Enhanced paper-based ELISA for simultaneous EVs/exosome isolation and detection using streptavidin agarose-based immobilization†
Junwoo
Lee‡
a,
Hyerin
Kim‡
a,
Youhee
Heo
b,
Yong Kyoung
Yoo
a,
Sung Il
Han
a,
Cheonjung
Kim
a,
Don
Hur
a,
Hyungsuk
Kim
a,
Ji Yoon
Kang
*b and
Jeong Hoon
Lee
 *a
*a
aDepartment of Electrical Engineering, Kwangwoon University, Seoul 01897, Republic of Korea. E-mail: jhlee@kw.ac.kr
bCenter for BioMicrosystems, Korea Institute of Science and Technology (KIST), Seoul 02792, Republic of Korea. E-mail: jykang@kist.re.kr
First published on 21st October 2019
Abstract
EVs/exosomes are considered as the next generation of biomarkers, including for liquid biopsies. Consequently, the quantification of EVs/exosomes is crucial for facilitating EV/exosome research and applications. Paper-based enzyme-linked immunosorbent assay (p-ELISA) is a portable diagnostic system with low cost that is simple and easy to use; however, it shows low sensitivity and linearity. In this study, we develop p-ELISA for targeting EVs/exosomes by using streptavidin agarose resin-based immobilization (SARBI). This method reduces assay preparation times, provides strong binding, and retains good sensitivity and linearity. The time required for the total assay, including preparation steps and surface immobilization, was shortened to ∼2 h. We evaluated SARBI p-ELISA systems with/without CD63 capture Ab and then with fetal bovine serum (FBS) and EVs/exosome-depleted fetal bovine serum (dFBS). The results provide evidence supporting the selective capture ability of SARBI p-ELISA. We obtain semiquantitative p-ELISA results using an exosome standard (ES) and human serum (HS), with R2 values of 0.95 and 0.92, respectively.
1. Introduction
EVs (extracellular vesicles)/exosomes with a size of 50–500 nm play an important role in information exchange between cells.1–4 The isolation, collection, and quantification of EVs/exosomes are crucial for facilitating the study of EVs/exosomes for diagnostics and therapy, as molecules in EVs/exosomes have been found to be associated with certain diseases, such as cancers, and their tumorigenesis and metastasis.5–7 EVs/exosomes have a small size and low density in biofluids. They are usually collected from biofluids (e.g., spinal fluid, blood, saliva, and urine8–10) and cell culture media. Furthermore, they are selectively isolated mainly by differential ultracentrifugation;11,12 other isolating techniques include density gradient ultracentrifugation, size exclusion chromatography, filtration, polymer-based precipitation, sieving isolation, and immunological separation.11,13Among the various isolation methods, immunological separation approaches have the advantage of selectively collecting EVs/exosomes using immuno-affinity.14,15 Moreover, one can simultaneously obtain assay results using an enzyme-linked immunosorbent assay (ELISA) technique.16 Commercial ELISA usually has good selectivity, sensitivity, and reliability.17,18 Although this method is popular, it requires a long processing time and large sample volumes; moreover, it is costly, involves labor-intensive work that must be performed by a well-trained person, and needs expensive plate reading equipment.19,20
The Whiteside group21 suggested paper-based ELISA (p-ELISA); unfortunately, it has reproducibility and sensitivity issues. Nonetheless, p-ELISA needs lower reagent and sample quantities and lower processing time, leading to reduced analysis costs.22–24 In addition, it needs only cheap desktop scanners or mobile cameras instead of plate readers, thereby minimizing equipment restrictions.18,25,26 p-ELISA is a powerful platform that meets the ASSURED criteria (affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to end users) set by the World Health Organization for disease diagnostics in resource-limited regions.
Studies have suggested sandwich ELISA immobilized capture antibody (Ab) on the well surface.27,28 We used sandwich ELISA for achieving enhanced sensitivity and selectivity.29,30 To realize sandwich p-ELISA, it is critical to immobilize the Ab onto the paper; otherwise, ∼40% of the Ab may be desorbed from the cellulose fiber.31,32 Various methods for immobilizing Ab onto a paper have been studied; however, a current barrier to their practical use is the difficulty in achieving shortened assay times and strong immobilization.20–22,27
To overcome these critical challenges, we propose a p-ELISA platform that uses streptavidin agarose resin-based immobilization (SARBI). The SARBI p-ELISA assay provided strong binding owing to the strong affinity of biotinylated Ab and streptavidin agarose. Furthermore, we propose an isolation and detection platform for EV/exosome diagnostics and demonstrate its use for the exosome standard (ES) and human serum (HS), respectively. We also evaluate the selective capture ability of SARBI p-ELISA.
