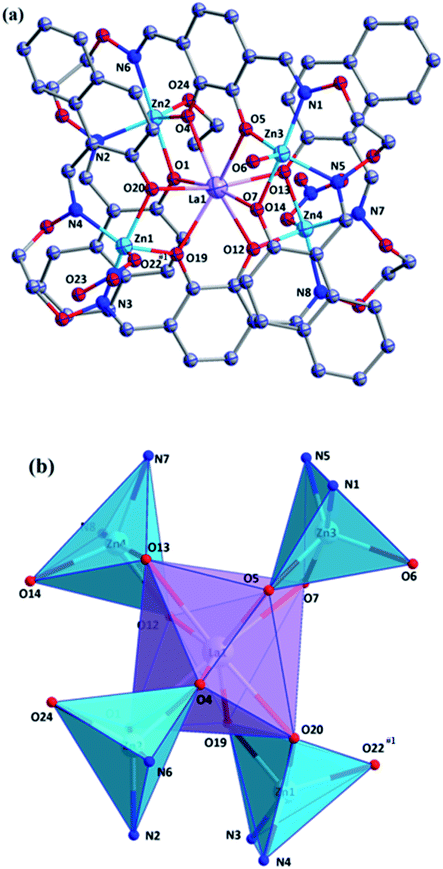
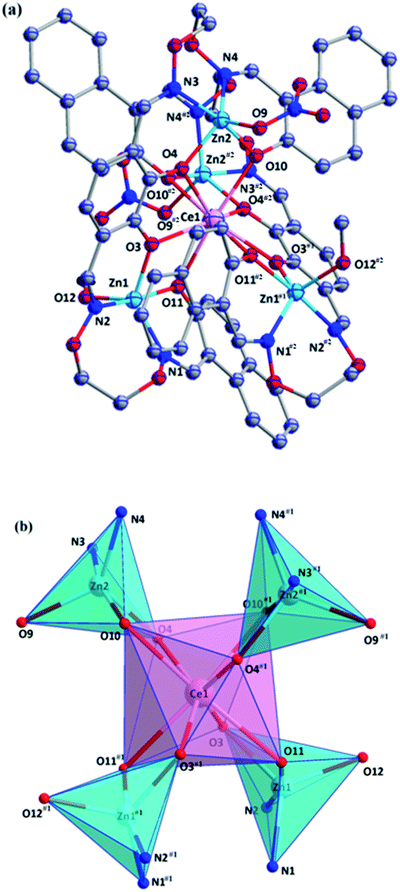
Zn1 and Zn2 ions were located in penta-coordinated spheres and adopted trigonal bipyramid coordination environments (τ1 = 0.704 and τ2 = 0.67).17 The coordination number of CeIII ion is 8, consisting of eight phenolic oxygen atoms from two full deprotonated (L)4− units and adopted a distorted square antiprismatic coordination environment (Fig. 5).18
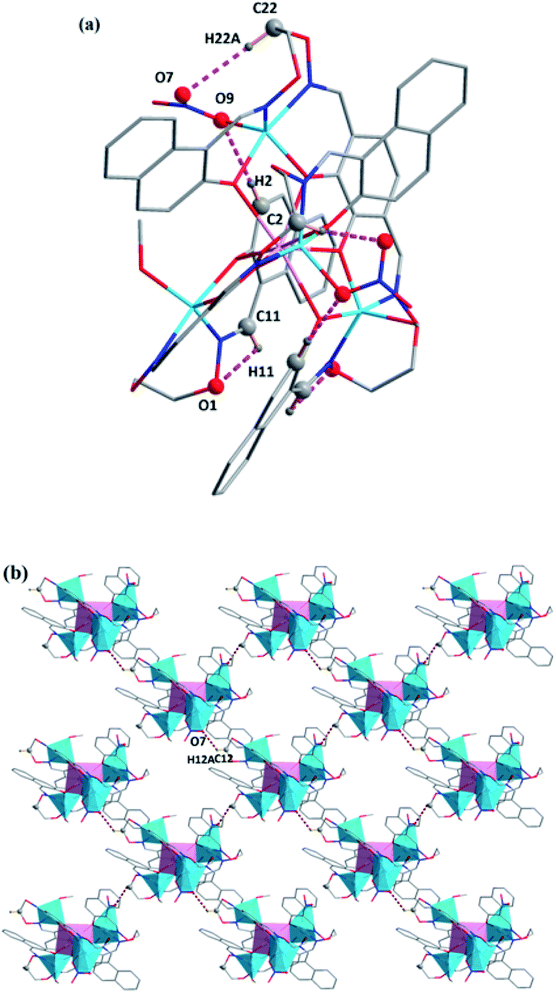
In the crystal structure of complex 2, there were many intramolecular hydrogen bonds (C2–H2⋯O9, C11–H11⋯O1 and C22–H22A⋯O7).19 As shown in Fig. 6. Moreover, the 2D supramolecular structure was formed by C12–H12A⋯O7 hydrogen bonding interactions in complex 2 (ref. 21) (Fig. 6).
The ZnII ions also were located in the N2O2 sites, and four ZnII ions are also penta-coordinated. The ZnII ions (Zn1 and Zn2) adopted trigonal bipyramid coordination environment (τ1 = 0.67 and τ2 = 0.72).17 The PrIII ion was also located in the O8 site that consists of eight phenoxo oxygen atoms, forming a distorted square antiprismatic coordination environment.18
The main interactions in complex 3 are listed in Table 3, four pairs of intramolecular hydrogen bonds (C13–H13B⋯O10, C24–H24⋯O7, C27–H27⋯O9 and C35–H35B⋯O11) were formed.19 Besides, The O10 atom of nitrate group as acceptor formed a hydrogen bond with the donor (C23H23B–) in complex 3, which adopted a 2D supramolecular structure22 (Fig. S3†).
| D–H⋯A | d(D–H) | d(H–A) | d(D–A) | ∠D–X–A | Sum |
|---|---|---|---|---|---|
| Complex 1 | |||||
| C7–H7A⋯O14 | 0.93 | 2.55 | 3.436(5) | 159 | |
| C47–H47B⋯O21 | 0.97 | 1.89 | 2.653(12) | 134 | |
| C56–H56A⋯O15 | 0.97 | 2.54 | 3.507(8) | 174 | |
| C12–H12A⋯O3 | 0.97 | 2.44 | 3.214(5) | 137 | 1 − x, 1 − y, 1 − z |
| C47–H47A⋯O15 | 0.97 | 2.56 | 3.244(8) | 128 | −1/2 + x, 1/2 + y, z |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Complex 2 | |||||
| C2–H2⋯O9 | 0.95 | 2.51 | 3.451(5) | 173 | 1 − x, y, 1/2 − z |
| C22–H22A⋯O7 | 0.99 | 2.51 | 3.490(6) | 172 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Complex 3 | |||||
| C13–H13B⋯O10 | 0.99 | 2.53 | 3.516(4) | 173 | |
| C23–H23B⋯O10 | 0.99 | 2.54 | 3.148(4) | 120 | 1/2 + x, 1/2 + y, z |
| C27–H27⋯O9 | 0.95 | 2.50 | 3.448(4) | 175 | −x, y, 1/2 − z |
| C35–H35B⋯O11 | 0.98 | 2.58 | 3.438(8) | 146 | −x, y, 1/2 − z |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Complex 4 | |||||
| C2–H2A⋯O9 | 0.95 | 2.51 | 3.456(6) | 172 | 1 − x, y, 1/2 − z |
| C6–H6⋯O8 | 0.95 | 2.56 | 3.361(11) | 142 | −1/2 + x, 1/2 − y, −1/2 + z |
| C22–H22A⋯O7 | 0.99 | 2.52 | 3.499(6) | 172 | |
All ZnII ions lay in N2O3 coordination spheres. The Zn1 and Zn2 (Zn1#4 and Zn2#4) ions were all made of the N2O2 cavities and one coordinated nitrate group, which assumed trigonal bipyramid coordination environments (τ1 = 0.69 and τ2 = 0.66).17 The NdIII ion exhibited an O8 coordination sphere, which is made of eight phenoxo donors (O3, O4, O10, O11, O3#4, O4#4, O10#4 and O11#4) from two completely deprotonated (L)4− units, while the central NdIII ion is octa-coordinated with a distorted square antiprismatic coordination environment.18
In complex 4, three pairs of significant intramolecular hydrogen bonds (C2–H2A⋯O9, C11–H11⋯O1 and C22–H22A⋯O7) were formed19 (Fig. S5(a)†). Meanwhile, complex 4 molecules formed a 2D supramolecular structure by intermolecular hydrogen bonds (C6–H6⋯O8 and C12–H12A⋯O7)23 (Fig. S5(b)†).
3.4. Spectroscopic properties
The free ligand H4L and its corresponding complexes 1–4 were excited at 385 nm (λex) respectively (Fig. 7). The emission spectrum of H4L exhibited a broad emission band, and the emission maximum at 454 nm, which can be assigned to the π–π* electronic transitions in the ligand.24 Compared to H4L, the absorption peaks of complexes 1–3 are bathochromically-shifted, which is may originated from the LMCT emission.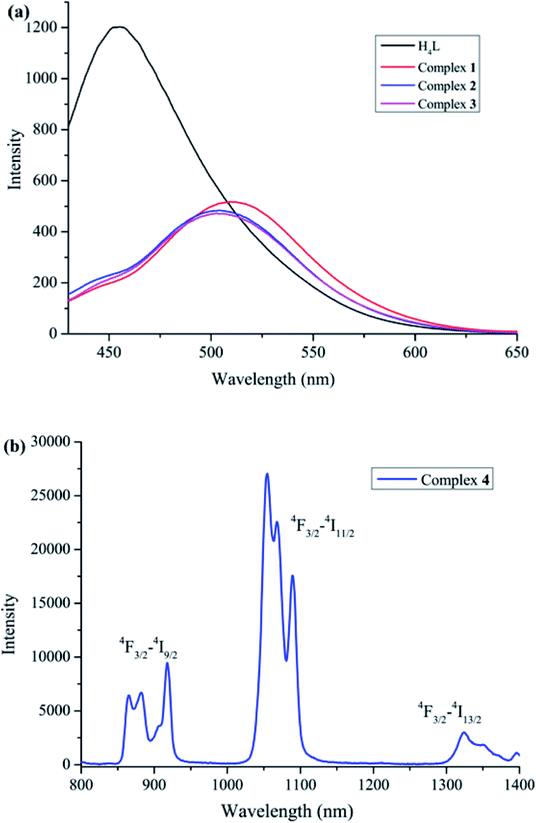 | ||
| Fig. 7 (a) Visible luminescence spectra of complexes 1–3. (b) NIR luminescence spectrum of complex 4. | ||
Due to energy mismatch, complexes 1–3 have no NIR luminescence. The NIR luminescence spectrum of complex 4 excited at 385 nm showed the characteristic emitting peaks at ca. 871, 917, 1055 and 1324 nm (Fig. 7(b)). These emission peaks are typical peaks of NdIII ions, and correspond to 4F3/2 → 4I9/2, 4F3/2 → 4I11/2 and 4F3/2 → 4I13/2 transitions.25 The ligand (L)4− units could serve as sensitizing agent for NdIII luminescence in the NIR region.
4 Conclusions
In this work, four rare hetero-pentanuclear 3d–4f complexes of a bis(salamo)-type ligand (H4L) have been synthesized and structurally characterized. In complexes 1–4, all four ZnII ions presented N2O3 coordination spheres. The LnIII ion exhibited an O8 coordination sphere, and assumed a distorted square antiprismatic coordination environment. In a conclusion, the studies demonstrated that incorporation of salamo-like ligand was an optimistic approach to build ZnII–LnIII complexes which can display excellent spectroscopic resting with the lanthanide ions used.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21761018), Science and Technology Program of Gansu Province (18YF1GA054) and the Program for Excellent Team of Scientific Research in Lanzhou Jiaotong University (201706), three of which are gratefully acknowledged.References
- (a) Y. X. Sun, L. Xu, T. H. Zhao, S. H. Liu, G. H. Liu and X. T. Dong, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2013, 43, 509–513 CrossRef CAS; (b) Y. X. Sun, S. T. Zhang, Z. L. Ren, X. Y. Dong and L. Wang, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2013, 43, 995–1000 CrossRef CAS; (c) Y. X. Sun and X. H. Gao, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2011, 41, 973–978 CrossRef CAS; (d) Y. X. Sun, L. Wang, X. Y. Dong, Z. L. Ren and W. S. Meng, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2013, 43, 599–603 CrossRef CAS; (e) Y. X. Sun, R. E. Lu, X. R. Li, Y. Y. Zhao and C. Y. Li, Chin. J. Inorg. Chem., 2015, 31, 1055–1062 CAS; (f) G. Murugavel, P. Sadhu and T. Punniyamurthy, Chem. Rec., 2016, 16, 1906–1917 CrossRef CAS PubMed; (g) A. B. Canaj, M. Siczek, M. Otręba, T. Lis, G. Lorusso, M. Evangelisti and C. J. Milioset, Dalton Trans., 2016, 45, 18591–18602 RSC.
- (a) L. Q. Chai, J. J. Huang, J. Y. Zhang and Y. X. Li, J. Coord. Chem., 2015, 68, 1224–1237 CrossRef CAS; (b) L. Q. Chai, K. Y. Zhang, L. J. Tang, J. Y. Zhang and H. S. Zhang, Polyhedron, 2017, 130, 100–107 CrossRef CAS; (c) L. Q. Chai, L. J. Tang, L. C. Chen and J. J. Huang, Polyhedron, 2017, 122, 228–240 CrossRef CAS; (d) L. Q. Chai, G. Liu, J. Y. Zhang, J. J. Huang and J. F. Tong, J. Coord. Chem., 2013, 66, 3926–3938 CrossRef CAS; (e) L. Q. Chai, J. J. Huang and H. S. Zhang, Spectrochim. Acta, Part A, 2014, 131, 526–533 CrossRef CAS PubMed.
- (a) Q. Zhao, Z. L. Wei, Q. P. Kang, H. Zhang and W. K. Dong, Spectrochim. Acta, Part A, 2018, 203, 472–480 CrossRef CAS PubMed; (b) L. Chen, W. K. Dong, H. Zhang, Y. Zhang and Y. X. Sun, Cryst. Growth Des., 2017, 17, 3636–3648 CrossRef CAS; (c) Y. D. Peng, F. Wang, L. Gao and W. K. Dong, J. Chin. Chem. Soc., 2018, 65, 893–899 CrossRef CAS; (d) X. Y. Dong, Q. P. Kang, X. Y. Li, J. C. Ma and W. K. Dong, Crystals, 2018, 8, 139 CrossRef; (e) L. Z. Liu, M. Yu, X. Y. Li, Q. P. Kang and W. K. Dong, Chin. J. Inorg. Chem., 2019, 35, 1283–1294 CAS.
- (a) X. Y. Li, L. Chen, L. Gao, Y. Zhang, S. F. Akogun and W. K. Dong, RSC Adv., 2017, 7, 35905–35916 RSC; (b) L. H. Li, W. K. Dong, Y. Zhang, S. F. Akogun and L. Xu, Appl. Organomet. Chem., 2017, e3818 CrossRef.
- (a) X. Q. Song, P. P. Liu, Z. R. Xiao, X. Li and Y. A. Liu, Inorg. Chim. Acta, 2015, 438, 232–244 CrossRef CAS; (b) Y. A. Liu, C. Y. Wang, M. Zhang and X. Q. Song, Polyhedron, 2017, 127, 278–286 CrossRef CAS; (c) P. P. Liu, L. Sheng, X. Q. Song, W. Y. Xu and Y. A. Liu, Inorg. Chim. Acta, 2015, 434, 252–257 CrossRef CAS.
- (a) H. L. Wu, G. L. Pan, Y. C. Bai, H. Wang, J. Kong, F. R. Shi, Y. H. Zhang and X. L. Wang, J. Chem. Res., 2014, 38, 211–217 CrossRef CAS; (b) H. L. Wu, Y. C. Bai, Y. H. Zhang, G. L. Pan, J. Kong, F. R. Shi and X. L. Wang, Z. Anorg. Allg. Chem., 2014, 640, 2062–2071 CrossRef CAS; (c) H. L. Wu, G. L. Pan, Y. C. Bai, Y. H. Zhang, H. Wang, F. R. Shi, X. L. Wang and J. Kong, J. Photochem. Photobiol., B, 2014, 135, 33–43 CrossRef CAS PubMed; (d) H. L. Wu, C. P. Wang, F. Wang, H. P. Peng, H. Zhang and Y. C. Bai, J. Chin. Chem. Soc., 2015, 62, 1028–1034 CrossRef CAS; (e) H. L. Wu, Y. C. Bai, Y. H. Zhang, Z. Li, M. C. Wu, C. Y. Chen and J. W. Zhang, J. Coord. Chem., 2014, 67, 3054–3066 CrossRef CAS; (f) H. L. Wu, G. L. Pan, Y. C. Bai, H. Wang, J. Kong, F. R. Shi, Y. H. Zhang and X. L. Wang, Res. Chem. Intermed., 2015, 41, 3375–3383 CrossRef CAS; (g) C. Y. Chen, J. W. Zhang, Y. H. Zhang, Z. H. Yang, H. L. Wu, G. L. Pana and Y. C. Bai, J. Coord. Chem., 2015, 68, 1054–1071 CrossRef CAS; (h) H. L. Wu, Y. Bai, J. K. Yuan, H. Wang, G. L. Pan, X. Y. Fan and J. Kong, J. Coord. Chem., 2012, 65, 2839–2851 CrossRef CAS.
- (a) L. W. Zhang, L. Z. Liu, F. Wang and W. K. Dong, Molecules, 2018, 23, 1141 CrossRef PubMed; (b) F. Wang, L. Gao, Q. Zhao, Y. Zhang, W. K. Dong and Y. J. Ding, Spectrochim. Acta, Part A, 2018, 190, 111–115 CrossRef CAS PubMed; (c) J. Hao, X. Y. Li, Y. Zhang and W. K. Dong, Materials, 2018, 11, 523 CrossRef PubMed; (d) F. Wang, L. Z. Liu, L. Gao and W. K. Dong, Spectrochim. Acta, Part A, 2018, 203, 56–64 CrossRef CAS PubMed; (e) L. Z. Liu, L. Wang, M. Yu, Q. Zhao, Y. Zhang, Y. X. Sun and W. K. Dong, Spectrochim. Acta, Part A, 2019, 222, 117209 CrossRef CAS PubMed.
- (a) S. S. Zheng, W. K. Dong, Y. Zhang, L. Chen and Y. J. Ding, New J. Chem., 2017, 41, 4966–4973 RSC; (b) Q. P. Li, Y. Peng, J. J. Qian, T. Yan, L. Du and Q. H. Zhao, Dalton Trans., 2019, 48, 12880–12887 RSC; (c) J. J. Qian, T. T. Li, Y. Hua and S. M. Huang, Chem. Commun., 2017, 53, 13027–13030 RSC; (d) J. J. Qian, Q. P. Li, L. F. Liang, Y. Yang, Z. Cao, P. P. Yu, S. M. Huang and M. C. Hong, Chem. Commun., 2016, 52, 9032–9035 RSC.
- X. Y. Dong, Q. Zhao, Q. P. Kang, X. Y. Li and W. K. Dong, Crystals, 2018, 8, 230 CrossRef.
- (a) S. Akine, T. Taniguchi, W. K. Dong, S. Masubuchi and T. Nabeshima, J. Org. Chem., 2005, 70, 1704–1711 CrossRef CAS PubMed; (b) J. Hao, X. Y. Li, L. Wang, Y. Zhang and W. K. Dong, Spectrochim. Acta, Part A, 2018, 204, 388–402 CrossRef CAS.
- (a) G. M. Sheldrick, SHELXS-2016, Program for crystal structure solution, University of Göttingen, Göttingen Germany, 2016 Search PubMed; (b) G. M. Sheldrick, SHELXL-2016, Program for crystal structure refinement, University of Göttingen, Göttingen Germany, 2016 Search PubMed.
- (a) X. Y. Dong, Y. X. Sun, L. Wang and L. Li, J. Chem. Res., 2012, 36, 387–390 CrossRef CAS; (b) Z. L. Ren, J. Hao, P. Hao, X. Y. Dong, Y. Bai and W. K. Dong, Z. Naturforsch., 2018, 73b, 203–210 Search PubMed; (c) L. Xu, L. C. Zhu, J. C. Ma, Y. Zhang, J. Zhang and W. K. Dong, Z. Anorg. Allg. Chem., 2015, 641, 2520–2524 CrossRef CAS.
- (a) L. Wang, J. Hao, L. X. Zhai, Y. Zhang and W. K. Dong, Crystals, 2017, 7, 277 CrossRef; (b) J. Chang, H. J. Zhang, H. R. Jia and Y. X. Sun, Chin. J. Inorg. Chem., 2018, 34, 2097–2107 CAS.
- (a) L. Gao, F. Wang, Q. Zhao, Y. Zhang and W. K. Dong, Polyhedron, 2018, 139, 7–16 CrossRef CAS; (b) L. W. Zhang, X. Y. Li, Q. P. Kang, L. Z. Liu, J. C. Ma and W. K. Dong, Crystals, 2018, 8, 173 CrossRef.
- (a) Q. P. Kang, X. Y. Li, Q. Zhao, J. C. Ma and W. K. Dong, Appl. Organomet. Chem., 2018, 32, e4379 CrossRef; (b) X. Y. Li, Q. P. Kang, L. Z. Liu, J. C. Ma and W. K. Dong, Crystals, 2018, 8, 43 CrossRef; (c) Y. D. Peng, X. Y. Li, Q. P. Kang, G. X. An, Y. Zhang and W. K. Dong, Crystals, 2018, 8, 107 CrossRef.
- (a) L. Gao, C. Liu, F. Wang and W. K. Dong, Crystals, 2018, 8, 77 CrossRef; (b) P. Wang and L. Zhao, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2016, 46, 1095–1101 CrossRef CAS; (c) L. Zhao, L. Wang, Y. X. Sun, W. K. Dong, X. L. Tang and X. H. Gao, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2012, 42, 1303–1308 CrossRef CAS.
- P. Seth, S. Ghosh, A. Figuerola and A. Ghosh, Dalton Trans., 2013, 43, 990–998 RSC.
- (a) X. Y. Dong, Q. Zhao, L. W. Zhang, H. R. Mu, H. Zhang and W. K. Dong, Molecules, 2018, 23, 1006 CrossRef; (b) Q. Zhao, X. X. An, L. Z. Liu and W. K. Dong, Inorg. Chim. Acta, 2019, 490, 6–15 CrossRef CAS.
- (a) X. X. An, Q. Zhao, H. R. Mu and W. K. Dong, Crystals, 2019, 9, 101 CrossRef CAS; (b) L. Wang, X. Y. Li, Q. Zhao, L. H. Li and W. K. Dong, RSC Adv., 2017, 7, 48730–48737 RSC; (c) H. J. Zhang, J. Chang, H. R. Jia and Y. X. Sun, Chin. J. Inorg. Chem., 2018, 34, 2261–2270 CAS.
- (a) Y. J. Dong, X. Y. Dong, W. K. Dong, Y. Zhang and L. S. Zhang, Polyhedron, 2017, 123, 305–315 CrossRef CAS; (b) L. W. Zhang, L. Z. Liu, F. Wang and W. K. Dong, Molecules, 2018, 23, 1141 CrossRef PubMed.
- B. Yu, C. Y. Li, Y. X. Sun, H. R. Jia, J. Q. Guo and J. Li, Spectrochim. Acta, Part A, 2017, 184, 249–254 CrossRef CAS PubMed.
- (a) G. Li, J. Hao, L. Z. Liu, W. M. Zhou and W. K. Dong, Crystals, 2017, 7, 217 CrossRef; (b) Y. Zhang, L. Z. Liu, Y. D. Peng, N. Li and W. K. Dong, Transition Met. Chem., 2019, 44, 627–639 CrossRef CAS.
- (a) Q. P. Kang, X. Y. Li, L. Wang, Y. Zhang and W. K. Dong, Appl. Organomet. Chem., 2019, 32, e5013 CrossRef; (b) Y. X. Sun, Y. Y. Zhao, C. Y. Li, B. Yu, J. Q. Guo and J. Li, Chin. J. Inorg. Chem., 2016, 32, 913–920 CAS.
- (a) X. Y. Li, Q. P. Kang, C. Liu, Y. Zhang and W. K. Dong, New J. Chem., 2019, 43, 4605–4619 RSC; (b) H. R. Jia, J. Chang, H. J. Zhang, J. Li and Y. X. Sun, Crystals, 2018, 8, 272 CrossRef; (c) J. Li, H. J. Zhang, J. Chang, H. R. Jia, Y. X. Sun and Y. Q. Huang, Crystals, 2018, 8, 176 CrossRef; (d) Q. P. Kang, X. Y. Li, Z. L. Wei, Y. Zhang and W. K. Dong, Polyhedron, 2019, 165, 38–50 CrossRef CAS.
- W. K. Dong, J. C. Ma, L. C. Zhu and Y. Zhang, Cryst. Growth Des., 2016, 16, 6903–6914 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1894560, 1894557, 1894559 and 1894558 for complexes 1–4. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ra07423f |
| This journal is © The Royal Society of Chemistry 2019 |