Open Access Article
Open Access ArticleFlavin-mediated extracellular electron transfer in Gram-positive bacteria Bacillus cereus DIF1 and Rhodococcus ruber DIF2
Tian Tiana,
Xiaoyang Fana,
Man Fengb,
Lin Su a,
Wen Zhang
a,
Wen Zhang ac,
Huimei Chi
ac,
Huimei Chi *ab and
Degang Fua
*ab and
Degang Fua
aState Key Laboratory of Bioelectronics, School of Biomedical Science and Engineering, Southeast University, Sipailou 2, Nanjing, 210096, China. E-mail: hmchi@seu.edu.cn
bState Key Laboratory of Palaeobiology and Stratigraphy, Nanjing Institute of Geology & Palaeontology, CAS, 210008, China
cCollege of Materials and Environmental Engineering, Hangzhou Dianzi University, Hangzhou 310018, People's Republic of China
First published on 11th December 2019
Abstract
Flavin-mediated extracellular electron transfer was studied in two Gram-positive bacteria: Bacillus cereus strain DIF1 and Rhodococcus ruber strain DIF2. The electrochemical activities of these strains were confirmed using amperometric I–t curves and cyclic voltammetry (CV). Spent anodes with biofilms in fresh anolytes showed no redox peaks, while new anodes in the spent broth showed relative redox peaks using CV measurements, indicating the presence of a redox electron mediator secreted by bacteria. Adding riboflavins (RF) and flavin mononucleotide (FMN) improved the electron transfer of the microbial fuel cells inoculated with the two strains. The redox peaks indicated that flavins existed in the anolyte, and HPLC analysis showed that RF and FMN were secreted by the two bacterial strains. The concentration of RF increased until the bacteria grew to the log phase in microbial fuel cells. The concentration of RF decreased and that of FMN increased after the log phase. The two strains secreted FMN only in the microbial fuel cell. These results confirmed that the electrochemical activity mediated by flavins and FMN is essential in the extracellular electron transfer process in the strains DIF1 and DIF2.
1 Introduction
Electrochemically active bacteria can respire using extracellular electron acceptors like electrodes and minerals via extracellular electron transfer (EET).1–3 There is a variety of solid surfaces to and from which microbes can deliver electrons by EET processes via outer-membrane c-type cytochromes.4 The EET process is important for microbial respiration in natural environments and industrial applications.1–4 Some exoelectrogens like Geobacter, Shewanella, Pseudomonas, and Rhodoferax species have been widely studied.5–8 A variety of microbial phyla have exoelectrogenic members, including Proteobacteria, Firmicutes, Acidobacteria, Actinobacteria, Eumycota, and Chlorophyta.9Even if Gram-negative bacteria possess the largest numbers in all electrochemically active bacteria, more electrochemically active Gram-positive bacteria were found in MFCs.10–15 The proteins involved in the electron transfer are localized in the outer membrane of Gram-negative bacteria or in the exosporium of Gram-positive spores.16–18 Despite their importance in iron redox cycles and bioenergy production, the underlying physiological, genetic, and biochemical mechanisms of extracellular electron transfer by Gram-positive bacteria remain poorly understood.13
Contact-dependent (direct) mechanisms transfer electrons via multiheme c-type cytochromes or nanowires across the bacterial cell envelope in Shewanella and Geobacter species.19–23 C-type cytochromes can also play a role in the charge transfer across the Gram-positive bacterial cell envelope during metal reduction.13 Contact independent mechanisms (indirect) involve soluble redox-active mediators that shuttle electrons from the electron transport chain to the insoluble electron acceptor.24–26 Extracellular redox-active compounds, flavins and other quinones have been shown to play a major role in delivering electrons from the cellular metabolic systems to extracellular insoluble substrates via a diffusion-based shuttling electron transfer mechanism.15,24–28 A synthetic flavin biosynthesis pathway of Bacillus subtilis was heterologously expressed in Shewanella oneidensis MR-1, resulting in a current increase of about 15 times.29
Flavins regulate extracellular electron transfer (EET) by reducing electron acceptors through outer-membrane (OM) cytochrome C in Shewanella oneidensis.25,29 The bio-reduction of U(VI) by the Shewanella species can be catalyzed by FMN secreted from the cells.30 Due to the thicker cell wall (10 to 80 nm), more Gram-positive bacteria seem to adopt the independent EET mechanism. Flavin-mediated electron transfer has been found in Gram-positive bacteria.10,11,31 Therefore, Gram-positive bacteria are involved in direct electron transfer and as indirect shuttle mediators.32
Here, the EET mechanism was investigated on two new isolated electrochemically active Gram-positive bacteria: the Bacillus cereus strain DIF1 and the Rhodococcus ruber strain DIF2. Two chambered microbial fuel cells were constructed to study the electrochemical character of the two bacteria. Scanning electron microscopy was used to observe the morphology of the bacterial biofilm on the anode. Amperometric I–t curves, CV and HPLC were used to confirm the extracellular components secreted by bacteria.
2 Experimental methods
2.1 Microbial fuel cell configuration and operation
The bacterial strain Bacillus cereus strain DIF1 (deposited in CCTCC as M2018274) and the Rhodococcus ruber strain DIF2 (deposited in CCTCC as M2018274) were isolated and cultivated in our lab. Before inoculation, bacteria were harvested using refrigerated centrifugation and re-suspended in DM medium. Two-chambered electrochemical fuel cells were designed and fabricated using glass. The total working volume of one chamber was 150 mL. A carbon paper (2 cm × 2 cm) was used as the electrode after soaking overnight in acetone and ultrasonic cleaning with alcohol and deionized water.Nafion-117 (DuPont) was used as the proton exchange membrane (PEM). The anolyte was DM medium and the catholyte was 50 mmol L−1 K3[Fe(CN)6] dissolved in PBS buffer. A 1000 ohm resistor was connected between the anode and cathode. All microbial fuel cells were placed in an incubator at 25 °C in the dark. The bacterial growth was monitored daily by measuring the OD at 600 nm with a spectrophotometer (Shimadzu, UV-vis spectrophotometer, UV-2600).
2.2 Scanning electron microscopy (SEM)
A Zeiss Ultra Plus Field Emission Scanning Electron Microscope (SEM) was used to observe the morphological characteristics of the bacteria on carbon paper anodes pre-treated as follows. First, small pieces of the carbon paper from anodes were fixed in 2.5% glutaraldehyde solution for 2 hours, washed three times with 0.2 mM PBS buffer, and sequentially washed with deionized water three times. Next, they were dehydrated successively for 5 minutes in 50%, 70%, and 90% t-butanol mixed with ethanol. The samples were then dehydrated continuously 3 times with 100% t-butanol for 5 minutes before drying overnight.2.3 Bioelectrochemical experiments
A three-electrode configuration with an Ag/AgCl reference electrode (R0303, Tianjin) on a CHI600E (Chenhua, Shanghai) multichannel potentiostat was adapted to our experiment. Meanwhile, the anode served as the working electrode and the cathode served as the counter electrode. The cyclic voltammetry scan rate was 10 mV S−1 ranging from −0.7 V to 0.3 V. Amperometric I–t curves were performed in the two-chamber MFCs to evaluate the contribution of flavins (adding 1 μM RF and 1 μM FMN successively).2.4 Quantitative analysis of flavins by HPLC
A reversed-phase HPLC system (Alliance 2695, USA) was used to analyze the concentration of riboflavin. The samples were collected from the anode chamber and centrifuged at 5000 rpm for 10 min to obtain the supernatant. Filtration through a microporous membrane (pore diameter 0.22 um) was conducted before the concentration measurement. The reference standard samples are commercially available, including RF, FMN and FAD, which were the main representatives of flavins. The wavelengths of excitation/emission for fluorescence spectra acquisition were set at 525/468 nm, respectively. All samples were completed with a C18 column (Eclipse Plus, 4.6 × 250 mm, 5 μm) at a flow rate of 1 mL min−1. The mobile phase was a solution of methanol (chromatographic grade, phase A) and acetic acid (1%, chromatographic grade, phase B). The reference standard samples were detected with isocratic elution using a solution of methanol–water–acetic acid (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70
70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v).
1, v/v).
3 Results
3.1 Morphological characterization
The SEM morphological image of the Bacillus cereus strain DIF1 shows that they are short rod-like bacterial cells 2–3 μm long and 1 μm wide that assemble together. They have no flagella or cilia (Fig. 1A). The SEM image of the Rhodococcus ruber strain DIF2 on the anode shows bacterial cells that are smaller than the Bacillus cereus strain DIF1; they are 1–2 μm long and 0.5 μm wide and they have flagella (Fig. 1B).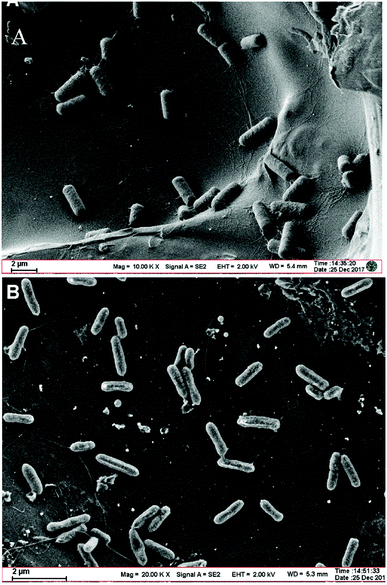 | ||
| Fig. 1 (A) Scanning electron micrograph of the Bacillus cereus strain DIF1 on a carbon paper anode. (B) Scanning electron micrograph of the Rhodococcus ruber strain DIF2 on a carbon paper anode. | ||
3.2 Cyclic voltammetry (CV) analysis of biofilms and anolytes
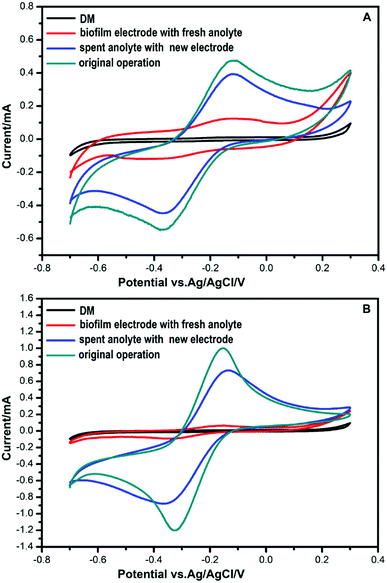
CV analysis was used to study the electrochemical behavior. Electrodes were UV-sterilized before being transferred to the culture medium. As can be seen from Fig. 2, no redox peaks are found when a DM medium without bacteria is used as the anolyte. Anolytes with bacteria in DM had an oxidation peak in the forward scan of the voltammogram at −0.13 V. During the reverse scan, a reduction peak was found at −0.36 V. These results indicated that redox activity was caused by bacteria that play roles in electrochemical activity.When a new anode was immersed in the spent anolyte with bacteria, the redox peaks were nearly as high as that of the original anode operated steadily in MFCs. This suggested that there might be redox components in the anolyte that were secreted by the bacteria. In contrast, weak redox peaks were obtained when a spent anode with a biofilm was used in a fresh anolyte. This indicated that bacteria accumulated on the spent electrodes. A small amount of secretion may attach and mediate extracellular electron transport. Contact-dependent direct electron transfer may also cause the weak redox peaks.
3.3 Electrochemical activity analysis of the flavins in anolyte
To investigate the components secreted by bacteria, further CV analysis was conducted by adding riboflavin (RF) solutions. A couple of obvious redox peaks were observed clearly in the CV curves (Fig. 3) at nearly the same potential. The oxidation peaks in the forward scan of the voltammogram were observed at −0.16 V. During the reverse scan, a reduction peak was found at −0.36 V. Then, 0.5 μm RF and 1 μm RF were separately added to the supernatant of the culture medium to verify the redox components. It could be seen that the potential of the redox peaks did not change. Since the potential of the peaks after adding riboflavin was almost consistent with that of the supernatant secreted by bacteria, we deduced that the redox components might be dissolved flavins. This result is in accordance with the finding that flavins mediate extracellular electron transfer that was reported by You et al.10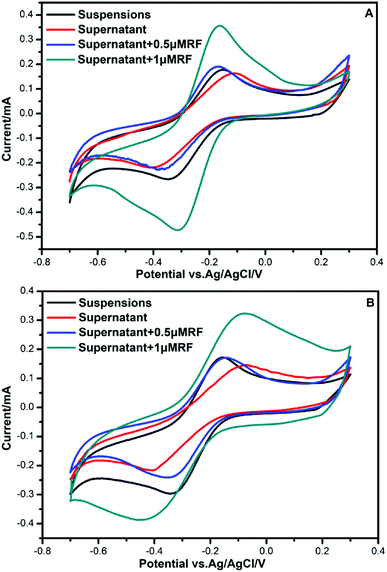 | ||
| Fig. 3 CVs for the different culture media. (A) Bacillus cereus strain DIF1. (B) Rhodococcus ruber strain DIF2. | ||
3.4 Amperometric I–t curves
Amperometric I–t curves showed current change with time of the MFCs inoculated by the strain DIF1 and strain DIF2 after adding RF (1 μM) and FMN (1 μM), and the addition points are indicated by arrows. When riboflavin was added to the three-electrode cell, MFCs produced maximum current outputs of 37.05 μA and 60.52 μA. Besides, the current outputs increased by about 1.48 times and 1.55 times (Fig. 4). The increasing current peaks (1.21 times and 1.31 times) were produced after adding FMN in succession (Fig. 4). We concluded that the strain DIF1 and strain DIF2 use flavins as the mediators of extracellular electron transfer in cell metabolism.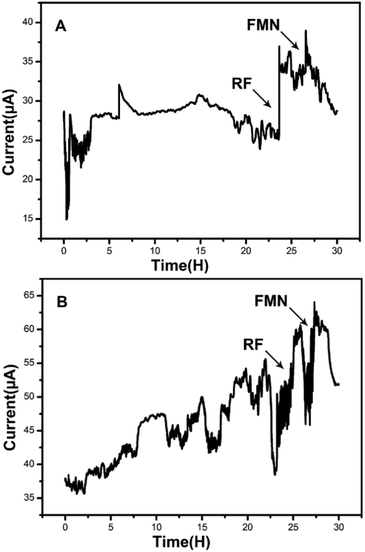 | ||
| Fig. 4 Amperometric I–t curves of the two bacteria when adding RF and FMN. (A) Bacillus cereus strain DIF1, (B) Rhodococcus ruber strain DIF2. | ||
3.5 HPLC chromatograms
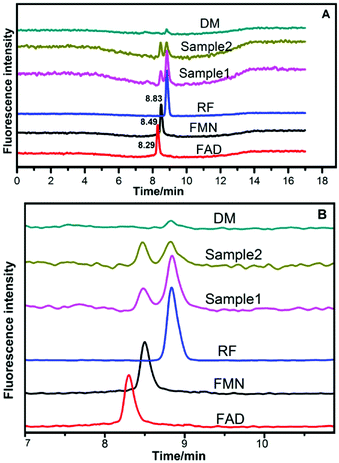
The HPLC chromatograms of RF, FMN, and FAD are shown in Fig. 5. Two samples of anolytes with components secreted by bacteria showed two obvious peaks, which were consistent with the peaks of the standard reagents of RF and FMN. The retention times in the spectra were 8.83 min, 8.50 min, and 8.30 min. The DM medium contained a very small amount of RF, but no FMN or FAD was found.3.6 HPLC analysis of flavins in anolytes
The bacterial growth was monitored daily by measuring OD with a spectrophotometer (SHIMADZU, UV-2600). Strains DIF1 and DIF2 were separately inoculated into the anode chamber of MFCs, which was placed in an incubator at 25 °C. The concentration curves of flavins and cell growth curves are clearly shown in Fig. 6. RF, FMN, and FAD were detected in MFC anolytes.11 In our experiments, only RF and FMN were detected. FAD was not detected during the experimental period. FMN began to increase at 36 h. Interestingly, FMN had a rapid increase when the cell growth reached the log phase. Meanwhile, RF began to slowly decrease. At about 168 h, the concentration of FMN from the B. cereus strain was even higher than that of RF.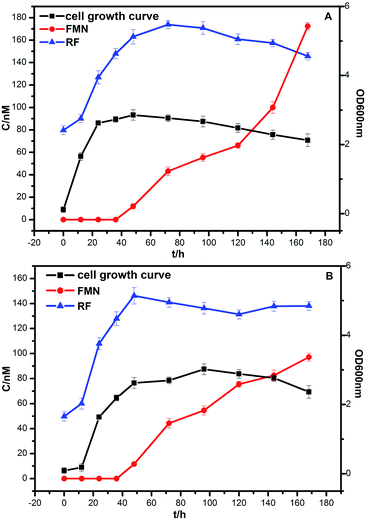 | ||
| Fig. 6 The growth curve of bacteria and the concentration curve of flavins. (A) Bacillus cereus strain DIF1, (B) Rhodococcus ruber strain DIF2. | ||
3.7 HPLC analysis of flavins inside and outside MFC
FMN and RF were also detected in the general DM medium without the MFC system. As indicated in Fig. 7, the two bacterial strains secrete FMN in the MFC system after the log phase. However, FMN could not be detected when the two strains were cultured in the general DM medium outside MFC. In Fig. 7, the HPLC peaks indicate RF and FMN in the log phase at different conditions. In MFC, the concentration of FMN increased clearly after the bacterial cell growth to the log phase, as shown in Fig. 6.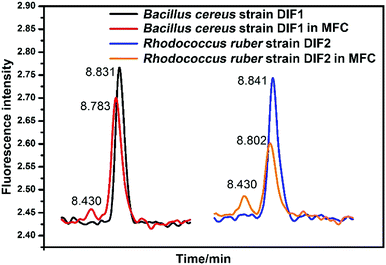 | ||
| Fig. 7 HPLC chromatograms of RF and FMN in anolytes compared with those in general medium. Peaks of FMN indicating the anolytes from MFC. | ||
4 Discussion
Most prior EET studies focus on Gram-negative bacteria. Electrochemically active Gram-positive bacteria were found in succession.10,11,13–15,33 Physiological and genomic evidence indicated that c-type cytochromes play a role in the charge transfer across the Gram-positive bacterial cell envelope during metal reduction.13 Most Gram-positive bacteria secrete shuttle mediators to transfer electrons.10,11,14,15 Flavins are one of the shuttle mediators secreted by Gram-positive bacteria.10,11,14 Recently, NAD was reported as a redox mediator for EET of Bacillus subtilis in a harsh environment.36 In the Bacillus species, bacteria could respond to different environments to maintain viability under extreme conditions.36 In our experiment, FMN was secreted in the MFC system under normal conditions. To determine whether these two strains can secrete NAD in extreme conditions, more experiments need to be conducted.In our results, flavins influenced the electron shuttle secreted by the two Gram-positive bacteria, as shown in Fig. 3. The redox peaks in Fig. 3 indicate that flavins are secreted by the two bacterial strains. Shewanella species were also shown to secrete flavins as electron mediators, but direct contact-dependent electron transfer was still the main pattern of electron transfer.14,24,25,32,34 In our experiment, when the anode with the biofilm was placed into a fresh anolyte, the redox activity was fairly weak. When a new anode was immersed in the spent anolyte, the redox peaks were obvious, as shown in Fig. 2. These results indicated that the shuttle mediator was the main electron transfer pattern in the strain DIF1 and strain DIF2. Flavins have been shown to be electron shuttles in the Bacillus species.10,11 However, the mechanism of electron transfer has not been studied in Rhodococcus species.33
The reduction peak was found at −0.36 V, which represented the presence of an electron shuttle. This was almost consistent with the value reported in the research for Shewanella woodyi, which was −0.38 V.37 When we added RF to the supernatant of the anolytes, it was shown that the potential of redox peaks almost did not change. This indicated an electron transfer pathway underlying flavins. The normal potential of flavins was reported at −0.41 V.11 The cell-free supernatant of Bacillus sp. WS-XY1 showed redox peaks around 0.41 V.11 The shift in the reduction peak in our result may be due to different anolyte conditions, such as different pH values.
In previous studies, flavins were found to be electron mediators in Gram-positive bacteria.10,11,35 However, the secreted FAD concentration was higher than that of FMN after cell growth at the log phase.10 In our experiments, FAD was not detected by HPLC (Fig. 5), but RF and FMN were detected. The RF concentration curves agreed with the bacterial cell growth curves (Fig. 6). This may indicate that RF has an effect on cell growth. FAD is mainly involved in intercellular metabolism; thus, FAD can be used as an indicator for cell lysis.10 FAD could not be detected in our experiment, which indicated there was no cell lysis. The two bacterial strains secreted FMN in the MFC system but not in the general medium, which revealed that FMN was the essential electron shuttle in the EET process (Fig. 7).
Flavin molecules have been found as endogenous electron shuttles that can interact with the electron acceptor to promote ideal organism growth.34 In our experiments, the FMN concentration increased after the growth of bacterial cells to the log phase, which indicated that FMN plays an important role in extracellular electron transfer in the strain DIF1 and strain DIF2. This may also be shown by the current increase after adding RF and FMN (Fig. 4). RF and FMN have ever been indicated as a function of time in the experiment of interactions between the redox reaction of single cells and bioluminescence of group communication via the EET pathway.37 FMN secreted by bacteria can also catalyze the bioreduction of U(VI).30 RF and FMN have specific binding sites in OM c-Cyts in Shewanella oneidensis MR-1 and by controlling the rate of electron transfer to surfaces, they highlight the potential roles of these flavin–cytochrome complexes.5 The flavin/OM c-Cyt interaction regulates the extent of extracellular electron transport coupled to intracellular metabolic activity.24 Flavins were secreted by Shewanella cells living at redox boundaries, where these mineral phases can be significant electron acceptors for growth.26 In Gram-positive bacteria, the related mechanism has not been observed and discussed. Therefore, more attention should be paid to the flavin-mediated electron transfer mechanism in Gram-positive bacteria.
5 Conclusions
The flavin-mediated EET of Gram-positive Bacillus cereus strain DIF1 and Rhodococcus ruber strain DIF2 was studied in MFC. Electrochemical activity was observed in the two Gram-positive bacteria. Cyclic voltammograms and HPLC analysis demonstrated that riboflavins were secreted into anolytes and contributed as electron mediators in both bacteria. Meanwhile, FMN was secreted as another electron mediator together with riboflavin. Indirect electron transfer mediated by flavins played an important role in the two Gram-positive bacteria, but direct electron transfer did not exist or was very weak. This finding will provide more evidence of Gram-positive bacteria secreted redox mediators, which is valuable for biogeochemical and bioelectrical processes.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was funded by State Key Laboratory of Paleontology and Stratigraphy, Nanjing Institute of Geology & Palaeontology CAS (No. 173103).References
- A. L. Neal, K. M. Rosso, G. G. Geesey, Y. A. Gorby and B. J. Little, Geochim. Cosmochim. Acta, 2003, 67(23), 4489–4503 CrossRef CAS.
- B. E. Logan, Nat. Rev. Microbiol., 2009, 7(5), 375–381 CrossRef CAS PubMed.
- L. Shi, H. Dong, G. Reguera, H. Beyenal, A. Lu, J. Liu, H. Q. Yu and J. K. Fredrickson, Nat. Rev. Microbiol., 2016, 14(10), 651–662 CrossRef CAS PubMed.
- D. R. Bond and D. R. Lovley, Appl. Environ. Microbiol., 2003, 69(3), 1548–1555 CrossRef CAS PubMed.
- A. Okamoto, S. Kalathil, X. D. K. Hashimoto, R. Nakamura and K. H. Nealson, Sci. Rep., 2014, 4, 5628 CrossRef CAS PubMed.
- K. H. Nealson and A. R. Rowe, Microb. Biotechnol., 2016, 9(5), 595–600 CrossRef PubMed.
- X. Y. Yong, Z. Y. Yan, H. B. Shen, J. Zhou, X. Y. Wu, L. J. Zhang, T. Zheng, M. Jiang, P. Wei, H. H. Jia and Y. C. Yong, Bioresour. Technol., 2017, 241, 1191–1196 CrossRef CAS PubMed.
- S. K. Chaudhuri and D. R Lovley, Nat. Biotechnol., 2003, 21(10), 1229–1232 CrossRef CAS PubMed.
- M. Li, M. Zhou, X. Tian, C. Tan, C. T. McDaniel, D. J. Hassett and T. Gu, Biotechnol. Adv., 2018, 36, 1316–1327 CrossRef CAS PubMed.
- L. X. You, L. D. Liu, Y. Xiao, Y. F. Dai, B. L. Chen, Y. X. Jiang and F. Zhao, Bioelectrochemistry, 2018, 119, 196–202 CrossRef CAS PubMed.
- S. Wu, Y. Xiao, L. Wang, Y. Zheng, K. Chang, Z. Zheng, Z. Yang, J. R. Varcoe and F. Zhao, Electrochim. Acta, 2014, 146, 564–567 CrossRef CAS.
- K. C. Wrighton, P. Agbo, F. Warnecke, K. A. Weber, E. L. Brodie, T. Z. DeSantis, P. Hugenholtz, G. L. Andersen and J. D. Coates, ISME J., 2008, 2(11), 1146–1156 CrossRef CAS PubMed.
- K. C. Wrighton, J. C. Thrash, R. A. Melnyk, J. P. Bigi, K. G. Byrne-Bailey, J. P. Remis, D. Schichnes, M. Auer, C. J. Chang and J. D. Coates, Appl. Environ. Microbiol., 2011, 77(21), 7633–7639 CrossRef CAS PubMed.
- V. R. Nimjea, C. Chen, C. Chen, J. Jean, A. S. Reddy, C. Fan, K. Pan, H. Liu and J. Chen, J. Power Sources, 2009, 190(2), 258–263 CrossRef.
- S. Freguia, M. Masuda, S. Tsujimura and K. Kano, Bioelectrochemistry, 2009, 76(1–2), 14–18 CrossRef CAS PubMed.
- E. Dalla Vecchia, E. I. Suvorova, J. Maillard and R. Bernier-Latmani, Geobiology, 2014, 12(1), 48–61 CrossRef CAS PubMed.
- C. A. Francis, K. L. Casciotti and B. M. Tebo, Arch. Microbiol., 2002, 178, 450–456 CrossRef CAS PubMed.
- J. D. Gregory, Y. E. Lee and B. M. Tebo, Appl. Environ. Microbiol., 2006, 72(5), 3184–3190 CrossRef PubMed.
- D. Ding and X. Sun, Genes, 2018, 9(1), E41 CrossRef PubMed.
- C. M. Paquete and R. O. Louro, Dalton Trans., 2010, 39(18), 4259–4266 RSC.
- S. J. Field, P. S. Dobbin, M. R. Cheesman, N. J. Watmough, A. J. Thomson and D. J. Richardson, J. Biol. Chem., 2000, 275(12), 8515–8522 CrossRef CAS PubMed.
- G. Reguera, FEMS Microbiol. Ecol., 2018, 94(7), fiy086 CrossRef CAS PubMed.
- D. J. Walker, R. Y. Adhikari, D. E. Holmes, J. E. Ward, T. L. Woodard, K. P. Nevin and D. R. Lovley, ISME J., 2018, 12(1), 48–58 CrossRef CAS PubMed.
- A. Okamoto, K. Hashimoto, K. H. Nealson and R. Nakamura, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(19), 7856–7861 CrossRef CAS PubMed.
- G. Hong and R. Pachter, J. Phys. Chem. B, 2016, 120(25), 5617–5624 CrossRef CAS PubMed.
- H. von Canstein, J. Ogawa, S. Shimizu and J. R. Lloyd, Appl. Environ. Microbiol., 2008, 74, 615–623 CrossRef CAS PubMed.
- C. Wu, Y. Cheng, B. Li, W. Li, D. B. Li and H. Yu, Bioresour. Technol., 2013, 136, 711–714 CrossRef CAS PubMed.
- A. Okamoto, K. Hashimoto and K. H. Nealson, Angew. Chem., Int. Ed. Engl., 2014, 53(41), 10988–10991 CrossRef CAS PubMed.
- Y. Yang, Y. Ding, Y. Hu, B. Cao, S. A. Rice, S. Kjelleberg and H. Song, ACS Synth. Biol., 2015, 4(7), 815–823 CrossRef CAS PubMed.
- Y. Suzuki, Y. Kitatsuji, T. Ohnuki and S. Tsujimura, Phys. Chem. Chem. Phys., 2010, 12(34), 10081–10087 RSC.
- H. L. Ehrlich, Geobiology, 2008, 6(3), 220–224 CrossRef CAS PubMed.
- E. Marsili, D. B. Baron, I. D. Shikhare, D. Coursolle, J. A. Gralnick and D. R. Bond, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 3968–3973 CrossRef CAS PubMed.
- P. Cheng, R. Shan, H. Yuan, L. Deng and Y. Chen, Bioresour. Technol., 2018, 267, 774–777 CrossRef CAS PubMed.
- A. Prokhorova, K. Sturm-Richter, A. Doetsch and J. Gescher, Appl. Environ. Microbiol., 2017, 83(6), e03033-16 CrossRef PubMed.
- S. H. Light, L. Su, R. Rivera-Lugo, J. A. Cornejo, A. Louie, A. T. Iavarone, C. M. Ajo-Franklin and D. A. Portnoy, Nature, 2018, 562(7725), 140–144 CrossRef CAS PubMed.
- L. X. Chen, C. L. Cao, S. H. Wang, J. R. Varcoe, R. C. T. Slade, C. A. Rossa and F. Zhao, iScience, 2019, 12, 260–269 CrossRef CAS PubMed.
- X. T. Tian, F. Zhao, L. X. You, X. Wu, Z. Y. Zheng, R. R. Wu, Y. X. Jiang and A. G. Sun, Phys. Chem. Chem. Phys., 2017, 19, 1746–1750 RSC.
| This journal is © The Royal Society of Chemistry 2019 |