2. Materials and methods
2.1 Fabrication of p-ELISA wells
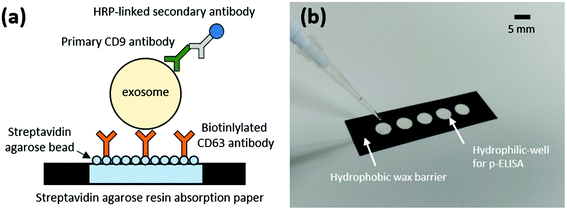
Fig. 1a shows a schematic of the proposed SARBI p-ELISA for EVs/exosome isolation/detection. Briefly, a wax-patterned microwell on cellulose paper is used for assaying EVs/exosomes. For this purpose, we first fabricated p-ELISA using a wax-patterning method. Cellulose paper (Whatman chromatography cellulose paper, Grade 1) was obtained from GE Healthcare Life Sciences (Pittsburgh PA, USA). The wells were designed using CorelDraw (Corel Co., Canada) software and printed on paper using a wax printer (Xerox ColorQube 8870, Japan) to make hydrophobic wax patterns.33 After wax printing, the paper was placed in an oven at 120 °C for 1 min 20 s to melt the wax; then, it was cooled to room temperature. This heating process causes the wax to melt through the paper, creating a hydrophobic barrier that serves as a 3D hydrophilic well (diameter: 5 mm, Fig. 1b).33 As reported previously in ref. 19, 34 and 35, the hydrophobic walls of p-ELISA act as a virtual wall that helps confine samples/reagents within the hydrophilic paper wells (reservoirs). This makes the sample reaction stable and prevents sample/reagent losses.2.2 Immobilization of p-ELISA
Fig. 2 shows the fabrication process and EVs/exosome assay. For linker formation on the paper substrate, we purchased streptavidin agarose resin (Thermo Scientific™) and prepared SARBI via biotinylated mouse anti-human anti-CD63 Ab (Abcam). After fabricating the hydrophilic paper wells, we added 5 μl of streptavidin agarose resin to the paper wells for immobilization of capture Ab and 5 μl of phosphate-buffered saline (PBS) containing 1% (w/v) bovine serum albumin (BSA). Then, we waited 10 min for it to block the surface, thereby preventing nonspecific binding. We repeated the BSA blocking process two more times. Next, we added 5 μl of biotinylated anti-CD63 Ab solution (20 μg ml−1) in PBS containing 1% (w/v) BSA, enabling a 10 min immobilization reaction on the paper surface. We repeated this process two more times.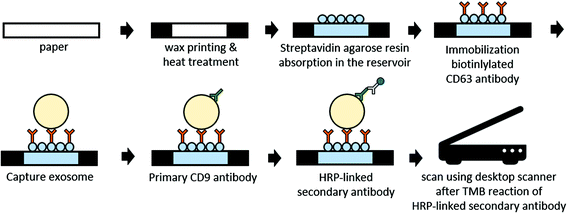 | ||
| Fig. 2 Schematic representation of the fabrication and assay procedure for this paper-based sandwich ELISA on a paper well. | ||
Agarose resins consist of agarose beads and aqueous slurries. The amount of agarose resin was determined by the volume of the paper wells. In addition, since the size of the agarose beads is larger than the pore size of the paper, they accumulate or stay on the surface (Fig. S2†).
2.3 Assay for EVs/exosomes using p-ELISA
For EVs/exosome preparation, we purchased a lyophilized ES and followed the procedure by the manufacturer for the reconstituted one (HansaBioMed; HBM-PEP, particles per ml: >1 × 1010, 100 μg per 100 μl) and used horseradish peroxidase (HRP)-linked secondary Ab (EXOAB-CD9A-1, System Biosciences). We prepared BSA (#SLBM7800 V, Sigma Aldrich, St Louis, MO), 3,3′,5,5′-tetramethylbenzidine (TMB, Thermo Scientific™), PBS (Gibco Life Technologies™), fluorescence-labeled ES (FL-exosome, HBM-F-PEP, HansaBioMed), human serum (Merck Millipore, USA), and fetal bovine serum (FBS, Corning, #35-015-CV). EVs/exosome-depleted FBS (dFBS) was prepared by 16 h ultracentrifugation of FBS at 4 °C and 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g using a Type 70Ti rotor (Beckman Ultracentrifuge Optima L-90 K, CA, USA).
000g using a Type 70Ti rotor (Beckman Ultracentrifuge Optima L-90 K, CA, USA).
After CD63 capture Ab was immobilized, 15 μl of PBS containing 1% (w/v) BSA was added to the wells, and we waited for 10 min to prevent nonspecific binding. We prepared EVs/exosome samples using 7.5 μl of ES or HS to bind the CD63 Ab. After a 10 min reaction, we added 15 μl of 1PBS to 1% (w/v) BSA for 10 min to prevent nonspecific binding. Then, we added 2.5 μl of anti-CD9 primary Ab (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution in PBS) for 1 min. To remove unreacted residues, we placed an absorbent pad at the bottom of the paper well. Then, 20 μl of PBS was added twice to the paper wells, and the paper wells were washed with PBS. For secondary Ab binding, 2.5 μl of HRP-linked secondary Ab (1
1000 dilution in PBS) for 1 min. To remove unreacted residues, we placed an absorbent pad at the bottom of the paper well. Then, 20 μl of PBS was added twice to the paper wells, and the paper wells were washed with PBS. For secondary Ab binding, 2.5 μl of HRP-linked secondary Ab (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution in PBS) was added for 1 min and rinsed off with PBS. After rinsing off, 2.5 μl of TMB was reacted for 20 min. In all experiments, we set a relative humidity of 35% and room temperature. CD9 and CD63 proteins that are commonly expressed on membranes in different cell types,36 and the co-expression of CD63 and CD9 and/or CD81 have been proposed in proteomic studies. Therefore, we demonstrated our p-ELISA platform by following the sandwich type ELISA that used CD9 and CD63.36–38
1000 dilution in PBS) was added for 1 min and rinsed off with PBS. After rinsing off, 2.5 μl of TMB was reacted for 20 min. In all experiments, we set a relative humidity of 35% and room temperature. CD9 and CD63 proteins that are commonly expressed on membranes in different cell types,36 and the co-expression of CD63 and CD9 and/or CD81 have been proposed in proteomic studies. Therefore, we demonstrated our p-ELISA platform by following the sandwich type ELISA that used CD9 and CD63.36–38
2.4 Data acquisition and quantification
We used a desktop scanner (Epson Perfection V39, Epson) for measuring the signal intensity from the ES and HS. Grayscale, RGB (red, green, blue), and HSL (hue, saturation, lightness) values are used to analyze the colorimetric assays in paper-based analytical devices.20,39,40 Among them, the saturation value in HSL was determined to be the optimal parameter for colorimetric analysis for our p-ELISA and was analyzed using Image-J (Wayne Rasband, National Institute of Health, Bethesda, MD). To obtain fluorescence images, we used a fluorescence microscope (IX-71, Olympus, Japan) with a thermo-electrically cooled charge coupled device (CCD) camera (Hamamatsu Co., Japan). Then, we analyzed the fluorescence intensity using Image-J software.33 For analyzing the EVs/exosome's size and distribution, we used nanoparticle tracking analysis (NTA, NanoSight LM10, NanoSight Technology, UK) and dynamic light scattering (DLS, Zetasizer Nano-S90, Malvern, UK). NTA is a light-scattering technique that is useful for the rapid sizing and enumeration of EVs. We used NTA 3.3 Dev Build 3.3.104 software (Malvern) with a CCD camera (camera level: 12, 30 FPS, temperature: 24.0–24.3 °C, viscosity: (water) 0.903–0.909 cP).3. Results
3.1 EVs/exosome assay preparation
Because biofluids contain a lot of biomolecules, a method for selectively capturing a target molecule is required. Sandwich p-ELISA is considered a good platform for such selective capture compared with direct/indirect p-ELISA; however, it requires preparation time for capture Ab for immobilization, which adds to the total sample preparation time (see Table 1). Table 1 lists previously reported immobilization methods for sandwich p-ELISA. Wang et al. suggested chitosan-glutaraldehyde functionalization targeting for AFP, CA125, and CEA with 2 h required for the immobilizing step.27,41 Similarly, Chen et al. used linker-NeutrAvidin for exosome detection with an immobilization time of 1 h 45 min.20 Shortening the immobilization process is a key factor in point-of-care testing applications. Recently, Khan et al. reported a 1 h immobilizing process using physisorption targeting for T7 bacteriophages.42 A powerful advantage of our proposed immobilization method over other p-ELISA techniques is its processing time. We used streptavidin agarose resin with only 35 min preparation for complete immobilization; therefore, we could complete the process within 2 h.| Author | Target biomolecule | Capture Ab immobilization method/chemical | Capture Ab immobilization time | Year/ref. |
|---|---|---|---|---|
| Wang et al. | AFP, CA125, CEA | Chitosan-glutaraldehyde | 2 h (whole process >3 h) | 2012/27 |
| Khan et al. | T7 bacteriophages | Physisorption | 1 h (whole process >3 h) | 2015/42 |
| Chen et al. | Exosomes | Linker-NeutrAvidin | 1 h 45 min (whole process <3 h) | 2014/20 |
| This work | Exosomes | Streptavidin agarose resin absorption | 35 min (whole process <2 h) |
3.2 EVs/exosome assay using ES samples
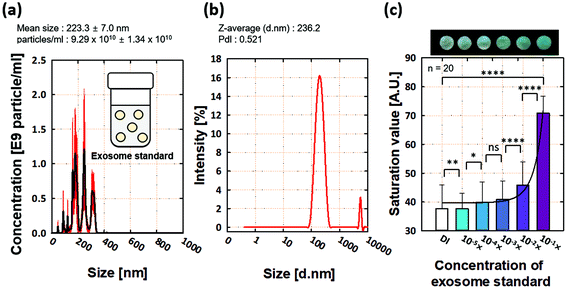
The ES sample was measured by NTA before capturing EVs/exosomes using our proposed p-ELISA (Fig. 3a), which can detect particles with a size of 30–1000 nm.43 For the 10−1× ES sample, the mean particle size and concentration were 223.3 ± 7 nm and 9.29 × 1010 particles per ml, respectively; this is consistent with the exosome size. Similarly, we used DLS (Fig. 3b) to confirm that the averaged particle size was 236.2 nm for the 10−1× ES sample. The observed peak with a size of ∼900 nm (Fig. 3b) could be attributed to ES aggregation.44We conducted EVs/exosome assays using ES samples (Fig. 3c). We first prepared ES concentrations of 10−1, 10−2, 10−3, 10−4, and 10−5×. We used deionized (DI) water as a reference (negative control). For all ES samples, we observed an increase in the change in saturation values for ES concentrations from 10−5 to 10−1×. We averaged 20 runs and showed error bars using standard deviations. Regression analysis showed that the R2 value was 0.95.
Downstream analysis such as phenotyping or RNA sequencing of exosomes is important for clinical diagnosis. To identify the availability of downstream analysis, we tried to show the extraction of an exosome from a paper substrate. For this purpose, we prepared a captured ES via biotinylated Ab on a streptavidin agarose resin paper. For extracting the captured ES, we disassociated the Ag–Ab interaction using 4 μl of 50 mM glycine–HCl (pH 3) followed by the addition 4 μl of 1 M Tris-HCl (pH 8.6). Then, the extracted samples were analyzed using a NanodropR (Thermo Fisher Scientific) (Fig. S5†). We found that the concentration of the extracted ES sample was significantly higher than that of the extracted control sample (DI water).
3.3 EVs/exosome assay using EVs/exosomes in human serum
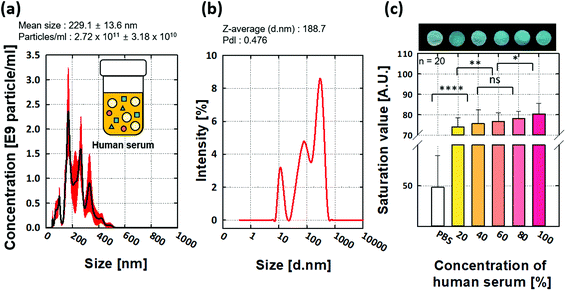
The particle size of EVs/exosomes from the 100% HS showed a mean size of 229.1 ± 13.6 nm (Fig. 4a). By using DLS analysis (Fig. 4b), we found that the average captured particle size was 188.7 nm; therefore, it is reasonable that HS contains EVs/exosomes.We conducted EVs/exosome assays using five different HS concentrations (Fig. 4c). We first prepared HS concentrations of 20%, 40%, 60%, 80%, and 100% by dilution and then prepared a negative control using PBS solution without HS. From 20 runs, we obtained error bars with standard deviations (Fig. 4c). Regression analysis showed that the R2 value was 0.92, indicating the feasibility of p-ELISA in EVs/exosome applications.
The error bars in SARBI p-ELISA are larger than those in commercial ELISA; similar problems have been observed previously because of the nonuniformity and large surface roughness of paper materials.35,45 By using SARBI p-ELISA, we reduced the immobilization time and whole assay time and acquired enhanced signals compared with a previous study.19 Technologies for achieving a high sensitivity and LOD are needed for the reliability and reproducibility of p-ELISA. Although p-ELISA still faces technical issues in terms of sensitivity and LOD, it is highly advantageous because it only requires cheap and convenient equipment such as a desktop scanner and a smartphone camera.
3.4 p-ELISA system verification
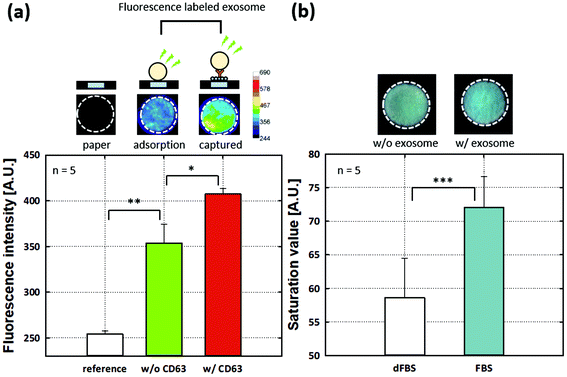
To evaluate SARBI's ability for p-ELISA, we used a commercial ELISA kit (System Bioscience, EXOEL-CD63A-1) that was used in previous studies.38,46,47 We expect it to provide more reliable data for validating our proposed p-ELISA platform. By using a commercial ELISA kit, we assayed the ES and HS. As shown in Fig. S1a (see the ESI†), 10−1× ES was assayed using commercial ELISA assays and DI water was tested for a negative control. Similarly, 100% HS was prepared for commercial ELISA assays and PBS was tested for a negative control (Fig. S1b†). From both commercial ELISA assays (Fig. S1a and S1b†), we found clear and consistent evidence of the presence of EVs/exosomes in both the ES and HS via the proposed SARBI p-ELISA.In addition, we verified the SARBI p-ELISA system with two additional strategies to realize specific binding and proper assays (Fig. 5). First, we estimated the adsorbed EVs/exosomes and specific bound on one of the wells in the SARBI p-ELISA system. The ES and HS data (Fig. 3c and 4c) are obtained with control experiments using DI water and PBS, respectively; however, we expect that observed signals could be attributed to adsorption of the HRP-conjugated secondary Ab and color changes due to TMB reactions. To closely examine the adsorption effects of EVs/exosomes on SARBI p-ELISA, we prepared an FL-exosome and allowed it to adsorb on SARBI p-ELISA wells for 10 min without CD63 capturing Ab (see Fig. 5a: w/o CD63) and washed it out with PBS. We excluded the effect of adsorption from HRP-conjugated secondary antibodies and acquired a fluorescence intensity of ∼354. Then, to check the CD63 capture Ab ability, we assayed SARBI p-ELISA with CD63 capturing Ab. We added 7.5 μl of FL-exosomes onto SARBI p-ELISA with CD63 capturing Ab; then, we washed it out with PBS (see Fig. 5a: w/CD63). After PBS washing, we measured a fluorescence intensity of ∼407 using a fluorescence microscope. For a reference (Fig. 5a), we also measured the fluorescence signal of bare paper materials as ∼254 without functionalization. All data were averaged from five runs, and error bars represented standard deviations. Considering the clear differences in fluorescence intensity from the results with/without CD63 capture Ab, we confirmed the CD63 capture Ab ability of SARBI p-ELISA. Nonspecific binding/absorption is a critical issue in sensitivity/LOD in paper-based analytical devices. Previous studies suggested surface treatment methods such as high-concentration NaCl, BSA blocking, and pH regulation to avoid nonspecific binding.48 Among them, we used BSA blocking in our proposed p-ELISA device; however, we expected that appropriate surface treatment would also impede nonspecific binding/absorption efficiently.
Second, we tested p-ELISA using FBS and dFBS (Fig. 5b). dFBS has been used for the samples that contain no EVs/exosomes.49 We prepared dFBS by 16 h ultracentrifugation of FBS at 4 °C under 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g using a Type 70Ti rotor according to previous protocols for depleting EVs/exosomes from the FBS.50 After adding 7.5 μl of FBS and dFBS onto paper wells immobilized with CD63 capture Ab, we reacted the CD9 primary Ab and then rinsed off the unreacted one. After adding the HRP-linked secondary Ab for reaction, we washed out with PBS and then reacted TMB. Note that previous studies have shown that antibodies reacted with rat and human proteins for measuring (or capturing) EVs/exosomes in FBS, whereas the manufacturer's specifications show that the CD63 capture antibody and the CD9 primary antibody only respond to human, mouse, and rat proteins.49,51,52 All data were averaged from five runs, and error bars represented standard deviations. The signal differences between FBS and dFBS were analyzed owing to the presence of EVs/exosomes. Thus, the results in Fig. 5a and b provide experimental evidence supporting the assay abilities for selectively capturing EVs/exosomes via the proposed SARBI p-ELISA.
000g using a Type 70Ti rotor according to previous protocols for depleting EVs/exosomes from the FBS.50 After adding 7.5 μl of FBS and dFBS onto paper wells immobilized with CD63 capture Ab, we reacted the CD9 primary Ab and then rinsed off the unreacted one. After adding the HRP-linked secondary Ab for reaction, we washed out with PBS and then reacted TMB. Note that previous studies have shown that antibodies reacted with rat and human proteins for measuring (or capturing) EVs/exosomes in FBS, whereas the manufacturer's specifications show that the CD63 capture antibody and the CD9 primary antibody only respond to human, mouse, and rat proteins.49,51,52 All data were averaged from five runs, and error bars represented standard deviations. The signal differences between FBS and dFBS were analyzed owing to the presence of EVs/exosomes. Thus, the results in Fig. 5a and b provide experimental evidence supporting the assay abilities for selectively capturing EVs/exosomes via the proposed SARBI p-ELISA.
4. Discussion
We proposed a sandwich p-ELISA that targets EVs/exosomes for isolation/detection functions. The proposed devices were easy to fabricate and cost-effective, and require minimal steps, in turn, reducing the processing time within 2 hours. For the ES and HS, we conducted EVs/exosome assays using five different ES and HS concentrations with R2 values of 0.95 and 0.92, indicating the feasibility of p-ELISA for EVs/exosome applications. Despite the inevitable deviation of p-ELISA compared with commercial ELISA, this device provides simple and cheap detection tools with isolation functions required for extracellular vesicle research, such as disease diagnosis/prognosis, immunotherapy, and therapeutics. The proposed SARBI p-ELISA could provide a cheap and convenient platform for point-of-care testing, especially in resource-limited regions/countries.Author contributions
J. Y. K. and J. H. L. conceived the idea and, together with D. H. and H. K (Hyungsuk Kim), designed the experiments. J. L., H. K. (Hyerin Kim), Y. H., Y. K. Y., S. I. H., and C. K. performed the experiments. J.L., Y.K.Y., J. Y. K., and J. H. L. co-wrote the manuscript, and all authors discussed the results and commented on the manuscript.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This research was funded by the National Research Foundation of Korea (NRF) grant no. NRF-2017M3A7B4049851. This work was also supported by “Human Resources Program in Energy Technology” of the Korea Institute of Energy Technology Evaluation and Planning (KETEP), granted financial resource from the Ministry of Trade, Industry & Energy, Republic of Korea (No. 20174010201620).References
- M. Wu, Y. Ouyang, Z. Wang, R. Zhang, P.-H. Huang, C. Chen, H. Li, P. Li, D. Quinn, M. Dao, S. Suresh, Y. Sadovsky and T. J. Huang, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 10584–10589 CrossRef CAS PubMed.
- C. Bang and T. Thum, Int. J. Biochem. Cell Biol., 2012, 44, 2060–2064 CrossRef CAS PubMed.
- J. Maia, S. Caja, M. C. Strano Moraes, N. Couto and B. Costa-Silva, Front. Cell Dev. Biol., 2018, 6, 18 CrossRef.
- C. Théry, K. W. Witwer, E. Aikawa, M. J. Alcaraz, J. D. Anderson, R. Andriantsitohaina, A. Antoniou, T. Arab, F. Archer, G. K. Atkin-Smith, D. C. Ayre, J.-M. Bach, D. Bachurski, H. Baharvand, L. Balaj, S. Baldacchino, N. N. Bauer, A. A. Baxter, M. Bebawy, C. Beckham, A. B. Zavec, A. Benmoussa, A. C. Berardi, P. Bergese, E. Bielska, C. Blenkiron, S. Bobis-Wozowicz, E. Boilard, W. Boireau, A. Bongiovanni, F. E. Borràs, S. Bosch, C. M. Boulanger, X. Breakefield, A. M. Breglio, M. Á. Brennan, D. R. Brigstock, A. Brisson, M. L. Broekman, J. F. Bromberg, P. Bryl-Górecka, S. Buch, A. H. Buck, D. Burger, S. Busatto, D. Buschmann, B. Bussolati, E. I. Buzás, J. B. Byrd, G. Camussi, D. R. Carter, S. Caruso, L. W. Chamley, Y.-T. Chang, C. Chen, S. Chen, L. Cheng, A. R. Chin, A. Clayton, S. P. Clerici, A. Cocks, E. Cocucci, R. J. Coffey, A. Cordeiro-da-Silva, Y. Couch, F. A. Coumans, B. Coyle, R. Crescitelli, M. F. Criado, C. D'Souza-Schorey, S. Das, A. D. Chaudhuri, P. de Candia, E. F. De Santana, O. De Wever, H. A. Del Portillo, T. Demaret, S. Deville, A. Devitt, B. Dhondt, D. Di Vizio, L. C. Dieterich, V. Dolo, A. P. D. Rubio, M. Dominici, M. R. Dourado, T. A. Driedonks, F. V. Duarte, H. M. Duncan, R. M. Eichenberger, K. Ekström, S. El Andaloussi, C. Elie-Caille, U. Erdbrügger, J. M. Falcón-Pérez, F. Fatima, J. E. Fish, M. Flores-Bellver, A. Försönits, A. Frelet-Barrand, F. Fricke, G. Fuhrmann, S. Gabrielsson, A. Gámez-Valero, C. Gardiner, K. Gärtner, R. Gaudin, Y. S. Gho, B. Giebel, C. Gilbert, M. Gimona, I. Giusti, D. C. Goberdhan, A. Görgens, S. M. Gorski, D. W. Greening, J. C. Gross, A. Gualerzi, G. N. Gupta, D. Gustafson, A. Handberg, R. A. Haraszti, P. Harrison, H. Hegyesi, A. Hendrix, A. F. Hill, F. H. Hochberg, K. F. Hoffmann, B. Holder, H. Holthofer, B. Hosseinkhani, G. Hu, Y. Huang, V. Huber, S. Hunt, A. G.-E. Ibrahim, T. Ikezu, J. M. Inal, M. Isin, A. Ivanova, H. K. Jackson, S. Jacobsen, S. M. Jay, M. Jayachandran, G. Jenster, L. Jiang, S. M. Johnson, J. C. Jones, A. Jong, T. Jovanovic-Talisman, S. Jung, R. Kalluri, S.-I. Kano, S. Kaur, Y. Kawamura, E. T. Keller, D. Khamari, E. Khomyakova, A. Khvorova, P. Kierulf, K. P. Kim, T. Kislinger, M. Klingeborn, D. J. Klinke II, M. Kornek, M. M. Kosanović, Á. F. Kovács, E.-M. Krämer-Albers, S. Krasemann, M. Krause, I. V. Kurochkin, G. D. Kusuma, S. Kuypers, S. Laitinen, S. M. Langevin, L. R. Languino, J. Lannigan, C. Lässer, L. C. Laurent, G. Lavieu, E. Lázaro-Ibáñez, S. Le Lay, M. Lee, Y. X. F. Lee, D. S. Lemos, M. Lenassi, A. Leszczynska, I. T. Li, K. Liao, S. F. Libregts, E. Ligeti, R. Lim, S. K. Lim, A. Linē, K. Linnemannstöns, A. Llorente, C. A. Lombard, M. J. Lorenowicz, Á. M. Lörincz, J. Lötvall, J. Lovett, M. C. Lowry, X. Loyer, Q. Lu, B. Lukomska, T. R. Lunavat, S. L. Maas, H. Malhi, A. Marcilla, J. Mariani, J. Mariscal, E. S. Martens-Uzunova, L. Martin-Jaular, M. C. Martinez, V. R. Martins, M. Mathieu, S. Mathivanan, M. Maugeri, L. K. McGinnis, M. J. McVey, D. G. Meckes, Jr., K. L. Meehan, I. Mertens, V. R. Minciacchi, A. Möller, M. M. Jørgensen, A. Morales-Kastresana, J. Morhayim, F. Mullier, M. Muraca, L. Musante, V. Mussack, D. C. Muth, K. H. Myburgh, T. Najrana, M. Nawaz, I. Nazarenko, P. Nejsum, C. Neri, T. Neri, R. Nieuwland, L. Nimrichter, J. P. Nolan, E. N. N. Hoen, N. N. Hooten, L. O'Driscoll, T. O'Grady, A. O'Loghlen, T. Ochiya, M. Olivier, A. Ortiz, L. A. Ortiz, X. Osteikoetxea, O. Østergaard, M. Ostrowski, J. Park, D. M. Pegtel, H. Peinado, F. Perut, M. W. Pfaffl, D. G. Phinney, B. C. Pieters, R. C. Pink, D. S. Pisetsky, E. P. v. Strandmann, I. Polakovicova, I. K. Poon, B. H. Powell, I. Prada, L. Pulliam, P. Quesenberry, A. Radeghieri, R. L. Raffai, S. Raimondo, J. Rak, M. I. Ramirez, G. Raposo, M. S. Rayyan, N. Regev-Rudzki, F. L. Ricklefs, P. D. Robbins, D. D. Roberts, S. C. Rodrigues, E. Rohde, S. Rome, K. M. Rouschop, A. Rughetti, A. E. Russell, P. Saá, S. Sahoo, E. Salas-Huenuleo, C. Sánchez, J. A. Saugstad, M. J. Saul, R. M. Schiffelers, R. Schneider, T. H. Schøyen, A. Scott, E. Shahaj, S. Sharma, O. Shatnyeva, F. Shekari, G. V. Shelke, A. K. Shetty, K. Shiba, P. R. M. Siljander, A. M. Silva, A. Skowronek, O. L. Snyder, II, R. P. Soares, B. W. Sódar, C. Soekmadji, J. Sotillo, P. D. Stahl, W. Stoorvogel, S. L. Stott, E. F. Strasser, S. Swift, H. Tahara, M. Tewari, K. Timms, S. Tiwari, R. Tixeira, M. Tkach, W. S. Toh, R. Tomasini, A. C. Torrecilhas, J. P. Tosar, V. Toxavidis, L. Urbanelli, P. Vader, B. W. van Balkom, S. G. van der Grein, J. Van Deun, M. J. van Herwijnen, K. Van Keuren-Jensen, G. van Niel, M. E. van Royen, A. J. van Wijnen, M. H. Vasconcelos, I. J. Vechetti Jr., T. D. Veit, L. J. Vella, É. Velot, F. J. Verweij, B. Vestad, J. L. Viñas, T. Visnovitz, K. V. Vukman, J. Wahlgren, D. C. Watson, M. H. Wauben, A. Weaver, J. P. Webber, V. Weber, A. M. Wehman, D. J. Weiss, J. A. Welsh, S. Wendt, A. M. Wheelock, Z. Wiener, L. Witte, J. Wolfram, A. Xagorari, P. Xander, J. Xu, X. Yan, M. Yáñez-Mó, H. Yin, Y. Yuana, V. Zappulli, J. Zarubova, V. Žėkas, J.-Y. Zhang, Z. Zhao, L. Zheng, A. R. Zheutlin, A. M. Zickler, P. Zimmermann, A. M. Zivkovic, D. Zocco and E. K. Zuba-Surma, J. Extracell. Vesicles, 2018, 7, 1535750 CrossRef PubMed.
- Y. H. Soung, S. Ford, V. Zhang and J. Chung, Cancers, 2017, 9, 8 CrossRef PubMed.
- M. S. Sinha, A. Ansell-Schultz, L. Civitelli, C. Hildesjö, M. Larsson, L. Lannfelt, M. Ingelsson and M. Hallbeck, Acta Neuropathol., 2018, 136, 41–56 CrossRef PubMed.
- G. K. Tofaris, J. Parkinson's Dis., 2017, 7, 569–576 Search PubMed.
- J. Lin, J. Li, B. Huang, J. Liu, X. Chen, X.-M. Chen, Y.-M. Xu, L.-F. Huang and X.-Z. Wang, Sci. World J., 2015, 2015, 657086 Search PubMed.
- A. Michael, S. D. Bajracharya, P. S. T. Yuen, H. Zhou, R. A. Star, G. G. Illei and I. Alevizos, Oral Dis., 2010, 16, 34–38 CrossRef CAS.
- J. L. Welton, S. Loveless, T. Stone, C. von Ruhland, N. P. Robertson and A. Clayton, J. Extracell. Vesicles, 2017, 6, 1369805–1369805 CrossRef.
- P. Li, M. Kaslan, S. H. Lee, J. Yao and Z. Gao, Theranostics, 2017, 7, 789–804 CrossRef CAS PubMed.
- M. A. Livshits, E. Khomyakova, E. G. Evtushenko, V. N. Lazarev, N. A. Kulemin, S. E. Semina, E. V. Generozov and V. M. Govorun, Sci. Rep., 2015, 5, 17319 CrossRef.
- R. T. Davies, J. Kim, S. C. Jang, E.-J. Choi, Y. S. Gho and J. Park, Lab Chip, 2012, 12, 5202–5210 RSC.
- J. C. Akers, D. Gonda, R. Kim, B. S. Carter and C. C. Chen, J. Neuro-Oncol., 2013, 113, 1–11 CrossRef.
- C. Lässer, M. Eldh and J. Lötvall, J. Visualized Exp., 2012, 3037, DOI:10.3791/3037.
- M. Logozzi, A. De Milito, L. Lugini, M. Borghi, L. Calabrò, M. Spada, M. Perdicchio, M. L. Marino, C. Federici, E. Iessi, D. Brambilla, G. Venturi, F. Lozupone, M. Santinami, V. Huber, M. Maio, L. Rivoltini and S. Fais, PLoS One, 2009, 4, e5219 CrossRef PubMed.
- H. J. Cho, S. A. Masri, D. Deregt, S. G. Yeo and E. J. Thomas, Can. J. Vet. Res., 1991, 55, 56–59 CAS.
- E. J. Ruitenberg, P. A. Steerenberg, B. J. M. Brosi and J. Buys, J. Immunol. Methods, 1976, 10, 67–83 CrossRef CAS.
- T. Mazzu-Nascimento, G. G. Morbioli, L. A. Milan, D. F. Silva, F. C. Donofrio, C. A. Mestriner and E. Carrilho, Anal. Methods, 2017, 9, 2644–2653 RSC.
- C. Chen, B.-R. Lin, M.-Y. Hsu and C.-M. Cheng, J. Visualized Exp., 2015, 52722, DOI:10.3791/52722.
- C.-M. Cheng, A. W. Martinez, J. Gong, C. R. Mace, S. T. Phillips, E. Carrilho, K. A. Mirica and G. M. Whitesides, Angew. Chem., Int. Ed., 2010, 49, 4771–4774 CrossRef CAS.
- M. Sher, R. Zhuang, U. Demirci and W. Asghar, Expert Rev. Mol. Diagn., 2017, 17, 351–366 CrossRef CAS.
- A. W. Martinez, Bioanalysis, 2011, 3, 2589–2592 CrossRef CAS.
- J. Hu, S. Wang, L. Wang, F. Li, B. Pingguan-Murphy, T. J. Lu and F. Xu, Biosens. Bioelectron., 2014, 54, 585–597 CrossRef CAS.
- X. Liu, C. M. Cheng, A. W. Martinez, K. Mirica, X. Li, S. Phillips, M. Sanchez and G. M. Whitesides, A portable microfluidic paper-based device for ELISA, 2011 Search PubMed.
- C.-K. Hsu, H.-Y. Huang, W.-R. Chen, W. Nishie, H. Ujiie, K. Natsuga, S.-T. Fan, H.-K. Wang, J. Y.-Y. Lee, W.-L. Tsai, H. Shimizu and C.-M. Cheng, Anal. Chem., 2014, 86, 4605–4610 CrossRef CAS.
- S. Wang, L. Ge, X. Song, J. Yu, S. Ge, J. Huang and F. Zeng, Biosens. Bioelectron., 2012, 31, 212–218 CrossRef CAS PubMed.
- Y.-H. P. Hsieh, Y.-T. Chen and K. Gajewski, J. Food Sci., 2009, 74, C602–C607 CrossRef CAS PubMed.
- J. Maertens, J. Verhaegen, H. Demuynck, P. Brock, G. Verhoef, P. Vandenberghe, J. Van Eldere, L. Verbist and M. Boogaerts, J. Clin. Microbiol., 1999, 37, 3223–3228 CAS.
- T.-H. Chen, F. Lee, Y.-L. Lin, A. Dekker, W.-B. Chung, C.-H. Pan, M.-H. Jong, C.-C. Huang, M.-C. Lee and H. Tsai, Differentiation of Foot-and-Mouth Disease-Infected Pigs from Vaccinated Pigs Using Antibody-Detecting Sandwich ELISA, 2011 Search PubMed.
- P. Jarujamrus, J. Tian, X. Li, A. Siripinyanond, J. Shiowatana and W. Shen, Analyst, 2012, 137, 2205–2210 RSC.
- S. Hosseini, P. Vazquez and S. O. Martinez-Chapa, Paper and Fiber-Based Bio-Diagnostic Platforms: Current Challenges and Future Needs, 2017 Search PubMed.
- S. I. Han, K. S. Hwang, R. Kwak and J. H. Lee, Lab Chip, 2016, 16, 2219–2227 RSC.
- M.-Y. Hsu, C.-Y. Yang, W.-H. Hsu, K.-H. Lin, C.-Y. Wang, Y.-C. Shen, Y.-C. Chen, S.-F. Chau, H.-Y. Tsai and C.-M. Cheng, Biomaterials, 2014, 35, 3729–3735 CrossRef CAS.
- B. Pang, C. Zhao, L. Li, X. Song, K. Xu, J. Wang, Y. Liu, K. Fu, H. Bao, D. Song, X. Meng, X. Qu, Z. Zhang and J. Li, Anal. Biochem., 2018, 542, 58–62 CrossRef CAS.
- D. Duijvesz, C. Y. L. Versluis, C. A. M. van der Fels, M. S. Vredenbregt-van den Berg, J. Leivo, M. T. Peltola, C. H. Bangma, K. S. I. Pettersson and G. Jenster, Int. J. Cancer, 2015, 137, 2869–2878 CrossRef CAS.
- A. B. Banizs, T. Huang, K. Dryden, S. S. Berr, J. R. Stone, R. K. Nakamoto, W. Shi and J. He, Int. J. Nanomed., 2014, 9, 4223–4230 CAS.
- M. Oliveira-Rodríguez, S. López-Cobo, H. T. Reyburn, A. Costa-García, S. López-Martín, M. Yáñez-Mó, E. Cernuda-Morollón, A. Paschen, M. Valés-Gómez and M. C. Blanco-López, J. Extracell. Vesicles, 2016, 5, 31803 CrossRef.
- C. Kim, Y. K. Yoo, S. I. Han, J. Lee, D. Lee, K. Lee, K. S. Hwang, K. H. Lee, S. Chung and J. H. Lee, Lab Chip, 2017, 17, 2451–2458 RSC.
- T.-T. Wang, C. k. Lio, H. Huang, R.-Y. Wang, H. Zhou, P. Luo and L.-S. Qing, Talanta, 2020, 206, 120211 CrossRef.
- W. Liu, Y. Guo, M. Zhao, H. Li and Z. Zhang, Anal. Chem., 2015, 87, 7951–7957 CrossRef CAS.
- M. S. Khan, T. Pande and T. G. M. van de Ven, Colloids Surf., B, 2015, 132, 264–270 CrossRef CAS.
- A. Malloy, Mater. Today, 2011, 14, 170–173 CrossRef CAS.
- R. Linares, S. Tan, C. Gounou, N. Arraud and A. R. Brisson, J. Extracell. Vesicles, 2015, 4, 29509–29509 CrossRef.
- T. Yu-Ting, W.-H. Sung, H.-C. Chen, M.-Y. Hsu and C.-M. Cheng, A Novel Diagnostic Approach for Monitoring Aqueous Humour VEGF Level in Ocular Diseases, in Paper-Based ELISA, IntechOpen Limited, London, UK, 2018 Search PubMed.
- D. Zocco and N. Zarovni, in Extracellular Vesicles: Methods and Protocols, ed. W. P. Kuo and S. Jia, Springer New York, New York, NY, 2017, pp. 269–285, DOI:10.1007/978-1-4939-7253-1_22.
- P.-S. Sung, T.-F. Huang and S.-L. Hsieh, Nat. Commun., 2019, 10, 2402 CrossRef.
- L. G. Lee, E. S. Nordman, M. D. Johnson and M. F. Oldham, Biosensors, 2013, 3, 360–373 CrossRef CAS.
- F. Angelini, V. Ionta, F. Rossi, F. Miraldi, E. Messina and A. Giacomello, BioImpacts, 2016, 6, 15–24 CrossRef CAS.
- C. Théry, S. Amigorena, G. Raposo and A. Clayton, Curr. Protoc. Cell Biol., 2006, 30, 3.22.21–23.22.29 Search PubMed.
- J. N. Caballero, G. Frenette, C. Belleannée and R. Sullivan, PLoS One, 2013, 8, e65364 CrossRef CAS.
- J. Kowal, G. Arras, M. Colombo, M. Jouve, J. P. Morath, B. Primdal-Bengtson, F. Dingli, D. Loew, M. Tkach and C. Théry, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E968–E977 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9an01140d |
| ‡ These authors have contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |