Open Access Article
Open Access ArticlePanax notoginseng saponins radiosensitize colorectal cancer cells by regulating the SNHG6/miR-137 axis
Caihui Xua,
Teng Liub,
Haiyan Liua,
Gongbin Chena and
Yinmou Guo *a
*a
aDepartment of Oncology, Shangqiu First People's Hospital, No. 292, South Kaixuan Road, Suiyang District, Shangqiu, 476100, Henan, China. E-mail: guoym18917@163.com; Tel: +86-0370-3255630
bXinxiang Medical University, Hongqi District, Xinxiang, Henan, China
First published on 26th November 2019
Abstract
Panax notoginseng saponins (PNS) have recently attracted great attention for their anti-cancer activity in colorectal cancer (CRC). The aim of this study was to explore the functional role and underlying mechanisms of PNS on CRC radiosensitivity. Cell viability was assessed by a Cell Counting kit-8 assay. Cell survival and apoptosis were determined using colony formation assay and flow cytometry, respectively. Quantitative real-time PCR was used to quantify the levels of SNHG6 and miR-137. The targeted correlation between SNHG6 and miR-137 was validated by dual-luciferase reporter and RNA immunoprecipitation assays. Our data supported that PNS weakened the viability of CRC cells. Moreover, PNS promoted the radiosensitivity of CRC cells. Mechanistically, PNS enhanced CRC cell radiosensitivity by upregulating SNHG6. SNHG6 directly targeted miR-137 and inhibited miR-137 expression. MiR-137 was involved in the regulatory effect of SNHG6 on CRC cell radiosensitivity. Furthermore, PNS increased miR-137 expression through SNHG6 in CRC cells. Our study suggested that PNS promoted radiosensitivity in CRC cells at least partly through regulating the SNHG6/miR-137 axis, providing a novel understanding of the anti-cancer mechanism of PNS in CRC.
1. Introduction
Colorectal cancer (CRC) is the third most common malignancy and the second cause of cancer-related death worldwide. Despite advances in diagnosis and treatment, CRC remains a prevalent cancer, with an estimated 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800
800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 new CRC cases and 881
000 new CRC cases and 881![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths in 2018 around the world.1 Although radiotherapy has been widely accepted as an important and effective therapy method, the development of radioresistance is still one of major obstacles for CRC management, resulting in CRC recurrence and an unfavorable prognosis.2–4 Therefore, identifying new effective targets to overcome the radioresistance is very important for improving CRC treatment.
000 deaths in 2018 around the world.1 Although radiotherapy has been widely accepted as an important and effective therapy method, the development of radioresistance is still one of major obstacles for CRC management, resulting in CRC recurrence and an unfavorable prognosis.2–4 Therefore, identifying new effective targets to overcome the radioresistance is very important for improving CRC treatment.
Panax notoginseng (Burk.) F. H. Chen (Araliaceae), a perennial herb, has been widely used as a traditional Chinese medicine in Asia for a long time.5–7 Panax notoginseng saponins (PNS) are the crucial bioactive components derived from the root of Panax notoginseng and exhibit a wide variety of pharmacological effects, such as anti-diabetic, anti-inflammatory, antioxidant, hepatoprotective, hypolipidemic, renoprotective and anti-hyperglycemic activities.6,8 PNS have recently attracted a great attention for their anti-cancer activity in many types of human cancers, such as breast cancer and lung cancer.9,10 Previous researches had reported that PNS repressed CRC progression and promoted CRC cell chemosensitivity.11,12 The purpose of this study was to investigate the detailed role and underlying mechanisms of PNS on CRC radiosensitivity.
Long non-coding RNAs (lncRNAs), a heterogeneous group of non-coding transcripts with >200 nucleotides in length, perform diverse roles in numerous biological processes across every branch of life.13 Accumulating evidence has suggested that CRC-related lncRNAs regulate CRC progression and radioresistance development by various mechanisms, including their functions as molecular sponges of microRNAs (miRNAs).14,15 Small nucleolar RNA host gene 6 (SNHG6) has been identified as a potential oncogenic lncRNA in CRC, offering a possibility of SNHG6 as a biomarker for CRC diagnosis, treatment and prognosis.16,17 Previous studies had demonstrated that PNS played a protective role in many human diseases through regulation of several miRNAs or signaling pathways.10,18,19 Nevertheless, the effect of interplay between PNS and SNHG6 on CRC radioresistance still is unclear.
In the present study, our data firstly demonstrated that PNS weakened cell viability and promoted the radiosensitivity in CRC cells. Whereafter, we further investigated whether SNHG6 was involved in PNS-mediated regulatory effect on CRC radioresistance.
2. Materials and methods
2.1 Clinical specimens and ethics statement
CRC tissues and corresponding noncancerous tissues were collected from 46 cases of CRC patients undergoing surgical resection without prior radiochemotherapy in the Oncology Department of Shangqiu First People's Hospital. All specimens were stored at −80 °C until RNA extraction. Written informed consent was signed by each participant. The study was approved by the Human Research Ethics Committee of Shangqiu First People's Hospital and all experimental processes were carried out in accordance with the local ethical guidelines.2.2 Cell culture, PNS treatment and irradiation
Human immortalized noncancerous NCM460 colon cells obtained from Bena Culture Collection (Beijing, China) and two CRC cell lines (SW620 and LoVo) purchased from ATCC (Manassas, VA, USA) were maintained in RPMI-1640 medium (Life Technologies, Bleiswijk, the Netherlands) containing 10% fetal calf serum (FCS, PAA Laboratories, Pasching, Australian), 1% penicillin/streptomycin (Life Technologies) in a humidified incubator with 5% CO2 at 37 °C.Cells were exposed to different concentration (0, 50, 100 and 200 μM) of PNS (95% purity, Kunming Pharmaceutical Corporation, Yunnan, China) or 0.1% DMSO vehicle (we verified that this DMSO concentration did not affect CRC cell viability) for different time point (0, 12, 24 and 48 h). In addition to cell viability assay, the PNS concentration in other experiments was 200 μM.
Cells were pretreated with 200 μM of PNS for 24 h and then irradiated with increasing dosage intervals (0, 2, 4, 6, and 8 Gy) of X-rays for 24 h by an Elekta™ linear accelerator (Elekta AB, Stockholm, Sweden) using 6 MV photons, followed by the measurement of cell survival and apoptosis.
2.3 Oligonucleotide and plasmid transfection
For SNHG6 overexpression, cells were introduced with SNHG6 overexpression plasmid (pcDNA-SNHG6), and nontarget pcDNA (pcDNA-con) was used as negative control. SNHG6 depletion was performed using siRNA against SNHG6 (si-SNHG6) or negative control nontarget siRNA (si-con). MiR-137 mimic or miR-137 inhibitor (anti-miR-137) was transfected into cells to elevate or reduce miR-137 expression, with miR-con mimic or anti-miR-con as negative control. FuGENE HD transfection reagent (Roche, Mannheim, Germany) was used for all transfections, referring to the protocols of manufacturers. All oligonucleotides and plasmids were purchased from GenePharma (Shanghai, China).2.4 Determination of IC50 value and cell viability
In cell viability assay, cells were seeded in 96-well plates and then were exposed to different concentration (0, 50, 100 and 200 μM) of PNS for different time point (0, 12, 24 and 48 h). In IC50 value assay, cells were exposed to various concentrations (0.01, 0.1, 1, 10, 100 and 1000 μM) of PNS for 48 h. Cell viability and IC50 value were determined using a Cell Counting kit-8 (CCK-8, Dojindo, Kumamoto, Japan) following the manufacturer's guidance. Absorbance at an emission wavelength of 450 nm was measured by a microplate reader (Synergy H1 Hybrid Reader, BioTek, Winooski, VT, USA). Cell viability was presented as a percentage of control (0.1% DMSO) cells.2.5 Colony formation assay
Cell survival was determined using a standard colony formation assay. After various treatments, individual cells were obtained using 0.1% trypsin (Takara, Beijing, China) and seeded in 6-well plates at a density of 200 cells. 2 weeks later, cells were fixed with 90% ethanol and stained with 0.1% crystal violet (Sigma-Aldrich, Tokyo, Japan). Colonies containing at least 50 cells was counted and the survival fraction was calculated based on the number of control cells.2.6 Flow cytometry of cell apoptosis
Cell apoptosis was assessed by flow cytometry using the Annexin V-FITC/PI Apoptosis Detection kit (Sigma-Aldrich) in accordance with the instructions of manufacturers. After various treatments, cells were trypsinized and washed three times with ice-cold PBS. Afterwards, cells were double stained with 0.02 μg ml−1 of Annexin V-FITC and 1 μg ml−1 of PI for 15 min in the dark at room temperature. Cell apoptosis was detected by a FACSCalibur flow cytometer (Becton Dickinson, Heidelberg, Germany) with CellQuestPro software.2.7 Reverse transcription and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from CRC tissues and cells with a PicoPure™ RNA isolation kit (Applied Biosystems, Darmstadt, Germany) following the instructions of manufacturers. For quantification of SNHG6, total RNA (1 μg) was reverse-transcribed using a ProtoScript II reverse transcription kit (New England Biolabs, Ipswich, MA, USA). For miR-137 determination, cDNA was synthesized from 1 μg of total RNA with TaqMan MicroRNA reverse transcription kit (Applied Biosystems). qRT-PCR was performed using Power SYBR™ Green PCR Master Mix (Applied Biosystems) in accordance with manufacturer's guidance on the ABI Prism 7000 Sequence Detection System (Applied Biosystems). Levels of SNHG6 and miR-137 were normalized to the geometric mean of housekeeping genes GAPDH and snRNA RNU6, respectively, using the 2−ΔΔCt.2.8 Bioinformatics
Online software LncBase v.2 was used to predict the target miRNA of SNHG6 at http://www.carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index.2.9 Dual-luciferase reporter assay
SNHG6 luciferase reporter (SNHG6-WT) containing the complementary sequence of SNHG6 and site-directed mutant of seeded sequence (SNHG6-MUT) were obtained from GenePharma. Cells at 50–70% confluence were cotransfected with SNHG6-WT or SNHG6-MUT and miR-con mimic or miR-137 mimic. After 48 h post-transfection, luciferase activity was detected using the dual-luciferase Reporter Assay System (Promega, Fitchburg, WI, USA) and the radio of Firefly/Renilla luciferase activity was calculated.2.10 RNA immunoprecipitation assay
RNA immunoprecipitation (RIP) experiment was implemented to verify whether RNA-induced silencing complex (RISC) contained SNHG6 and miR-137 in CRC cells with an Imprint RNA RIP kit (Merck Millipore, Milan, Italy). Cells were lysed with RIPA lysis buffer (Beyotime, Shanghai, China), and then incubated with magnetic beads-conjugated anti-argonaute 2 (anti-Ago2, Abcam, Cambridge, UK) or negative control IgG antibody. Beads were harvested by centrifugation and total RNA was extracted, followed by the determination of SNHG6 and miR-137 levels by qRT-PCR.2.11 Statistical analysis
All experimental data were expressed as mean ± SD. Differences between two groups were compared using a Student's t-test (two-tailed) or Mann–Whitney U test. Statistical significance with more than two groups was analyzed with one-way ANOVA. P values at 0.05 or smaller were considered significant.3. Results
3.1 PNS weakened the viability of CRC cells
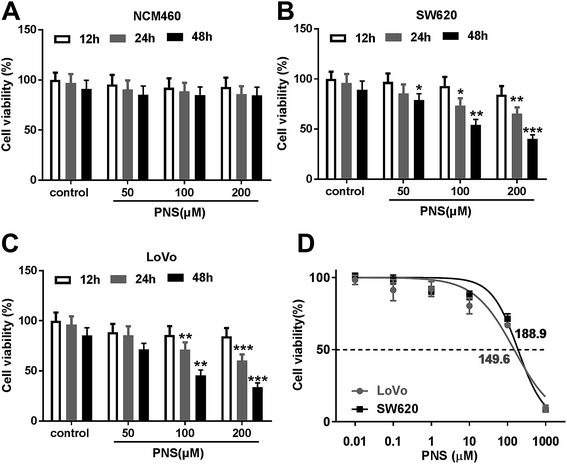
For a preliminary observation for the effect of PNS on CRC cell viability, CRC cells were exposed to different concentration (0, 50, 100 and 200 μM) of PNS for different time point (12, 24 and 48 h). CCK-8 assays revealed that compared with negative control, the viability of noncancerous NCM460 colon cells did not been affected by PNS (Fig. 1A). However, PNS treatment triggered a significant repression of cell viability in both SW620 and LoVo cells in dose- and time-dependent manners (Fig. 1B and C). Notably, when cells were treated with 50 μM of PNS for 48 h, SW620 cell viability was remarkably reduced by PNS treatment, while no significant change was found in LoVo cells (Fig. 1B and C). Additionally, CCK-8 results showed that the IC50 values for PNS in SW620 and LoVo cells were 188.9 and 149.6 μM, respectively (Fig. 1D). These results together suggested an inhibitory activity of PNS in CRC cell viability.3.2 PNS promoted the radiosensitivity of CRC cells
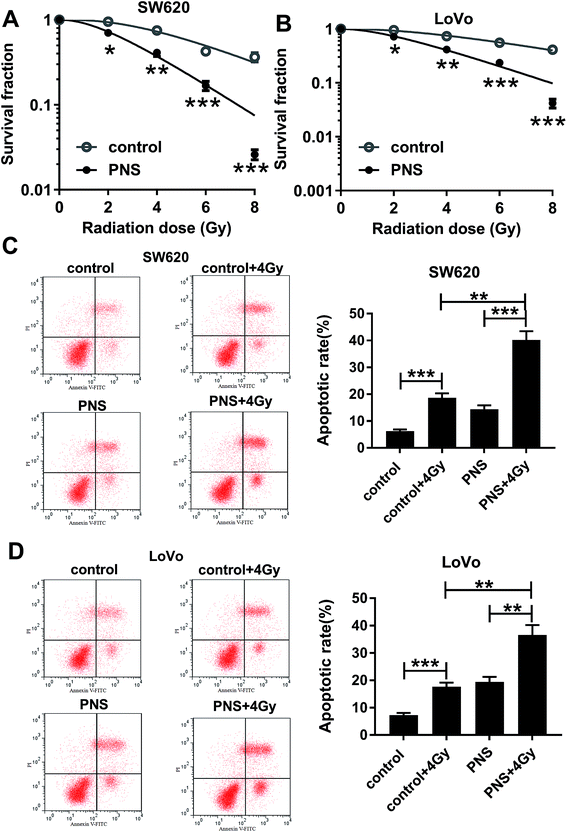
To investigate the effect of PNS on CRC radiosensitivity, CRC cells were exposed to 200 μM of PNS or 0.1% DMSO before radiation with increasing dosage intervals (0, 2, 4, 6 and 8 Gy) of X-rays. Colony formation assays showed that compared with DMSO group, PNS treatment profoundly repressed the colony formation ability after irradiation in the two CRC cells (Fig. 2A and B). Flow cytometry results demonstrated that irradiation (4 Gy) exposure significantly increased cell apoptosis when treatment with or without PNS (Fig. 2C and D). Moreover, simultaneous PNS treatment and irradiation exposure led to a more distinct promotion on cell apoptosis (Fig. 2C and D). All these data strongly point to the notion that PNS enhanced the sensitivity to radiotherapy in CRC cells.3.3 PNS repressed SNHG6 expression in CRC cells
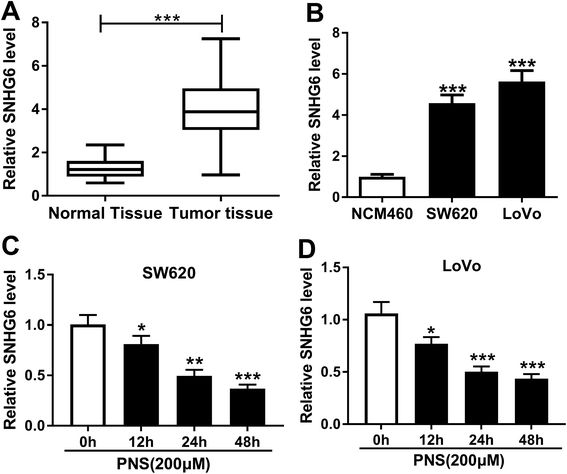
Previous researches had reported that SNHG6 accelerated CRC progression through promotion of cell proliferation, migration, invasion and epithelial–mesenchymal transition.16,17 Herein, we determined the expression of SNHG6 in CRC tissues and cell lines. As demonstrated by qRT-PCR, SNHG6 expression was significantly upregulated in CRC tissues and cells compared with their counterparts (Fig. 3A and B), supporting the involvement of SNHG6 in CRC progression. We next evaluated whether SNHG6 expression in the two CRC cells was influenced by PNS treatment. In contrast to negative control, PNS treatment resulted in decreased SNHG6 level in a time-dependent manner in both SW620 and LoVo cells (Fig. 3C and D). These data together implied that PNS treatment hindered the expression of SNHG6 in CRC cells.3.4 PNS enhanced CRC cell radiosensitivity by SNHG6
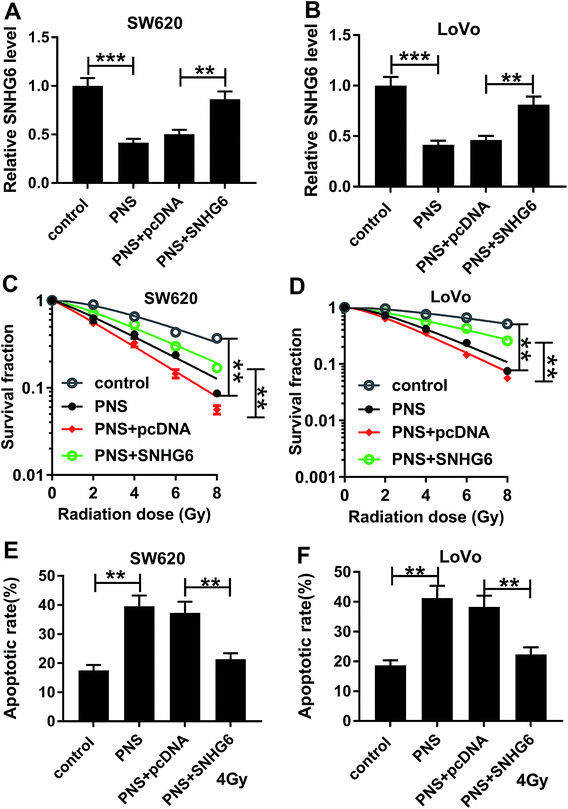
Given our data that PNS suppressed SNHG6 expression in CRC cells, we further investigated whether SNHG6 was involved in the promotional effect of PNS on radiosensitivity of CRC cells. As shown by qRT-PCR, transient transfection of pcDNA-SNHG6, but not a negative plasmid, significantly reversed the decreased effect of PNS on SNHG6 expression in the two CRC cells (Fig. 4A and B). Subsequent experiments revealed that the repressive effect of PNS on colony formation capacity was highly abated by cotransfection of pcDNA-SNHG6 after irradiation treatment (Fig. 4C and D). Moreover, PNS-mediated pro-apoptosis effect was markedly abolished by pcDNA-SNHG6 introduction after 4 Gy of X-rays treatment (Fig. 4E and F). Together, these results strongly established that SNHG6 was involved in PNS-mediated increased radiosensitivity in CRC cells.3.5 SNHG6 directly targeted miR-137 and inhibited miR-137 expression
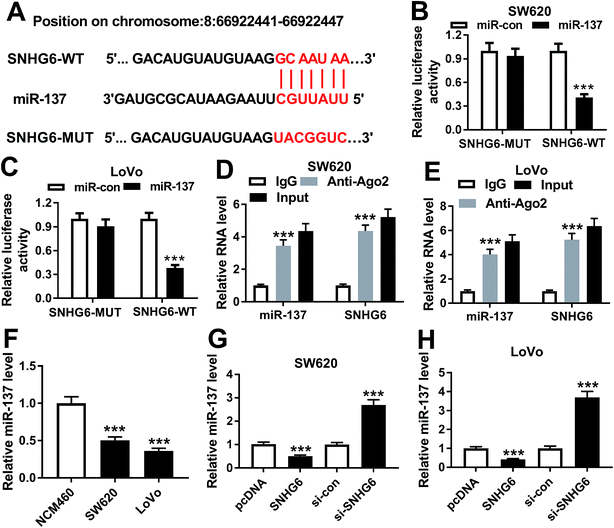
In order to further understand the molecular mechanisms by which SNHG6 influenced CRC cell radiosensitivity, we performed a detailed analysis for the targeted miRNAs of SNHG6. Using LncBase v.2 software, the predicted data revealed that SNHG6 harbored a putative complementary sequence for miR-137 (Fig. 5A). To confirm this, we carried out dual-luciferase reporter assays and RIP assays. When we performed the dual-luciferase reporter assays, cotransfection of SNHG6 luciferase reporter and miR-137 mimic into SW620 and LoVo cells produced lower luciferase activity than in cells cotransfected with miR-con mimic (Fig. 5B and C). However, site-directed mutant of seeded sequence remarkably abolished the effect of miR-137 on reporter gene expression (Fig. 5B and C), indicating that the miR-137-binding sites were functional in the two cells. Ago2 is a core component of the RISC, where the mature miRNAs direct the complex to target mRNAs.20 Thus, anti-Ago2 antibody was used for RIP assay. RIP experiments revealed that compared with IgG control, the levels of miR-137 and SNHG6 were simultaneously strongly enriched in the RISC by anti-Ago2 antibody (Fig. 5D and E), eliciting an endogenous interaction between miR-136 and SNHG6 in the two CRC cells. qRT-PCR assays also showed a downregulation of miR-137 level in CRC cells compared with normal control (Fig. 5F). After that, we further investigated whether SNHG6 modulated miR-137 expression in SW620 and LoVo cells. In contrast to their counterparts, miR-137 expression was significantly decreased by transfection of pcDNA-SNHG6, while it was strikingly increased after si-SNHG6 introduction (Fig. 5G and H). All these results strongly pointed to a role of SNHG6 as a molecular sponge for miR-137.3.6 MiR-137 was involved in the regulatory effect of SNHG6 on CRC cell radiosensitivity
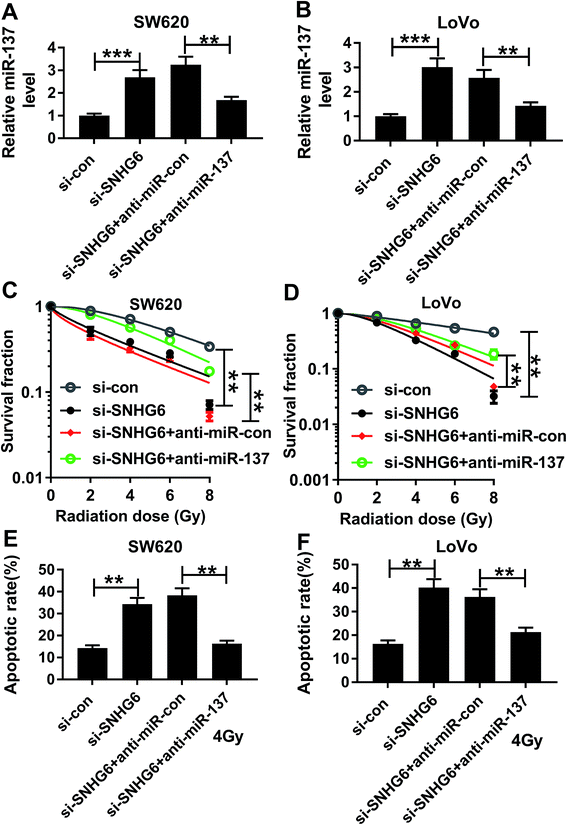
To provide further mechanistic insight into the link between SNHG6 and miR-137 on radiosensitivity of CRC cells, siRNA against SNHG6 (si-SNHG6) and anti-miR-137 were cotransfected into SW620 and LoVo cells. Cotransfection of anti-miR-137, but not a scrambled control sequence, significantly abated si-SNHG6-mediated increased miR-137 expression in the two CRC cells (Fig. 6A and B). Subsequent experiments revealed that in comparison to negative group, transfection of si-SNHG6 remarkably enhanced the sensitivity to radiotherapy in the two CRC cells, as evidenced by the significant inhibition of colony formation capacity and the prominent promotion of cell apoptosis after irradiation treatment (Fig. 6C–F). However, this effect of si-SNHG6 was strongly abolished by cotransfection of anti-miR-137 (Fig. 6C–F). Taken together, these results suggested that miR-137 was involved in the regulatory effect of SNHG6 on radiosensitivity in CRC cells.3.7 PNS increased miR-137 expression through SNHG6 in CRC cells
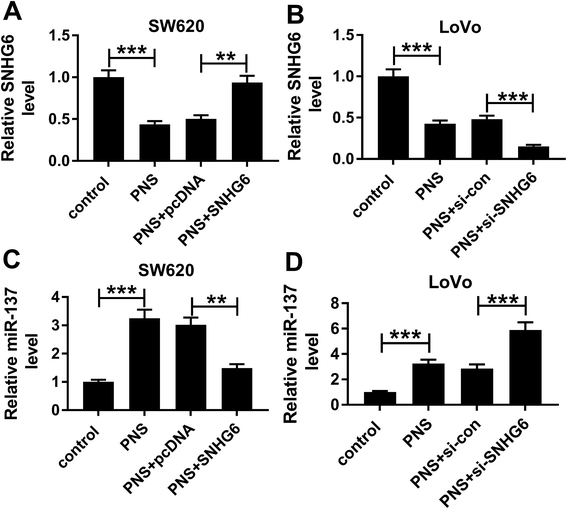
Next, we observed whether, if so, how PNS influenced miR-137 expression in CRC cells. qRT-PCR results revealed that in contrast to their counterparts, SNHG6 expression was significantly repressed by PNS treatment in the two CRC cells, while the effect was highly abated by pcDNA-SNHG6 transfection and strikingly enhanced after introduction of si-SNHG6 (Fig. 7A and B). Moreover, PNS treatment triggered a significant upregulation of miR-137 expression in the two CRC cells (Fig. 7C and D). However, this effect was markedly abolished by restored SNHG6 expression, while it was significantly enhanced after SNHG6 depletion (Fig. 7C and D). These data together established that PNS increased the expression of miR-137 through repressing SNHG6 level in CRC cells.4. Discussion
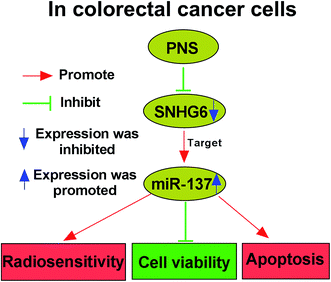
Although PNS are widely acknowledged to exhibit various health benefits, their anti-cancer activity and underlying molecular mechanisms are not well understood, particularly in cancer radiosensitivity. Hence, in this study, we aimed to explore the functional role and molecular mechanism of PNS in CRC radiosensitivity. Our results indicated that PNS promoted the sensitivity to radiotherapy in CRC cells. Mechanistically, PNS radiosensitized CRC cells through regulating SNHG6/miR-137 axis (Fig. 8).Previous studies had demonstrated that PNS hindered breast cancer metastasis and repressed lung cancer cell growth, highlighting their anti-cancer activity.9,10 PNS were also reported to play anti-proliferative activity in CRC cells through accelerating the apoptosis and hampering cell cycle progression.11,21 Wang et al. also manifested that PNS enhanced CRC cell sensitivity to chemotherapeutic agents. Therefore, our study started from the hypothesis that PNS elevated the radiosensitivity in CRC cells. To validate this, we firstly determined the impact of PNS on CRC cell viability, and our results demonstrated that PNS weakened the viability of CRC cells in time- and dose-dependent manners. Moreover, our data firstly established that PNS promoted the sensitivity to radiotherapy in CRC cells, as evidenced by the decreased survival and increased apoptosis after irradiation treatment. More interestingly, the viability of noncancerous NCM460 colon cells did not been affected by PNS, possibly due to lower PNS concentration or the certain addiction of CRC cells to PNS.
Up to now, several Chinese herb extracts, such as astragaloside IV and Huaier extract, have been illuminated to attenuate cancer growth via regulating the expression of special lncRNAs.22,23 Moreover, curcumin was described to promote the radiosensitivity in nasopharyngeal carcinoma cells through modulation of six lncRNAs.24 However, whether PNS could enhance CRC cell radiosensitivity by lncRNAs has not been reported. In the present study, our data indicated a significant upregulation of SNHG6 in CRC tissues and cells, in accordance with former works.25–27 These documents also manifested that SNHG6 facilitated the proliferation, invasion and migration of CRC cells, illuminating its oncogenic role in CRC. Therefore, we further explored whether PNS radiosensitized CRC cells via regulating SNHG6. Our results firstly demonstrated that PNS reduced the expression of SNHG6 in a time-dependent manner in CRC cells. Furthermore, SNHG6 was involved in PNS-mediated increased radiosensitivity in CRC cells. In a word, PNS promoted CRC cell radiosensitivity by downregulating SNHG6 expression.
It is widely accepted that lncRNAs exert their biological functions partly through acting as molecular sponges of special miRNAs. Thus, we used online software LncBase v.2 to search for the targeted miRNAs of SNHG6. Among these candidates, miR-137 was fascinating in the present study because of its tumor-suppressive property in a lot of human cancers, such as breast cancer, non-small cell lung cancer and ovarian cancer.28–30 MiR-137 was also reported to hamper CRC progression by suppression of cell proliferation, invasion and migration.31–33 Additionally, miR-137 enhanced the chemosensitivity in ovarian cancer and colon cancer.34,35 In the present study, we firstly validated that SNHG6 directly targeted miR-137 and repressed miR-137 expression. Moreover, miR-137 mediated the regulatory effect of SNHG6 on radiosensitivity in CRC cells. More importantly, our results substantiated that PNS increased miR-137 expression via SNHG6 in CRC cells. Previous documents had demonstrated that miR-137 inhibited CRC progression and chemosensitized CRC cells through targeting several miRNAs, such as formin-like 2 (FMNL2) and Y-box-binding protein 1 (YBX1).35,36 More researches about the relevant targets of miR-137 in CRC radioresistance will be performed in further work.
In conclusion, our results suggested that PNS promoted radiosensitivity in CRC cells at least partly through regulating SNHG6/miR-137 axis. The clinical significance of PNS and its potential value as a promising adjuvant in CRC radiotherapy should be further explored.
Conflicts of interest
There is no conflict of interest regarding the publication of this paper.References
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Ca-Cancer J. Clin., 2018, 68, 394–424 CrossRef PubMed.
- H. E. Barker, J. T. Paget, A. A. Khan and K. J. Harrington, Nat. Rev. Cancer, 2015, 15, 409–425 CrossRef CAS PubMed.
- E. Y. Huang, J. C. Chang, H. H. Chen, C. Y. Hsu, H. C. Hsu and K. L. Wu, BMC Cancer, 2018, 18, 321 CrossRef PubMed.
- X. C. Wang, X. Yue, R. X. Zhang, T. Y. Liu, Z. Z. Pan, M. J. Yang, Z. H. Lu, Z. Y. Wang, J. H. Peng, L. Y. Le, G. Y. Wang, Q. H. Peng, Y. Meng, W. Huang and R. Y. Liu, Clin. Cancer Res., 2019, 25, 4567–4579 CrossRef PubMed.
- W. Z. Yang, Y. Hu, W. Y. Wu, M. Ye and D. A. Guo, Phytochemistry, 2014, 106, 7–24 CrossRef CAS PubMed.
- T. Ng, J. Pharm. Pharmacol., 2006, 58, 1007–1019 CrossRef CAS PubMed.
- J. Zhang and X. Fang, Zhonghua Yixue Shijian Zazhi, 2004, 34, 16–20 Search PubMed.
- R. Uzayisenga, P. A. Ayeka and Y. Wang, Phytother. Res., 2014, 28, 510–516 CrossRef PubMed.
- P. Wang, J. Cui, X. Du, Q. Yang, C. Jia, M. Xiong, X. Yu, L. Li, W. Wang and Y. Chen, J. Ethnopharmacol., 2014, 154, 663–671 CrossRef CAS PubMed.
- Q. Yang, P. Wang, J. Cui, W. Wang, Y. Chen and T. Zhang, J. Ethnopharmacol., 2016, 193, 255–265 CrossRef CAS PubMed.
- C. Z. Wang, J. T. Xie, A. Fishbein, H. H. Aung, H. He, S. R. Mehendale, T. C. He, W. Du and C. S. Yuan, Phytother. Res., 2009, 23, 6–13 CrossRef CAS PubMed.
- C. Z. Wang, X. Luo, B. Zhang, W. X. Song, M. Ni, S. Mehendale, J. T. Xie, H. H. Aung, T. C. He and C. S. Yuan, Cancer Chemother. Pharmacol., 2007, 60, 69–79 CrossRef CAS PubMed.
- J. J. Quinn and H. Y. Chang, Nat. Rev. Genet., 2016, 17, 47–62 CrossRef CAS PubMed.
- M. Xu, X. Chen, K. Lin, K. Zeng, X. Liu, B. Pan, X. Xu, T. Xu, X. Hu and L. Sun, Mol. Cancer, 2018, 17, 141 CrossRef PubMed.
- Y. Zou, S. Yao, X. Chen, D. Liu, J. Wang, X. Yuan, J. Rao, H. Xiong, S. Yu and X. Yuan, Eur. J. Cell Biol., 2018, 97, 369–378 CrossRef CAS PubMed.
- X. Wang, Q. Lai, J. He, Q. Li, J. Ding, Z. Lan, C. Gu, Q. Yan, Y. Fang, X. Zhao and S. Liu, Int. J. Med. Sci., 2019, 16, 51–59 CrossRef CAS PubMed.
- M. Zhang, W. Duan and W. Sun, OncoTargets Ther., 2019, 12, 3349–3360 CrossRef PubMed.
- R. Wei, D. Qiu, I. W. Wilson, H. Zhao, S. Lu, J. Miao, S. Feng, L. Bai, Q. Wu, D. Tu, X. Ma and Q. Tang, BMC Genomics, 2015, 16, 835 CrossRef PubMed.
- S. Chen, J. Liu, X. Liu, Y. Fu, M. Zhang, Q. Lin, J. Zhu, L. Mai, Z. Shan, X. Yu, M. Yang and S. Lin, J. Ethnopharmacol., 2011, 137, 263–270 CrossRef CAS PubMed.
- S. M. Hammond, Adv. Drug Delivery Rev., 2015, 87, 3–14 CrossRef CAS PubMed.
- C. Z. Wang, J. T. Xie, B. Zhang, M. Ni, A. Fishbein, H. H. Aung, S. R. Mehendale, W. Du, T. C. He and C. S. Yuan, Int. J. Oncol., 2007, 31, 1149–1156 CAS.
- Y. Li, Y. Ye and H. Chen, Biomed. Pharmacother., 2018, 99, 134–141 CrossRef PubMed.
- J. Wang, X. Wang, T. Chen, L. Jiang and Q. Yang, Cell. Physiol. Biochem., 2017, 44, 581–593 CrossRef PubMed.
- Q. Wang, H. Fan, Y. Liu, Z. Yin, H. Cai, J. Liu, Z. Wang, M. Shao, X. Sun and J. Diao, Int. J. Oncol., 2014, 44, 858–864 CrossRef CAS PubMed.
- Z. Li, R. Qiu, X. Qiu and T. Tian, Cell. Physiol. Biochem., 2018, 49, 463–478 CrossRef CAS PubMed.
- Y. Zhu, Y. Xing, F. Chi, W. Sun, Z. Zhang and D. Piao, OncoTargets Ther., 2018, 11, 5743–5752 CrossRef CAS PubMed.
- M. Xu, X. Chen, K. Lin, K. Zeng, X. Liu, X. Xu, B. Pan, T. Xu, L. Sun, B. He, Y. Pan, H. Sun and S. Wang, J. Hematol. Oncol., 2019, 12, 3 CrossRef PubMed.
- F. Chen, N. Luo, Y. Hu, X. Li and K. Zhang, Cell. Physiol. Biochem., 2018, 47, 2147–2158 CrossRef CAS PubMed.
- S. Nuzzo, S. Catuogno, M. Capuozzo, A. Fiorelli, P. Swiderski, S. Boccella, F. de Nigris and C. L. Esposito, Mol. Ther.--Nucleic Acids, 2019, 17, 256–263 CrossRef CAS PubMed.
- P. Dong, Y. Xiong, H. Watari, S. J. Hanley, Y. Konno, K. Ihira, T. Yamada, M. Kudo, J. Yue and N. Sakuragi, J. Exp. Clin. Cancer Res., 2016, 35, 132 CrossRef PubMed.
- X. Liu, L. Cui and D. Hua, Oncol. Res., 2018, 27, 99–106 CrossRef PubMed.
- M. Liu, N. Lang, M. Qiu, F. Xu, Q. Li, Q. Tang, J. Chen, X. Chen, S. Zhang, Z. Liu, J. Zhou, Y. Zhu, Y. Deng, Y. Zheng and F. Bi, Int. J. Cancer, 2011, 128, 1269–1279 CrossRef CAS PubMed.
- Y. Sun, X. Zhao, Y. Zhou and Y. Hu, Oncol. Rep., 2012, 28, 1346–1352 CrossRef CAS PubMed.
- J. Sun, X. Cai, M. M. Yung, W. Zhou, J. Li, Y. Zhang, Z. Li, S. S. Liu, A. N. Y. Cheung, H. Y. S. Ngan, Y. Li, Z. Dai, Y. Kai, A. Tzatsos, W. Peng, D. W. Chan and W. Zhu, Oncogene, 2019, 38, 564–580 CrossRef CAS PubMed.
- Y. Guo, Y. Pang, X. Gao, M. Zhao, X. Zhang, H. Zhang, B. Xuan and Y. Wang, Cancer Biomarkers, 2017, 18, 1–9 CAS.
- L. Liang, X. Li, X. Zhang, Z. Lv, G. He, W. Zhao, X. Ren, Y. Li, X. Bian, W. Liao, W. Liu, G. Yang and Y. Ding, Gastroenterology, 2013, 144, 624–635.e624 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |