Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Model peptide for anti-sigma factor domain HHCC zinc fingers: high reactivity toward 1O2 leads to domain unfolding†
Valentin
Chabert‡
 a,
Vincent
Lebrun‡
a,
Colette
Lebrun
a,
Vincent
Lebrun‡
a,
Colette
Lebrun
 b,
Jean-Marc
Latour
b,
Jean-Marc
Latour
 a and
Olivier
Sénèque
a and
Olivier
Sénèque
 *a
*a
aUniv. Grenoble Alpes, CNRS, CEA, BIG, LCBM (UMR 5249), F-38000 Grenoble, France. E-mail: olivier.seneque@cea.fr
bUniv. Grenoble Alpes, CEA, CNRS, INAC-SyMMES, F-38000 Grenoble, France
First published on 21st February 2019
Abstract
All organisms have to cope with the deleterious effects of reactive oxygen species. Some of them are able to mount a transcriptional response to various oxidative stresses, which involves sensor proteins capable of assessing the redox status of the cell or to detect reactive oxygen species. In this article, we describe the design, synthesis and characterization of Zn·LASD(HHCC), a model for the Zn(Cys)2(His)2 zinc finger site of ChrR, a sensor protein involved in the bacterial defence against singlet oxygen that belongs to the family of zinc-binding anti-sigma factors possessing a characteristic H/C–X24/25–H–X3–C–X2–C motif. The 46-amino acid model peptide LASD(HHCC) was synthetized by solid phase peptide synthesis and its Zn2+-binding properties were investigated using electronic absorption, circular dichroism and NMR. LASD(HHCC) forms a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex with Zn2+, namely Zn·LASD(HHCC), that adopts a well-defined conformation with the Zn2+ ion capping a 3-helix core that reproduces almost perfectly the fold of the ChrR in the vicinity of its zinc site. H2O2 reacts with Zn·LASD(HHCC) to yield a disulfide with a second order rate constant of 0.030 ± 0.002 M−1 s−1. Zn·LASD(HHCC) reacts rapidly with singlet oxygen to yield sulfinates and sulfonates. A lower limit of the chemical reaction rate constant between Zn·LASD(HHCC) and 1O2 was determined to be 3.9 × 106 M−1 s−1. Therefore, the Zn(Cys)2(His)2 site of Zn·LASD(HHCC) appears to be at least 5 times more reactive toward these two oxidants than that of a classical ββα zinc finger. Consequences for the activation mechanism of ChrR are discussed.
1 complex with Zn2+, namely Zn·LASD(HHCC), that adopts a well-defined conformation with the Zn2+ ion capping a 3-helix core that reproduces almost perfectly the fold of the ChrR in the vicinity of its zinc site. H2O2 reacts with Zn·LASD(HHCC) to yield a disulfide with a second order rate constant of 0.030 ± 0.002 M−1 s−1. Zn·LASD(HHCC) reacts rapidly with singlet oxygen to yield sulfinates and sulfonates. A lower limit of the chemical reaction rate constant between Zn·LASD(HHCC) and 1O2 was determined to be 3.9 × 106 M−1 s−1. Therefore, the Zn(Cys)2(His)2 site of Zn·LASD(HHCC) appears to be at least 5 times more reactive toward these two oxidants than that of a classical ββα zinc finger. Consequences for the activation mechanism of ChrR are discussed.
Introduction
Organisms living in an aerobic environment have to cope with the deleterious effects of oxidative stress, which is caused by a family of molecules produced from dioxygen in the living cell, the so-called Reactive Oxygen Species (ROS).1 ROS are classified into two categories according to their production pathways: Type I for those arising from electron(s) addition to O2 (e.g.: superoxide anion O2˙−, hydrogen peroxide H2O2, hypochlorous acid HOCl, hydroxyl radical HO˙…) and Type II for electronically excited states of dioxygen arising from a physical activation of the ground (triplet) state (3O2).2–4 The lowest energy excited state of O2, commonly called singlet oxygen (1O2), is produced mainly in photosynthetic organisms. These organisms, as well as others like mammalians, have been shown to elicit a specific response to 1O2, demonstrating that sensing pathways exist for this ROS.5–7 One of the best characterized is that of Rhodobacter sphaeroides. It involves the protein σE, a group IV sigma factor up-regulating the expression of the response against 1O2 stress.8–11 σE is negatively regulated by its cognate anti-sigma factor, ChrR. In normal conditions, the anti-sigma factor sequesters its cognate sigma factor (Fig. 1A). Under 1O2 stress, the anti-sigma factor undergoes a structural change and releases its sigma factor, which can then bind RNA polymerase, up-regulating the expression of specific genes involved in the response to 1O2 stress. The anti-sigma factor interacts with the sigma factor mainly via its N-terminal domain, named anti-sigma factor domain (ASD).10 It is estimated that 33% of the group IV sigma factors are regulated by an anti-sigma factor containing an anti-sigma factor domain. Among the predicted cytoplasmic group IV anti-sigma factors, 92% contain a conserved zinc binding motif H–X3–C–X2–C, classifying them in the zinc-binding anti-sigma factor (ZAS) subfamily.8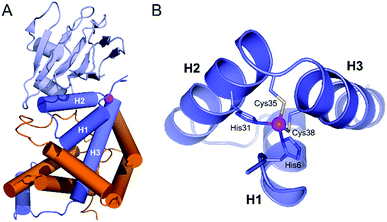 | ||
| Fig. 1 (A) Crystallographic structure of the ChrR–σE complex (pdb 2Z2S)8 showing the sigma factor σE in orange and the anti-sigma factor ChrR in blue with anti-sigma factor domain in dark blue and cupin-like domain in light blue. The zinc ion is shown in magenta. (B) View of the anti-sigma factor domain zinc finger site embedded within helices H1, H2 and H3, showing Zn2+-binding histidines and cysteines of the H–X24–H–X3–C–X2–C motif. | ||
In 2011, Andreini et al. classified 93% of the 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 763 known structures of protein zinc sites into a minimum set of structural motifs.12 The two existing structures of ChrR10 could not enter in any of the families defined by Andreini, which demonstrates the singularity of the zinc finger site of their anti-sigma factor domain. ChrR contains an anti-sigma factor domain featuring a HHCC zinc finger site with a H–X24–H–X3–C–X2–C motif (Fig. 1B). Interestingly, RslA and RsrA, two ChrR homologs featuring also a zinc finger site – HHCC within a H–X24–H–X3–C–X2–C motif for RslA13 and CHCC within a C–X25–H–X3–C–X2–C motif for RsrA14 – are sensitive to oxidative conditions and trigger a response against oxidative stress.13–16 Noteworthy, the zinc site of RsrA is described as a redox switch, acting as a detection module for disulfide stress. A similar role was proposed for the zinc site of RslA to sense oxidative stress.13–16 It has been proposed that the oxidation of the ZAS protein zinc finger induces Zn2+ release and unfolding of the anti-sigma factor domain, leading to dissociation of the ZAS protein from its cognate sigma factor. However, it was recently demonstrated that oxidation of zinc-binding cysteine of the ZAS zinc finger is not always sufficient to dissociate the anti-sigma factor–sigma factor complex.17 Similarly, it has been hypothesized that the zinc site of ChrR could sense 1O2via zinc-bound thiol oxidation inducing dissociation of the anti-sigma factor domain from σE, which is supported by the demonstration that (i) formation of the zinc finger site in the N-ter domain anti-sigma factor domain of ChrR is essential for the ChrR–σE complex formation, and (ii) the Zn-loaded anti-sigma factor domain alone is sufficient to sequester σE. Additionally, it was also shown that the C-terminal cupin-like domain of ChrR is required for 1O2 response in vivo,10,11 but there is no data ruling out the involvement of the zinc finger site – and its oxidation – in these studies. The full mechanism of activation of ChrR by 1O2 remains to be elucidated.
763 known structures of protein zinc sites into a minimum set of structural motifs.12 The two existing structures of ChrR10 could not enter in any of the families defined by Andreini, which demonstrates the singularity of the zinc finger site of their anti-sigma factor domain. ChrR contains an anti-sigma factor domain featuring a HHCC zinc finger site with a H–X24–H–X3–C–X2–C motif (Fig. 1B). Interestingly, RslA and RsrA, two ChrR homologs featuring also a zinc finger site – HHCC within a H–X24–H–X3–C–X2–C motif for RslA13 and CHCC within a C–X25–H–X3–C–X2–C motif for RsrA14 – are sensitive to oxidative conditions and trigger a response against oxidative stress.13–16 Noteworthy, the zinc site of RsrA is described as a redox switch, acting as a detection module for disulfide stress. A similar role was proposed for the zinc site of RslA to sense oxidative stress.13–16 It has been proposed that the oxidation of the ZAS protein zinc finger induces Zn2+ release and unfolding of the anti-sigma factor domain, leading to dissociation of the ZAS protein from its cognate sigma factor. However, it was recently demonstrated that oxidation of zinc-binding cysteine of the ZAS zinc finger is not always sufficient to dissociate the anti-sigma factor–sigma factor complex.17 Similarly, it has been hypothesized that the zinc site of ChrR could sense 1O2via zinc-bound thiol oxidation inducing dissociation of the anti-sigma factor domain from σE, which is supported by the demonstration that (i) formation of the zinc finger site in the N-ter domain anti-sigma factor domain of ChrR is essential for the ChrR–σE complex formation, and (ii) the Zn-loaded anti-sigma factor domain alone is sufficient to sequester σE. Additionally, it was also shown that the C-terminal cupin-like domain of ChrR is required for 1O2 response in vivo,10,11 but there is no data ruling out the involvement of the zinc finger site – and its oxidation – in these studies. The full mechanism of activation of ChrR by 1O2 remains to be elucidated.
We have previously shown that small metallopeptides modelling zinc finger sites are useful tools to gain interesting insights into the reactivity of zinc fingers toward ROS at the molecular level (oxidation products, kinetic rates).18–20 In particular, we have shown that a Zn(Cys)4 zinc finger of the treble clef family of zinc fingers could be efficiently oxidized by 1O2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21 whereas 1O2 oxidation of the Zn(Cys)2(His)2 site of classical ββα zinc fingers is far less efficient.22 This questions the role of the Zn(Cys)2(His)2 zinc finger in ChrR. To address this question, we developed a 46-amino acid peptide modelling the anti-sigma factor domain zinc finger site of ChrR and we studied its reactivity towards H2O2 and 1O2. We show that its oxidation leads to Zn2+ release and peptide unfolding. Remarkably, the cysteines of this uncommon Zn(Cys)2(His)2 site found in ZAS are rapidly oxidized by 1O2 compared to classical ββα zinc fingers, in agreement with their putative involvement in 1O2 detection.
21 whereas 1O2 oxidation of the Zn(Cys)2(His)2 site of classical ββα zinc fingers is far less efficient.22 This questions the role of the Zn(Cys)2(His)2 zinc finger in ChrR. To address this question, we developed a 46-amino acid peptide modelling the anti-sigma factor domain zinc finger site of ChrR and we studied its reactivity towards H2O2 and 1O2. We show that its oxidation leads to Zn2+ release and peptide unfolding. Remarkably, the cysteines of this uncommon Zn(Cys)2(His)2 site found in ZAS are rapidly oxidized by 1O2 compared to classical ββα zinc fingers, in agreement with their putative involvement in 1O2 detection.
Results
Peptide design
To date, two crystal structures of anti-sigma factor domain-containing proteins displaying a zinc finger site (HHCC type) have been elucidated: ChrR10 and RslA.13 Anti-sigma factor domains display four α-helices and among them, three are involved in the constitution of the zinc finger site, which is displayed in Fig. 1B. These three helices (H1, H2 and H3) are held together by the chelation of a Zn2+ ion and by closely packed hydrophobic side chains within the heart of the 3-helix core. Helices H2 and H3 (Fig. 1B) form a knuckle bearing the H–X3–C–X2–C conserved motif, coordinating the Zn2+ ion. The fourth zinc ligand is a histidine residue located at the N-terminus resulting in an overall H–X24–H–X3–C–X2–C Zn2+-binding motif (Fig. 2). In order to reproduce the reactivity of a given zinc finger site, it is important to perfectly reproduce the various features of the site that can influence its reactivity. This includes the coordination sphere of the Zn2+ ion, the folding of the peptide around and the hydrogen bonds that are established with the Zn2+-bound sulfurs.18,20,23 It has been shown with RsrA that the anti-sigma factor domain can adopt different folds depending whether it is bound to its sigma-factor or not.17 Since our aim is to better understand if the reactivity of ChrR's zinc site could play a role in the sensing of 1O2, we aim here at reproducing the fold as complexed with σE. Hydrophobic cores proximal to the zinc finger site can be important to enhance Zn2+ affinity to the model peptide and to ensure its proper folding.24 For these reasons, we decided to include the three helices in our model. | ||
| Fig. 2 Sequence alignment of the N-terminal domain of ChrR (top) and the model peptides presented in this article, Zn2+-binding amino acids underlined. | ||
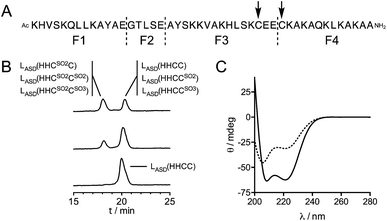
The design of the model peptides was based on the sequence and X-ray crystallographic structure of ChrR anti-sigma factor domain (Fig. 2),10 which was re-engineered as follows. (i) In the HH motif present at the N-terminus, only the second residue is involved in Zn2+ coordination as part of the canonical H–X24–H–X3–C–X2–C motif. In order to avoid scrambling of Zn2+ ligand, the first histidine of the HH motif was changed for a lysine. (ii) Helices H1, H2 and H3 present several hydrophobic side chains that point toward the exterior of the structure because they are involved in the interaction with the sigma factor σE. In order to favour the correct fold of our model peptide and to ensure solubility, these hydrophobic amino acids were randomly changed for polar amino acids (E, Q or K). (iii) As β-branched amino acids such as threonine or valine may destabilize α-helices,25 two threonines located in helices H1 and H2, and pointing toward the exterior of the structure were changed for lysines. Similarly, glycines in helix H3, which may destabilize a helical fold, were changed for glutamine and alanines. (iv) The C-terminus of the peptide was amidated to remove the carboxylate charge that could destabilize the C-terminus of helix H3.26 (v) Non-essential aspartates and arginines were changed for glutamates and lysines to avoid side reactions during solid phase peptide synthesis (SPPS) and maximize synthesis yields. This led to peptide L1, whose sequence is displayed in Fig. 2. As a first try to model the anti-sigma factor domain zinc finger, this 46-amino acid peptide was synthetized by SPPS. Unfortunately, when dissolved in various buffers at pH around 7, L1 precipitated when Zn2+ was added. Mapping charged amino acids of the L1 sequence onto the 3-helix motif of ChrR using Pymol27 revealed that most of the negatively charged glutamate residues are clustered on one face of the structure whereas positively charged lysines are clustered on another face. Suspecting that this could be the reason for precipitation of the Zn2+ complex, the charged amino acids were re-distributed in the sequence for random disposition onto the 3-helix surface. This led to peptide LASD(HHCC) (Fig. 2), which was synthetized by SPPS. Note that four pseudoproline dipeptides28 were used to avoid aggregation of the elongating chain during synthesis (see ESI† for details).
Zinc binding and folding properties
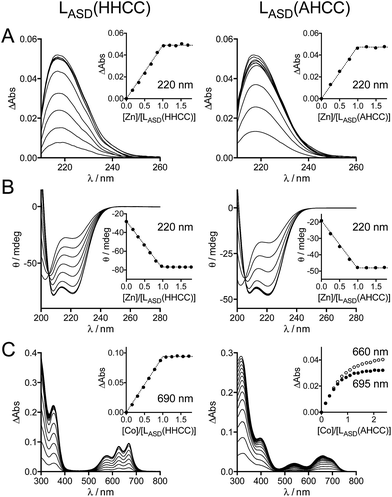
The metal binding properties of LASD(HHCC) were investigated by combination of UV-Vis absorption, circular dichroism (CD) and NMR. Titrations monitored by UV absorption were used to determine the stoichiometry of the peptide–metal complexes. Upon addition of Zn2+ in a buffered peptide solution, an increasing absorption band is observed at ca. 220 nm, which corresponds to Cys·S− → Zn2+ ligand-to-metal charge transfer (LMCT) transitions (Fig. 3A).24 This signal increases linearly up to one equivalent of zinc and plateaus afterwards (inserts Fig. 3A). This indicates the formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex only, i.e. Zn·LASD(HHCC). The intensity of this band is Δε = 6600 M−1 cm−1, which is in agreement with two zinc-bound cysteinates.20,24,29 To gain further insight into the coordination sphere of the Zn2+ ion, LASD(HHCC) was titrated with Co2+, a Zn2+ ion surrogate commonly used to probe its geometry and coordination sphere. Absorption bands characteristic of Cys·S− → Co2+ LMCT (in the range 220–400 nm) and d–d (in the range 500–700 nm) transitions appear upon coordination of Co2+ by LASD(HHCC) (Fig. 3C). Similarly to the Zn2+ titration, only the 1
1 complex only, i.e. Zn·LASD(HHCC). The intensity of this band is Δε = 6600 M−1 cm−1, which is in agreement with two zinc-bound cysteinates.20,24,29 To gain further insight into the coordination sphere of the Zn2+ ion, LASD(HHCC) was titrated with Co2+, a Zn2+ ion surrogate commonly used to probe its geometry and coordination sphere. Absorption bands characteristic of Cys·S− → Co2+ LMCT (in the range 220–400 nm) and d–d (in the range 500–700 nm) transitions appear upon coordination of Co2+ by LASD(HHCC) (Fig. 3C). Similarly to the Zn2+ titration, only the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Co·LASD(HHCC) complex is detected. The wavelengths (576, 627 and 670 nm) and intensity (ε = 620 M−1 cm−1 at 627 nm, i.e. ε > 300 M−1 cm−1) of the d–d transitions are in agreement with a tetrahedral Co(Cys)2(His)2 site.20,24,30,31 From these data, we can reasonably infer the formation of a tetrahedral 1
1 Co·LASD(HHCC) complex is detected. The wavelengths (576, 627 and 670 nm) and intensity (ε = 620 M−1 cm−1 at 627 nm, i.e. ε > 300 M−1 cm−1) of the d–d transitions are in agreement with a tetrahedral Co(Cys)2(His)2 site.20,24,30,31 From these data, we can reasonably infer the formation of a tetrahedral 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Zn·LASD(HHCC) complex with (Cys)2(His)2 coordination. Thereafter, the folding of the peptide was investigated by CD (Fig. 3B). The CD spectrum of the metal-free LASD(HHCC) shows an intense negative signal with minimum at ca. 200 nm and a shoulder at ca. 222 nm. This corresponds to a random coil peptide with a small content of helical fold. In agreement with absorption studies, upon titration of LASD(HHCC) by Zn2+, evolution of the CD spectrum is observed up to 1 eq. Zn2+ with a clean isodichroic point at 205 nm, thereby confirming the formation of a complex with 1
1 Zn·LASD(HHCC) complex with (Cys)2(His)2 coordination. Thereafter, the folding of the peptide was investigated by CD (Fig. 3B). The CD spectrum of the metal-free LASD(HHCC) shows an intense negative signal with minimum at ca. 200 nm and a shoulder at ca. 222 nm. This corresponds to a random coil peptide with a small content of helical fold. In agreement with absorption studies, upon titration of LASD(HHCC) by Zn2+, evolution of the CD spectrum is observed up to 1 eq. Zn2+ with a clean isodichroic point at 205 nm, thereby confirming the formation of a complex with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry only. The spectrum of Zn·LASD(HHCC) is characteristic of a peptide with major helix content, as expected for this model. In order to determine the Zn2+ binding constant of LASD(HHCC) at pH 7.0, KZnLASD(HHCC) = [Zn·LASD(HHCC)]/([Zn2+][LASD(HHCC)]), CD titrations were performed in competition with EDTA and TPEN (1 eq.), two high-affinity Zn2+ chelators (KZnEDTA = 1013.1 M−1 and KZnTPEN = 1014.9 M−1 at pH 7.0). In the case of EDTA, ca. 95% of Zn2+ is bound to LASD(HHCC) after addition of 1.0 eq. Zn2+vs. peptide (ESI†), indicating that LASD(HHCC) binds Zn2+ tighter than EDTA but also that EDTA is not suitable for precise determination of the binding constant.32,33 In the case of TPEN, we noticed a slow precipitation of the Zn2+-free peptide in presence of this chelator, which precluded any competition experiment. Thus, only a lower estimate of the binding constant can be drawn from the EDTA competition, that is KZnLASD(HHCC) ≥ 1015.0 M−1.
1 stoichiometry only. The spectrum of Zn·LASD(HHCC) is characteristic of a peptide with major helix content, as expected for this model. In order to determine the Zn2+ binding constant of LASD(HHCC) at pH 7.0, KZnLASD(HHCC) = [Zn·LASD(HHCC)]/([Zn2+][LASD(HHCC)]), CD titrations were performed in competition with EDTA and TPEN (1 eq.), two high-affinity Zn2+ chelators (KZnEDTA = 1013.1 M−1 and KZnTPEN = 1014.9 M−1 at pH 7.0). In the case of EDTA, ca. 95% of Zn2+ is bound to LASD(HHCC) after addition of 1.0 eq. Zn2+vs. peptide (ESI†), indicating that LASD(HHCC) binds Zn2+ tighter than EDTA but also that EDTA is not suitable for precise determination of the binding constant.32,33 In the case of TPEN, we noticed a slow precipitation of the Zn2+-free peptide in presence of this chelator, which precluded any competition experiment. Thus, only a lower estimate of the binding constant can be drawn from the EDTA competition, that is KZnLASD(HHCC) ≥ 1015.0 M−1.
In order to confirm that the N-terminal histidine, which is remote from the core H–X3–C–X2–C binding motif in the sequence, is bound to the Zn2+ ion in our Zn·LASD(HHCC) model, a peptide variant with the N-terminal histidine replaced by an alanine, namely LASD(AHCC), was synthesized (Fig. 2 and ESI†). Upon Zn2+ titration, the formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex only is evidenced by both UV-Vis absorption and CD spectroscopies (Fig. 3A and B). The spectral features are very similar for both peptides, including LMCT absorption and CD spectra, either in their Zn-free and Zn-loaded forms. The intensity of the LMCT band (Δε = 8400 M−1 cm−1) is compatible with two zinc-bound cysteinates. Noteworthy, the CD spectrum of Zn·LASD(AHCC) is very similar to that of Zn·LASD(HHCC), indicating a similar helix content in the Zn-loaded form. Regarding Co2+ binding, analysis of the UV-Vis Co2+ titration of LASD(AHCC) shows a completely different d–d transition pattern compared to LASD(HHCC) with a two-step growing phase (the bands at 660 and 695 nm have the same intensity at the beginning of the titration (<0.5 eq.), then the band at 655 nm becomes the most intense, Fig. 3C), indicating the formation of both 2
1 complex only is evidenced by both UV-Vis absorption and CD spectroscopies (Fig. 3A and B). The spectral features are very similar for both peptides, including LMCT absorption and CD spectra, either in their Zn-free and Zn-loaded forms. The intensity of the LMCT band (Δε = 8400 M−1 cm−1) is compatible with two zinc-bound cysteinates. Noteworthy, the CD spectrum of Zn·LASD(AHCC) is very similar to that of Zn·LASD(HHCC), indicating a similar helix content in the Zn-loaded form. Regarding Co2+ binding, analysis of the UV-Vis Co2+ titration of LASD(AHCC) shows a completely different d–d transition pattern compared to LASD(HHCC) with a two-step growing phase (the bands at 660 and 695 nm have the same intensity at the beginning of the titration (<0.5 eq.), then the band at 655 nm becomes the most intense, Fig. 3C), indicating the formation of both 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Co/peptide species during the titration, with tetrahedral geometry as attested by the ε values above 300 M−1 cm−1 for the d–d transitions. Additionally, no plateau is observed after 1.0 eq., indicating a less stable 1
1 Co/peptide species during the titration, with tetrahedral geometry as attested by the ε values above 300 M−1 cm−1 for the d–d transitions. Additionally, no plateau is observed after 1.0 eq., indicating a less stable 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex compared to Co·LASD(HHCC). Finally, the binding affinity of LASD(AHCC) for Zn2+ was assessed by competition with EDTA monitored by CD. When a 1
1 complex compared to Co·LASD(HHCC). Finally, the binding affinity of LASD(AHCC) for Zn2+ was assessed by competition with EDTA monitored by CD. When a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 LASD(AHCC)/EDTA/Zn2+ mixture is prepared, ca. 25% of Zn2+ is bound to LASD(AHCC) versus 95% for LASD(HHCC) in the same conditions (ESI†). This corresponds to a value of ca. 1012 for KZnLASD(AHCC), indicating that the replacement of the N-terminal histidine by an alanine lowers the Zn2+ affinity by at least 3 orders of magnitude.
1 LASD(AHCC)/EDTA/Zn2+ mixture is prepared, ca. 25% of Zn2+ is bound to LASD(AHCC) versus 95% for LASD(HHCC) in the same conditions (ESI†). This corresponds to a value of ca. 1012 for KZnLASD(AHCC), indicating that the replacement of the N-terminal histidine by an alanine lowers the Zn2+ affinity by at least 3 orders of magnitude.
Further insights into the structure of Zn·LASD(HHCC) and Zn·LASD(AHCC) were obtained by 1H NMR. The 1D 1H NMR spectra of metal-free LASD(HHCC) and LASD(AHCC) in H2O/D2O 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 display broad peaks with amide NH in the range 7.2–8.5 ppm indicating that these peptides are mostly random coil, in agreement with CD. Regarding Zn2+ complexes, the 1H NMR spectrum of Zn·LASD(HHCC) displays sharp amide NH resonances spread over a wider range from 6.9 to 9.3 ppm (Fig. 4A, top, and ESI†). Many of them present 3JHN,Hα < 6 Hz indicative of helical folding (Table S2 and Fig. S4 of ESI†). The 2D NOESY spectrum shows numerous correlation peaks corresponding to non-sequential NOEs that are characteristic of helical folding (Fig. S4 of ESI†). Additionally, several long-range NOEs between hydrophobic amino acid remote in the sequence indicate the formation of a hydrophobic core. This suggests the formation of a Zn2+ complex with a well-defined stable conformation. On the contrary, the 1H NMR spectrum of Zn·LASD(AHCC) (Fig. 4A, bottom) shows very broad resonances in the amide NH region although this complex displays a similar helix content as Zn·LASD(HHCC) as judged from its CD spectrum. This suggests a conformational equilibrium for Zn·LASD(AHCC). Indeed, the coordination of the N-terminal histidine to the Zn2+ ion is not necessary to fold the three helices, as indicated by CD but it plays a major role in freezing the conformation of the peptide in HHCC variant. Finally, the structure of Zn·LASD(HHCC) was calculated using X-PLOR with 466 H–H distance constraints (142 intraresidue, 138 sequential and 186 medium- and long-range) extracted from the NOESY spectrum and 35 ϕ dihedral constraints derived from 3JHN,Hα values. The superimposition of the ten lowest energy structures, which is depicted on Fig. 4B shows that Zn·LASD(HHCC) adopts a well-defined conformation with three helices (the backbone root mean square deviation over the ten structures is 0.83 Å). The zinc finger site caps this three-helix domain (Fig. 4C). The superimposition of the lowest energy structure of Zn·LASD(HHCC) with the zinc finger site taken from the crystallographic structure of ChrR anti-sigma factor domain (Fig. 4D) shows that the model reproduces almost perfectly the fold of the native protein, including helices H1, H2 and H3 as well as the loops between the helices. To summarize, LASD(HHCC) is able to bind only one Zn2+ ion to form a tetrahedral Zn(Cys)2(His)2 site that folds the peptide into a unique 3-helix conformation reproducing almost perfectly that of ChrR anti-sigma factor domain.
1 display broad peaks with amide NH in the range 7.2–8.5 ppm indicating that these peptides are mostly random coil, in agreement with CD. Regarding Zn2+ complexes, the 1H NMR spectrum of Zn·LASD(HHCC) displays sharp amide NH resonances spread over a wider range from 6.9 to 9.3 ppm (Fig. 4A, top, and ESI†). Many of them present 3JHN,Hα < 6 Hz indicative of helical folding (Table S2 and Fig. S4 of ESI†). The 2D NOESY spectrum shows numerous correlation peaks corresponding to non-sequential NOEs that are characteristic of helical folding (Fig. S4 of ESI†). Additionally, several long-range NOEs between hydrophobic amino acid remote in the sequence indicate the formation of a hydrophobic core. This suggests the formation of a Zn2+ complex with a well-defined stable conformation. On the contrary, the 1H NMR spectrum of Zn·LASD(AHCC) (Fig. 4A, bottom) shows very broad resonances in the amide NH region although this complex displays a similar helix content as Zn·LASD(HHCC) as judged from its CD spectrum. This suggests a conformational equilibrium for Zn·LASD(AHCC). Indeed, the coordination of the N-terminal histidine to the Zn2+ ion is not necessary to fold the three helices, as indicated by CD but it plays a major role in freezing the conformation of the peptide in HHCC variant. Finally, the structure of Zn·LASD(HHCC) was calculated using X-PLOR with 466 H–H distance constraints (142 intraresidue, 138 sequential and 186 medium- and long-range) extracted from the NOESY spectrum and 35 ϕ dihedral constraints derived from 3JHN,Hα values. The superimposition of the ten lowest energy structures, which is depicted on Fig. 4B shows that Zn·LASD(HHCC) adopts a well-defined conformation with three helices (the backbone root mean square deviation over the ten structures is 0.83 Å). The zinc finger site caps this three-helix domain (Fig. 4C). The superimposition of the lowest energy structure of Zn·LASD(HHCC) with the zinc finger site taken from the crystallographic structure of ChrR anti-sigma factor domain (Fig. 4D) shows that the model reproduces almost perfectly the fold of the native protein, including helices H1, H2 and H3 as well as the loops between the helices. To summarize, LASD(HHCC) is able to bind only one Zn2+ ion to form a tetrahedral Zn(Cys)2(His)2 site that folds the peptide into a unique 3-helix conformation reproducing almost perfectly that of ChrR anti-sigma factor domain.
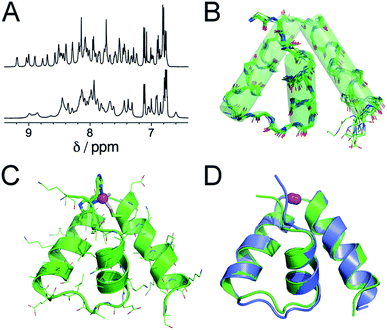 | ||
Fig. 4 (A) 1H NMR spectra (500 MHz, H2O/D2O 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 pH 6.4, 298 K) of Zn·LASD(HHCC) (top) and Zn·LASD(AHCC) (bottom). (B) Superimposition of the 10 lowest energy structures of Zn·LASD(HHCC) calculated using X-PLOR with NMR-derived distance and dihedral angle constraints. (C) Lowest energy structure of Zn·LASD(HHCC). (D) Superimposition of the solution structure of Zn·LASD(HHCC) (green) and the corresponding sub-domain of the crystallographic structure of ChrR (blue, pdb 2Z2S).10 The Zn2+ ion is shown in magenta. 1 pH 6.4, 298 K) of Zn·LASD(HHCC) (top) and Zn·LASD(AHCC) (bottom). (B) Superimposition of the 10 lowest energy structures of Zn·LASD(HHCC) calculated using X-PLOR with NMR-derived distance and dihedral angle constraints. (C) Lowest energy structure of Zn·LASD(HHCC). (D) Superimposition of the solution structure of Zn·LASD(HHCC) (green) and the corresponding sub-domain of the crystallographic structure of ChrR (blue, pdb 2Z2S).10 The Zn2+ ion is shown in magenta. | ||
Oxidation of Zn·LASD(HHCC) by H2O2
The reactivity of Zn·LASD(HHCC) toward H2O2 and 1O2 was investigated in order to assess the propensity of the anti-sigma factor domain zinc finger site to be oxidized by these two oxidants in comparison with other zinc fingers. The reaction of Zn·LASD(HHCC) (20 μM) with H2O2 (1 mM) in phosphate buffer (10 mM, pH 7.0) was monitored by CD (Fig. S3C of ESI†). This reaction is slow and after 15 h, the CD spectrum resembles that of zinc-free LASD(HHCC), indicating that the peptide unfolds upon reaction with H2O2. The product of the reaction was identified by ESI/MS as a disulfide (loss of two mass units). A similar CD spectrum was obtained by reacting Zn·LASD(HHCC) with 2.5 eq. HOCl, a more efficient oxidant for zinc-bound thiolates known to form disulfides.34 The kinetics of the reaction of Zn·LASD(HHCC) with H2O2 at 298 K was determined using previously described procedures,19 monitoring either the loss of the LMCT absorption in the UV or zinc release by using 4-(2-pyridylazo)resorcinol (PAR), which forms the Zn(PAR)2 complex with intense absorption in the visible when Zn2+ is released from the peptide upon oxidation. The PAR assay is not recommended to follow the oxidation of Zn(Cys)3(His) or Zn(Cys)4 because partially oxidized peptides displaying a reduced cysteine may retain Zn2+,19 but for this Zn(Cys)2(His)2 zinc finger peptide, both methods gave the same result. In condition of excess H2O2, a mono-exponential evolution of the absorption signal (either LMCT or Zn(PAR)2 band at 220 and 494 nm, respectively) is observed, indicating an apparent pseudo-first order reaction. Varying the concentration of H2O2 revealed a linear dependence on [H2O2] of the apparent first-order rate constant kobs (Fig. S3 of ESI†). Thus, the rate-determining step of the reaction is second order, first order in H2O2 and first order in Zn·LASD(HHCC), as previously observed for other zinc fingers.18–20 Indeed, the rate-determining step corresponds to the nucleophilic attack of H2O2 by the zinc-bound thiolate and its rate is given by r = k × [H2O2][Zn·LASD(HHCC)] with k = 0.030 ± 0.002 M−1 s−1 at 297 K.Oxidation of Zn·LASD(HHCC) by 1O2
In previous studies,21,22 we have shown that the reaction of Zn(Cys)4 and Zn(Cys)2(His)2 zinc finger models with 1O2 yields sulfinate species as major products, and disulfides in a lesser extent. Additionally, in a classical ββα Zn(Cys)2(His)2 zinc finger, Zn2+ coordination inhibits photooxidation of histidines. Oxidation of Zn·LASD(HHCC) by 1O2 was investigated as previously described for other Zn(Cys)4 and Zn(Cys)2(His)2 zinc finger models:21,22 the oxidation products were identified by combination of HPLC and ESI-MS analyses and the reaction rate was assessed in competition experiments with a reference compound. Rose bengal or methylene blue were used as photosensitizers to produce 1O2 in this study. Zn·LASD(HHCC) was photooxidized in D2O buffered with phosphate or ammonium acetate. HPLC analyses were performed with acidic eluent (0.1% TFA) so that Zn2+ is removed from the peptide during analysis. The HPLC chromatogram of a solution of Zn·LASD(HHCC) containing the photosensitizer but maintained in the dark showed a single peak at tR = 20.3 min corresponding to the reduced peptide LASD(HHCC) (Fig. 5B). Upon irradiation, a second peak appears at a shorter retention time (tR = 18.1 min). Prolongated irradiation shows an increase of the peak at 18.1 min at the expense of the one at 20.3 min. The main product detected by ESI-MS analysis of the crude corresponds to the addition of two oxygen atoms to the peptide, suggesting the formation of sulfinates. Other peaks corresponding to the addition of three and four oxygen atoms are also observed. The two HPLC peaks were collected, digested with glutamate carboxypeptidase (GluC, a peptidase that cleaves peptides at carboxylic side of glutamates or aspartates), and digestion mixtures were analysed by ESI-MS (Fig. 5A and Table S1†). GluC digestion of LASD(HHCC) can give four fragments: Ac-KHVSKQLLKAYAE (F1), GTLSE (F2), AYSKKVAKHLSKC31E(E) (F3) and C34KAKAQKLKAKAA-NH2 (F4). Digestion of both HPLC fractions gave unaltered N-terminal fragments F1 and F2, meaning that neither His2 nor Tyr11 are photooxidized. However, different patterns were observed for the two cysteine-containing fragments F3 and F4 (Table S1 of ESI†). For both HPLC fractions, no mass peak corresponding to the possible F3–F4 disulfide was detected. For the fraction eluting at 20.3 min, fragment F3 is unaltered but fragment F4 was detected in three distinct chemical forms: unaltered and with increase of 32 and 48 mass units. The product with a 32 mass unit increase was ascribed to the oxidation of Cys34 into a sulfinate by comparison to previous studies.21,22 This was confirmed by the loss of 66 mass units corresponding to H2SO2 upon MS/MS fragmentation. The formation of a sulfinate upon photooxidation of metal-bound thiolate has already been reported in many instances for Pt,35–37 Ni,38 Pd,38 Co39–41 and Cd42 complexes thereby supporting the formation of a sulfinate for Zn-bond cysteines in Zn·LASD(HHCC) but the product with a 48-mass unit increase is more intriguing. As F4 does not contain any amino acid prone to oxygen incorporation upon photooxidation other than Cys34 (i.e. His, Tyr, Trp or Met), this product could only be ascribed to the formation of a sulfonate on Cys34. This hypothesis is supported by the formation of glutathione sulfonate in the case of photooxidation of glutathione.43 Nevertheless, the mechanism of the formation of this product, especially the breaking of the O–O bond in the putative RS(O)(O)–OO− intermediate remains unclear. Altogether, the fraction eluting at 20.3 min contains the unoxidized peptide LASD(HHCC) as expected but also LASD(HHCCSO2) and LASD(HHCCSO3) oxidation products in which Cys34 was photooxidized into a sulfinate and a sulfonate, respectively (Fig. 5A). For the fraction eluting at 18.1 min, only a 32-mass unit increase was detected for F3, revealing photooxidation of Cys31 into a sulfinate (confirmed by loss of 66 mass units upon MS/MS fragmentation). Photooxidation of Cys31 rather than Tyr20 or His27 in F3 was supported by unaltered F1 fragment, which contains also histidine and tyrosine, and was further demonstrated by the loss of 66 Da corresponding to H2SO2 upon MS–MS fragmentation. F4 appears within three different forms: unaltered and with +32 and +48 mass units. This indicates the presence of Cys34 in reduced, sulfinate and sulfonate forms in the fraction eluting at 18.1 min. Therefore, this fraction contains three peptides: the primary oxidation product LASD(HHCSO2C) and the overoxidation products LASD(HHCSO2CSO2) and LASD(HHCSO2CSO3) (Fig. 5B). Noteworthy, mass analysis always revealed the presence of overoxidation products, even at short irradiation times, indicating that the rate of formation of overoxidation products is at least comparable to the rate of formation of primary oxidation products LASD(HHCCSO2) and LASD(HHCSO2C).Then, the rate of chemical reaction between Zn·LASD(HHCC) and 1O2 (kr) was assessed by competitive photooxidation with the peptide EGWGK as a competitor (kr(EGWGK) = (4.6 ± 0.5) × 106 M−1 s−1)21 following a procedure described previously.21,22 The method consists in comparing the consumption of the zinc finger peptide and the reference compound by HPLC (ESI). In such a competitive photooxidation experiment, the ratio of the rate constants kr1 and kr2 of two compounds C1 and C2 is given by eqn (1) where [Ci]0 and [Ci]f are the concentrations of compound Ci before and after photooxidation, respectively.44,45
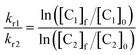 | (1) |
Since we were not able to separate unreacted LASD(HHCC) and LASD(HHCCSO2) by HPLC and as overoxidation are rapidly formed, this method underestimates the consumption of LASD(HHCC) and yields a sizeable underestimation of kr. Nevertheless, a lower limit of 3.9 × 106 M−1 s−1 for kr(Zn·LASD(HHCC)) was calculated from the competition experiments. Finally, CD was used to assess the consequences of 1O2 oxidation of Zn·LASD(HHCC) on the peptide fold. As shown in Fig. 5C, photooxidation causes dramatic loss of helical structure, attesting of peptide unfolding.
Discussion
Design and characterization of the anti-sigma factor domain zinc finger model
For several years, we have been developing peptidic zinc finger models of small size in order to study the reactivity of zinc finger sites toward reactive oxygen species such as H2O2,18–20 HOCl34 or 1O2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21,22 at the molecular level. Our approach involves the use of cyclic and branched peptides, named CPLT (Cyclic Peptides with Linear Tail),29 to create shortcuts in native zinc finger sequences. This affords models that reproduce almost perfectly the fold of native zinc finger domains around the Zn2+ ion with a minimal set of amino acids. The small size of these models allowed us to characterize oxidation products and to provide reliable kinetic data to describe the reactivity of zinc fingers. The CPLT design is well suited to zinc finger sites harbouring a CXXC motif in a β-hairpin loop. However, the elaboration of a CPLT model was not possible for the anti-sigma factor domain due to its 3-helix core that supports the four Zn2+-binding amino acids. Therefore, we decided to keep this core but to reduce the size of the anti-sigma factor domain sequence as much as possible without altering its fold around Zn2+. Re-engineering of the amino acid sequence allowed us to get a soluble 46-amino acid protein that folds upon Zn2+ binding and adopts the same conformation as that found in the sigma factor/anti-sigma factor complex. The design strategy used to elaborate this model revealed some important features: (i) it is necessary to avoid clustering of negatively or positively charged amino acids on the surface to provide sufficient solubility to the model, (ii) once the Zn2+ ion coordinated to the H–X3–C–X2–C motif, the constitution of a hydrophobic core within the heart of the 3-helix structure is most probably the driving force for the protein to get the proper folding; and (iii) the N-terminal histidine is absolutely required for conformational stability, providing at least 4 kcal mol−1 stabilization to the system as shown by the comparison between Zn·LASD(HHCC) and Zn·LASD(AHCC).
21,22 at the molecular level. Our approach involves the use of cyclic and branched peptides, named CPLT (Cyclic Peptides with Linear Tail),29 to create shortcuts in native zinc finger sequences. This affords models that reproduce almost perfectly the fold of native zinc finger domains around the Zn2+ ion with a minimal set of amino acids. The small size of these models allowed us to characterize oxidation products and to provide reliable kinetic data to describe the reactivity of zinc fingers. The CPLT design is well suited to zinc finger sites harbouring a CXXC motif in a β-hairpin loop. However, the elaboration of a CPLT model was not possible for the anti-sigma factor domain due to its 3-helix core that supports the four Zn2+-binding amino acids. Therefore, we decided to keep this core but to reduce the size of the anti-sigma factor domain sequence as much as possible without altering its fold around Zn2+. Re-engineering of the amino acid sequence allowed us to get a soluble 46-amino acid protein that folds upon Zn2+ binding and adopts the same conformation as that found in the sigma factor/anti-sigma factor complex. The design strategy used to elaborate this model revealed some important features: (i) it is necessary to avoid clustering of negatively or positively charged amino acids on the surface to provide sufficient solubility to the model, (ii) once the Zn2+ ion coordinated to the H–X3–C–X2–C motif, the constitution of a hydrophobic core within the heart of the 3-helix structure is most probably the driving force for the protein to get the proper folding; and (iii) the N-terminal histidine is absolutely required for conformational stability, providing at least 4 kcal mol−1 stabilization to the system as shown by the comparison between Zn·LASD(HHCC) and Zn·LASD(AHCC).
H2O2 and 1O2 oxidation
In previous studies on the oxidation of model zinc fingers, we have shown that H2O2 yields mainly disulfides as products at physiological pH conditions whereas 1O2 yields mainly sulfinates. Regarding kinetics, we have observed that neutral Zn(Cys)2(His)2 zinc finger sites react slower with H2O2 than negatively charged Zn(Cys)3(His) and Zn(Cys)4 zinc finger sites.19,20 The reactivity of Zn·LASD(HHCC) toward H2O2 was studied using the methodology previously described.19 Reaction with H2O2 leads to disulfide formation within the CEEC motif with a second order rate constant of 0.030 ± 0.002 M−1 s−1 at 297 K. This is within the range of second order rate constants determined for other Zn(Cys)2(His)2 zinc fingers (k = 0.008–0.037 M−1 s−1) but significantly lower than for Zn(Cys)3(His) and Zn(Cys)4 zinc fingers (k = 0.1–1.5 M−1 s−1), confirming the trend previously observed.20 Among Zn(Cys)2(His)2 zinc fingers models, Zn·LASD(HHCC) reacts faster with H2O2 than Zn·CP-1(CCHH) (k = 0.008 M−1 s−1), a classical CCHH zinc finger with well-defined ββα fold. Regarding 1O2, reaction of Zn·LASD(HHCC) is very fast leading to sulfinates but overoxidation products (sulfonate, bis-sulfinate) are also formed very rapidly. No photooxidation of Zn2+-bound histidines was detected, confirming that histidines are protected from 1O2 oxidation by Zn2+ coordination, as previously observed in the case of a classical ββα zinc finger.22 Unfortunately, all photooxidation products could not be separated by HPLC precluding precise determination of the chemical reaction rate constant kr.21,22 This was not the case in previous studies. Indeed, the size of the Zn·LASD(HHCC) model is large compared to our other previously described models (46 versus ca. 25 amino acids), rendering HPLC separation less sensitive to changes occurring at a single amino acid position. Hence, the size of the model clearly matters for optimal separation and the smaller the model, the more detailed the molecular information. Nevertheless, a lower limit of 3.9 × 106 M−1 s−1 could be determined for kr. The observation of at least two overoxidation products – even at short irradiation time – in the peak eluting at 20.3 min (Fig. 5A) during analysis of photooxidation products of Zn·LASD(HHCC) indicates that this kr value is probably underestimated. Hence, Zn·LASD(HHCC) reacts with 1O2 at least as fast as, but most probably faster than, Zn·LTC (kr = (4.3 ± 0.4) × 106 M−1 s−1), a Zn(Cys)4 treble clef zinc finger model, on contrary to what is observed for H2O2. In the case of H2O2, negatively charged Zn(Cys)4 sites react faster than neutral Zn(Cys)2(His)2 sites. Therefore, the observed trend for H2O2 regarding the charge of the zinc finger site and its reactivity cannot be extrapolated to 1O2 reactivity. The lower limit value of kr determined for Zn·LASD(HHCC) is higher than the kr value determined for Zn·CPF (kr = (0.70 ± 0.07) × 106 M−1 s−1), a model peptide for the CCHH classical ββα zinc fingers, which differs from Zn·CP-1(CCHH) by a single Tyr to Phe mutation only. In order to understand the greater reactivity of Zn·LASD(HHCC) compared to the classical ββα zinc finger CP-1/CPF observed for H2O2 and 1O2, we have examined the structure of the Zn(Cys)2(His)2 sites in both models focusing on two important factors for cysteine sulfur reactivity: solvent accessible surface and number of NH⋯S hydrogen bonds, the latter being known to decrease the nucleophilic reactivity of Zn2+-bound sulfurs.23,46 Based on our NMR structure of Zn·LASD(HHCC) and the pdb file 1MEY47 for Zn·CP-1(CCHH), we found that the former has higher sulfur solvent accessible surface area (8.8 Å2vs. 4.1 Å2) and less NH⋯S hydrogen bonds (1 vs. 3). Therefore, these two factors support the greater reactivity of the anti-sigma factor domain zinc finger model compared to the classical ββα model.Biological relevance
Zinc binding to ChrR and formation of the Zn(Cys)2(His)2 zinc finger site of ChrR is needed to ensure its proper folding and sequestering of σE.8 Plus, it was demonstrated that the anti-sigma factor domain of ChrR is sufficient to sequester σE.10 Our data show that the Zn·LASD(HHCC) model can be oxidized by 1O2 leading to its unfolding. This supports the hypothesis that photooxidation of the cysteines of ChrR Zn(Cys)2(His)2 zinc finger site may be a key event in the 1O2 sensing mechanism by ChrR promoting the breakdown the ChrR–σE complex. Nevertheless, to be a sensing unit this zinc finger site would require fast reaction with 1O2 in order to trigger quickly the cellular response against 1O2 before too many oxidative damages are produced. The kinetic data gained in this study are also in favour of a sensing role for ChrR Zn(Cys)2(His)2 site: it reacts with 1O2 faster than other known structural zinc finger sites, either of the Zn(Cys)2(His)2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22 or Zn(Cys)4
22 or Zn(Cys)4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21 type. Indeed, it seems that the uncommon topology of the zinc finger site of ChrR12 provides very high zinc affinity while maintaining “naked” cysteinates, with a low number of NH⋯S hydrogen bonds, for higher reactivity. However, Rajasekar et al. have shown that oxidation of the HHCC zinc finger of ZAS proteins is not always sufficient to dissociate the anti-sigma factor from its cognate sigma factor.17 In the case of ChrR, it was clearly demonstrated that its C-terminal cupin-like domain is required for the 1O2 response.10,11,17 Therefore, it is possible that 1O2 oxidation of both the zinc finger of ChrR anti-sigma factor domain and essential histidine residues of the cupin-like domain are required to cause sufficient conformational change to allow the dissociation of the ChrR–σE complex. This would correspond to a dual activation process making this system responsive to 1O2 only among ROS, for a specific triggering of the transcriptional response. Alternatively, the essential histidine and glutamate residues of the cupin-like domain (H141, H143, E147 and H177) could provide alternative Zn2+ ligands to compensate for the oxidized cysteines of the anti-sigma factor domain. A mixed coordination set involving residues of both the anti-sigma factor domain and the cupin-like domain around Zn2+ would generate sufficient conformational rearrangement to destabilize the ChrR–σE complex.
21 type. Indeed, it seems that the uncommon topology of the zinc finger site of ChrR12 provides very high zinc affinity while maintaining “naked” cysteinates, with a low number of NH⋯S hydrogen bonds, for higher reactivity. However, Rajasekar et al. have shown that oxidation of the HHCC zinc finger of ZAS proteins is not always sufficient to dissociate the anti-sigma factor from its cognate sigma factor.17 In the case of ChrR, it was clearly demonstrated that its C-terminal cupin-like domain is required for the 1O2 response.10,11,17 Therefore, it is possible that 1O2 oxidation of both the zinc finger of ChrR anti-sigma factor domain and essential histidine residues of the cupin-like domain are required to cause sufficient conformational change to allow the dissociation of the ChrR–σE complex. This would correspond to a dual activation process making this system responsive to 1O2 only among ROS, for a specific triggering of the transcriptional response. Alternatively, the essential histidine and glutamate residues of the cupin-like domain (H141, H143, E147 and H177) could provide alternative Zn2+ ligands to compensate for the oxidized cysteines of the anti-sigma factor domain. A mixed coordination set involving residues of both the anti-sigma factor domain and the cupin-like domain around Zn2+ would generate sufficient conformational rearrangement to destabilize the ChrR–σE complex.
Conclusions
In this article, we have described the elaboration of a model for the Zn(Cys)2(His)2 zinc finger site of the anti-sigma factor domain of ChrR, a protein involved in transcriptional response to 1O2 in several bacteria. This site has a special topology among zinc fingers and its role in 1O2 sensing by ChrR is under debate. This model, namely Zn·LASD(HHCC), is a 46-amino acid peptide inspired from ChrR anti-sigma factor domain sequence. It forms a 3-helix core when Zn2+ binds to the four cysteine and histidine side chains. Zn·LASD(HHCC) adopts the same structure as the corresponding sequence in ChrR anti-sigma factor domain. We have shown that the cysteines of the Zn(Cys)2(His)2 site of Zn·LASD(HHCC) are oxidized by 1O2 into sulfinates and other overoxidation products. Photooxidation is faster than that of other zinc finger sites, especially compared to classical ββα Zn(Cys)2(His)2 zinc fingers. Additionally, 1O2 oxidation destabilizes the zinc finger leading to its unfolding. The data presented in this work indicate that photooxidation of the zinc finger site of ChrR may be a key event in its 1O2 sensing mechanism.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has been partially supported by Labex ARCANE and CBH-EUR-GS (ANR-17-EURE-0003).Notes and references
- C. C. Winterbourn, in Encyclopedia of Radicals in Chemistry, Biology and Materials, John Wiley & Sons, Ltd, 2012 Search PubMed.
- C. S. Foote, Photochem. Photobiol., 1991, 54, 659 CrossRef CAS PubMed.
- E. C. Ziegelhoffer and T. J. Donohue, Nat. Rev. Microbiol., 2009, 7, 856–863 CrossRef CAS PubMed.
- M. S. Baptista, J. Cadet, P. Di Mascio, A. A. Ghogare, A. Greer, M. R. Hamblin, C. Lorente, S. C. Nunez, M. S. Ribeiro, A. H. Thomas, M. Vignoni and T. M. Yoshimura, Photochem. Photobiol., 2017, 93, 912–919 CrossRef CAS PubMed.
- B. B. Fischer, E. Hideg and A. Krieger-Liszkay, Antioxid. Redox Signal., 2013, 18, 2145–2162 CrossRef CAS PubMed.
- J. Glaeser, A. M. Nuss, B. A. Berghoff and G. Klug, in Advances in Microbial Physiology, ed. R. K. Poole, Academic Press Ltd, Elsevier Science Ltd, London, 2011, vol. 58, pp. 141–173 Search PubMed.
- L. O. Klotz, K. D. Kröncke and H. Sies, Photochem. Photobiol. Sci., 2003, 2, 88–94 RSC.
- J. D. Newman, J. R. Anthony and T. J. Donohue, J. Mol. Biol., 2001, 313, 485–499 CrossRef CAS PubMed.
- J. R. Anthony, J. D. Newman and T. J. Donohue, J. Mol. Biol., 2004, 341, 345–360 CrossRef CAS PubMed.
- E. A. Campbell, R. Greenwell, J. R. Anthony, S. Wang, L. Lim, K. Das, H. J. Sofia, T. J. Donohue and S. A. Darst, Mol. Cell, 2007, 27, 793–805 CrossRef CAS PubMed.
- R. Greenwell, T.-W. Nam and T. J. Donohue, J. Mol. Biol., 2011, 407, 477–491 CrossRef CAS PubMed.
- C. Andreini, I. Bertini and G. Cavallaro, PLoS One, 2011, 6, e26325 CrossRef CAS PubMed.
- K. G. Thakur, T. Praveena and B. Gopal, J. Mol. Biol., 2010, 397, 1199–1208 CrossRef CAS PubMed.
- J. G. Kang, M. S. B. Paget, Y. J. Seok, M. Y. Hahn, J. B. Bae, J. S. Hahn, C. Kleanthous, M. J. Buttner and J. H. Roe, EMBO J., 1999, 18, 4292–4298 CrossRef CAS PubMed.
- K. Zdanowski, P. Doughty, P. Jakimowicz, L. O'Hara, M. J. Buttner, M. S. B. Paget and C. Kleanthous, Biochemistry, 2006, 45, 8294–8300 CrossRef CAS PubMed.
- Y.-G. Jung, Y.-B. Cho, M.-S. Kim, J.-S. Yoo, S.-H. Hong and J.-H. Roe, Nucleic Acids Res., 2011, 39, 7586–7597 CrossRef CAS PubMed.
- K. V. Rajasekar, K. Zdanowski, J. Yan, J. T. S. Hopper, M.-L. R. Francis, C. Seepersad, C. Sharp, L. Pecqueur, J. M. Werner, C. V. Robinson, S. Mohammed, J. R. Potts and C. Kleanthous, Nat. Commun., 2016, 7, 12194 CrossRef CAS PubMed.
- O. Sénèque, E. Bourlès, V. Lebrun, E. Bonnet, P. Dumy and J.-M. Latour, Angew. Chem., Int. Ed., 2008, 47, 6888–6891 CrossRef PubMed.
- E. Bourlès, M. Isaac, C. Lebrun, J.-M. Latour and O. Sénèque, Chem.–Eur. J., 2011, 17, 13762–13772 CrossRef PubMed.
- M. Isaac, J.-M. Latour and O. Sénèque, Chem. Sci., 2012, 3, 3409–3420 RSC.
- V. Lebrun, A. Tron, L. Scarpantonio, C. Lebrun, J.-L. Ravanat, J.-M. Latour, N. D. McClenaghan and O. Sénèque, Angew. Chem., Int. Ed., 2014, 53, 9365–9368 CrossRef CAS PubMed.
- V. Lebrun, A. Tron, C. Lebrun, J.-M. Latour, N. D. McClenaghan and O. Sénèque, Chem.–Eur. J., 2015, 21, 14002–14010 CrossRef CAS PubMed.
- A. T. Maynard and D. G. Covell, J. Am. Chem. Soc., 2001, 123, 1047–1058 CrossRef CAS PubMed.
- O. Sénèque and J.-M. Latour, J. Am. Chem. Soc., 2010, 132, 17760–17774 CrossRef PubMed.
- V. Cornish, M. Kaplan, D. Veenstra, P. Kollman and P. Schultz, Biochemistry, 1994, 33, 12022–12031 CrossRef CAS.
- J. Venkatraman, S. C. Shankaramma and P. Balaram, Chem. Rev., 2001, 101, 3131–3152 CrossRef CAS.
- The PyMOL Molecular Graphics System, Schrödinger, LLC, 2002 Search PubMed.
- P. White, J. W. Keyte, K. Bailey and G. Bloomberg, J. Pept. Sci., 2004, 10, 18–26 CrossRef CAS PubMed.
- A. Jacques, B. Mettra, V. Lebrun, J.-M. Latour and O. Sénèque, Chem.–Eur. J., 2013, 19, 3921–3931 CrossRef CAS PubMed.
- B. Krizek, D. Merkle and J. Berg, Inorg. Chem., 1993, 32, 937–940 CrossRef CAS.
- A. R. Reddi, T. R. Guzman, R. M. Breece, D. L. Tiemey and B. R. Gibney, J. Am. Chem. Soc., 2007, 129, 12815–12827 CrossRef CAS PubMed.
- D. A. Deranleau, J. Am. Chem. Soc., 1969, 91, 4044–4049 CrossRef CAS.
- D. A. Deranleau, J. Am. Chem. Soc., 1969, 91, 4050–4054 CrossRef CAS.
- V. Lebrun, J.-L. Ravanat, J.-M. Latour and O. Sénèque, Chem. Sci., 2016, 7, 5508–5516 RSC.
- Y. Zhang, K. D. Ley and K. S. Schanze, Inorg. Chem., 1996, 35, 7102–7110 CrossRef CAS PubMed.
- W. B. Connick and H. B. Gray, J. Am. Chem. Soc., 1997, 119, 11620–11627 CrossRef CAS.
- D. Zhang, Y. Bin, L. Tallorin, F. Tse, B. Hernandez, E. V. Mathias, T. Stewart, R. Bau and M. Selke, Inorg. Chem., 2013, 52, 1676–1678 CrossRef CAS PubMed.
- C. A. Grapperhaus, M. J. Maguire, T. Tuntulani and M. Y. Darensbourg, Inorg. Chem., 1997, 36, 1860–1866 CrossRef CAS PubMed.
- C. Galvez, D. G. Ho, A. Azod and M. Selke, J. Am. Chem. Soc., 2001, 123, 3381–3382 CrossRef CAS.
- B. Hernandez, Y. J. Wang, D. Zhang and M. Selke, Chem. Commun., 2006, 997–999 RSC.
- D. Zhang, B. Hernandez and M. Selke, J. Sulfur Chem., 2008, 29, 377–388 CrossRef CAS PubMed.
- D. A. Cagan, A. C. Garcia, K. Li, D. Ashen-Garry, A. C. Tadle, D. Zhang, K. J. Nelms, Y. Liu, J. R. Shallenberger, J. J. Stapleton and M. Selke, J. Am. Chem. Soc., 2019, 141, 67–71 CrossRef CAS PubMed.
- T. Devasagayam and A. Sundquist, J. Photochem. Photobiol., B, 1991, 9, 105–116 CrossRef CAS.
- R. Higgins, C. S. Foote and H. Cheng, Adv. Chem. Ser., 1968, 102–117 CrossRef CAS.
- F. Wilkinson, W. Helman and A. Ross, J. Phys. Chem. Ref. Data, 1995, 24, 663–1021 CrossRef CAS.
- Y.-M. Lee and C. Lim, J. Am. Chem. Soc., 2011, 133, 8691–8703 CrossRef CAS PubMed.
- C. A. Kim and J. M. Berg, Nat. Struct. Biol., 1996, 3, 940–945 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details for peptide synthesis and characterization, absorption and circular dichroism analyses of Zn2+ binding, H2O2 oxidation, 1O2 oxidation and solution structure determination by NMR. See DOI: 10.1039/c9sc00341j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |