Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Cheating on forensic hair testing? Detection of potential biomarkers for cosmetically altered hair samples using untargeted hair metabolomics†
Lisa
Eisenbeiss
a,
Tina M.
Binz
 b,
Markus R.
Baumgartner
b,
Markus R.
Baumgartner
 b,
Thomas
Kraemer
b,
Thomas
Kraemer
 a and
Andrea E.
Steuer
a and
Andrea E.
Steuer
 *a
*a
aDepartment of Forensic Pharmacology and Toxicology, Zurich Institute of Forensic Medicine, University of Zurich, Zurich, Switzerland. E-mail: andrea.steuer@irm.uzh.ch; Fax: +41446356852; Tel: +41 446355679
bCenter for Forensic Hair Analytics, Zurich Institute of Forensic Medicine, University of Zurich, Zurich, Switzerland
First published on 7th August 2020
Abstract
Hair analysis has become an integral part in forensic toxicological laboratories for e.g. assessment of drug or alcohol abstinence. However, hair samples can be manipulated by cosmetic treatments, altering drug concentrations which eventually leads to false negative hair test results. In particular oxidative bleaching of hair samples under alkaline conditions significantly affects incorporated drug concentrations. To date, current techniques to detect cosmetic hair adulterations bear limitations such as the implementation of cut-off values or the requirement of specialized instrumentations. As a new approach, untargeted hair metabolomics analysis was applied to detect altered, endogenous biomolecules that could be used as biomarkers for oxidative cosmetic hair treatments. For this, genuine hair samples were treated in vitro with 9% hydrogen peroxide (H2O2) for 30 minutes. Untreated and treated hair samples were analyzed using liquid-chromatography high-resolution time-of-flight mass spectrometry. In total, 69 metabolites could be identified as significantly altered after hair bleaching. The majority of metabolites decreased after bleaching, yet totally degraded metabolites were most promising as suitable biomarkers. The formation of biomarker ratios of metabolites decreasing and increasing in concentrations improved the discrimination of untreated and treated hair samples. With the results of this study, the high variety of identified biomarkers now offers the possibility to include single biomarkers or biomarker selections into routine screening methods for improved data interpretation of hair test results.
1. Introduction
Over the past years, hair analysis has become increasingly important. Because of its non-invasive collection, easy-storage and in particular its long-term detection window, hair is the matrix of choice in forensic toxicology to assess retrospective consumption behaviors. It is therefore routinely applied for the assessment of drugs of abuse (DOA) or alcohol abstinence, workplace drug testing and in childhood custody cases.1 Yet, drug or alcohol concentrations can significantly be affected by the application of cosmetic hair treatments. These treatments include “detox-shampoos”, tinting, semi-permanent/permanent hair dyeing or hair straightening,2–7 however the most effective being oxidative hair treatments with hydrogen peroxide (H2O2) under alkaline conditions which are used for permanent hair dyeing and hair bleaching.8,9 Especially the latter can decrease DOA or alcohol marker concentrations significantly and might potentially result in false negative hair testing results.10 This makes them popular for intended hair manipulation to avoid positive tests.A major challenge in routine hair analysis is the detection of such adulteration attempts. So far, visual inspections of hair samples and colored extracts can give rise to suspected cosmetic hair treatments. Objective markers would however improve confidence in adulteration confirmation, particularly in court. For that reason, more complex techniques such as fluorescence microscopy11 or infrared spectroscopy12 have been developed over time to detect cosmetically treated hair samples. Yet, the detection of adulteration attempts with routinely available procedures like liquid chromatography tandem mass spectrometry (LC-MS/MS) would be ideal. In a first study, 1H-pyrrole-2,3,5-tricarboxylic acid (PTCA) was analyzed by LC-MS/MS and described as a marker for oxidative hair treatments.13 However, the implementation of cut-off values for PTCA is indispensable. Recently, 1H-pyrrole-2,3,4,5-tetracarboxylic acid (PTeCA) was identified to be exclusively formed through oxidative hair treatment which would in theory facilitate the detection of bleached hair samples as cut-off values do not need to be determined.14 At the moment, the current lack of commercially available reference standards and its melanin dependency still hinders its routine applicability. Hence, the need for further reliable markers for cosmetic hair treatments is more than relevant to date and new concepts for biomarker search are required.
The detection of direct oxidation products of DOA could be thought of as has been proposed for 2-nitro-6-monoacetylmorphine and 2-nitro-morphine as markers for KNO2 adulterated urine samples.15,16 Yet, this would require evaluation for every drug once again. A drug-independent, endogenous marker or a set thereof would therefore be better suited. As oxidative hair treatments can significantly affect DOA incorporated into the hair matrix, an impact on endogenous compounds in the same or similar way is likely.
Metabolomic analyses aim to extensively study small endogenous metabolites of an organism under different conditions (e.g. healthy vs. disease) to describe physiological processes and identify biomarkers.17 An untargeted metabolomics approach in theory analyses all metabolites without prior knowledge and serves as a hypothesis-generating tool. In recent studies by Joo et al. and Grosvenor et al., the influence of hair bleaching on amino acids and lipids and proteins was investigated, respectively.18,19 The authors could show that oxidative hair treatment has a significant impact on the studied compounds. However, these studies were conducted for a small selection of endogenous compounds and aimed to analyze the impact of hair bleaching on hair damage. Hence, applying an untargeted metabolomics approach should broaden the number of metabolites that potentially reveal differences of untreated and oxidatively treated hair samples and detect endogenous biomarkers that indicate manipulation attempts within routine hair analysis.
Within this proof-of-concept study, we aimed to identify a variety of possible biomarkers or biomarker combinations for oxidative hair treatments which could then potentially be applied within standard routine screening methods. For this, we applied an untargeted hair metabolomics approach to find altered and stable (for normalization purposes) endogenous metabolites between cosmetically untreated and bleached hair samples.
2. Materials and methods
2.1 Chemicals and reagents
1-Methylhistidine, ε-acetyllysine, adenine, adenosine, arginine, aspartic acid, azelaic acid, butyrylcarnitine (C4), chenodeoxycholic acid, cholic acid, choline, citrulline, cortisol, cortisone, creatinine, cysteic acid, DL-indole-3-lactic acid, decanoylcarnitine (C10), deoxycholic acid, glutamic acid, glutaric acid, glycocholic acid, hexanoylcarnitine (C6), hippuric acid, histidine, hypoxanthine, inosine, isoleucine, L-carnitine (C0), L-cysteine-S-sulfate, leucine, lysine, methionine, methylmalonic acid, mevalonolactone, N-acetylneuraminic acid, N,N-dimethylglycine, nicotinamide, nicotinic acid, octanoylcarnitine (C8), ornithine, p-aminobenzoic acid, phenylalanine, proline, propionylcarnitine (C3), pyroglutamic acid, raffinose, riboflavin, serine, suberic acid, taurine, taurocholic acid, theobromine, theophylline, threonine, trimethyllysine, tryptophan, tyrosine, uracil, uric acid, urocanic acid and valine, were purchased as analytical standards from Sigma-Aldrich (Buchs, Switzerland). The analytical reference standard of PTCA was sourced from ChemPur (Karlsruhe, Germany) and PTeCA was synthesized and delivered by ReseaChem Life Science (Burgdorf, Switzerland). All heavy-labeled and deuterated internal standards (IS) adenosine ribose-D1, arginine-13C6, caffeine 3-methyl-13C, carnitine trimethyl-D9, deoxycholic acid-D4, D-fructose 13C, hippuric acid 15N, leucine-D10, lysine-D4, phenylalanine-D1, proline 15N and tryptophan-D5 were purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA, USA) and delivered by ReseaChem Life Science (Burgdorf, Switzerland) or Sigma-Aldrich (Buchs, Switzerland). Water, acetonitrile (ACN), dichloromethane (DCM) and methanol (MeOH) of HPLC grade were obtained from Fluka (Buchs, Switzerland). All other chemicals used were from Merck (Zug, Switzerland) and of the highest grade available.Bleaching reagents (Venice Oxydant & active lotion containing 9% H2O2; Venice bleaching oxidation powder) were obtained from DOBI (Suhr, Switzerland) and commercial shampoo (I am, Intense Moisture, Buchs, Switzerland) from a local store.
2.2 Hair samples
Hair samples without cosmetic hair treatment (n = 21, analysis of the proximal 3 cm segment only) were used within this study (14 male and 7 female hair samples aged between 19 and 54). Hair sample colors ranged from black (n = 4), over dark brown to medium brown (n = 7), light brown to dark blond (n = 5), to medium blond to light blond (n = 5). Hair samples were obtained from volunteers who provided written informed consent and from the Center for Forensic Hair Analytics (Zurich, Switzerland) from routine case work after anonymization. Samples taken proactively from volunteers did not require an additional ethical approval according to the Swiss law (Humanforschungsgesetz) if the research is not aiming to investigate diseases or functions of the human body as is the case in the current study. Reuse of authentic samples from human case work was in full conformance with Swiss laws according to the statement of Cantonal Ethics Board of the Canton of Zurich (BASEC-no. Req-2017-00946). Samples were collected close to the scalp and stored in aluminum foil at room temperature.2.3 General hair adulteration procedure
Cosmetically untreated hair samples were split into two portions of which one was left untreated and one was bleached in vitro according to the manufacturer's instructions with a mixture of 2 g bleaching oxidase (containing 9% H2O2) and 1 g bleaching oxidation powder during incubation for 30 min. After initial rinsing of bleached hair samples to reach a neutral pH of the washing solution, untreated and bleached hair samples were both rinsed under running water and subsequently with 500 μL shampoo solution (1 mL shampoo, 9 mL H2O). Finally, hair samples were dried at room temperature overnight and processed as described in the following section 2.4.2.4 Sample preparation
All hair samples were prepared as described in detail elsewhere20 with minor changes. Hair samples were successively washed with DCM, acetone, H2O and acetone and dried overnight. 31.5 ± 0.5 mg of hair was weighed into a 2 mL Eppendorf tube containing one tungsten carbide ball and pulverized (10 min at 30 Hz) using a bench-top mill (Mixer Mill MM 400, Retsch, Haan, Germany). Hair samples were extracted with 1 mL ACN/H2O (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, v/v) and 20 μL IS solution (adenosine ribose-D1 (0.075 mM), arginine-13C6 (1.5 mM), caffeine 3-methyl-13C (1 mM), carnitine trimethyl-D9 (0.5 mM), deoxycholic acid-D4 (0.009 mM), D-fructose 13C (0.6 mM), hippuric acid 15N (2.5 mM), leucine-D10 (1.5 mM), lysine-D4 (3.5 mM), phenylalanine-D1 (1.5 mM), proline 15N (3.5 mM) and tryptophan-D5 (1.25 mM)) during 16 h of incubation in an ultrasonic bath. Afterwards, samples were centrifuged (5 min, 9′000 rpm) and the supernatant (900 μL) was evaporated to dryness under nitrogen at 35 °C. After reconstitution of the dried extract with 250 μL ACN/H2O (2
8, v/v) and 20 μL IS solution (adenosine ribose-D1 (0.075 mM), arginine-13C6 (1.5 mM), caffeine 3-methyl-13C (1 mM), carnitine trimethyl-D9 (0.5 mM), deoxycholic acid-D4 (0.009 mM), D-fructose 13C (0.6 mM), hippuric acid 15N (2.5 mM), leucine-D10 (1.5 mM), lysine-D4 (3.5 mM), phenylalanine-D1 (1.5 mM), proline 15N (3.5 mM) and tryptophan-D5 (1.25 mM)) during 16 h of incubation in an ultrasonic bath. Afterwards, samples were centrifuged (5 min, 9′000 rpm) and the supernatant (900 μL) was evaporated to dryness under nitrogen at 35 °C. After reconstitution of the dried extract with 250 μL ACN/H2O (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, v/v), samples were further purified by combined centrifugation and filtration (5 min, 9000 rpm, VWR centrifugal filter, 0.45 μm pore size, VWR, Dietikon, Switzerland). A solvent blank was prepared in the same manner to correct for false positive results and additionally, pooled quality control (QC) samples were prepared by mixing 30 μL of each final filtrate.
8, v/v), samples were further purified by combined centrifugation and filtration (5 min, 9000 rpm, VWR centrifugal filter, 0.45 μm pore size, VWR, Dietikon, Switzerland). A solvent blank was prepared in the same manner to correct for false positive results and additionally, pooled quality control (QC) samples were prepared by mixing 30 μL of each final filtrate.
2.5 UHPLC-QTOF MS analysis
Hair samples were analyzed in randomized order to minimize effects of potential varying instrument performances over time. LC-MS analysis of hair samples was performed as described in detail elsewhere.21 Briefly, a Thermo Fischer Ultimate 3000 UHPLC system (Thermo Fischer Scientific, San Jose, CA) coupled to a high-resolution (HR) time-of-flight (TOF) instrument (TripleTOF 6600, Sciex, Concord, Ontario, Canada) was used in combination with reversed-phase (Waters XSelect HSST RP-C18 (150 mm × 2.1 mm i.d.; 2.5 μm particle size; Waters (Baden-Daettwil, Switzerland)) and hydrophilic interaction liquid chromatography (Merck SeQuant ZIC HILIC column, 150 mm × 2.1 mm, 3.5 μm particle size). Gradient elution was performed after injection of 5 μL sample with the following mobile phases: mobile phase A (10 mM ammonium formate in H2O containing 0.1% (v/v) formic acid), mobile phase B (MeOH containing 0.1% (v/v) formic acid), mobile phase C (25 mM ammonium acetate containing 0.1% (v/v) acetic acid in H2O) and mobile phase D (ACN containing 0.1% (v/v) acetic acid). Gradient elution on the reversed-phase column was as follows: 1 min 100% A; 1–15 min increase to 100% B, decrease to start conditions after 18 min and re-equilibration for 2 min. Furthermore, gradient conditions on the HILIC column were: 1 min 95% D; 1–10 min decrease to 40% D; 10–12 min decrease to 10% D and held for 1 min; and increase to start conditions and re-equilibration for 4 min. Acquisition of HR mass spectra (MS) and tandem mass spectrometry (MS/MS) data for each sample was achieved using TOF-MS only and information-dependent acquisition (IDA) in positive (HSST pos and HILIC pos) and negative (HSST neg and HILIC neg) electrospray ionization (ESI) mode. The MS analysis was performed with a DuoSpray ion source at a resolving power (full width at half maximum (fwhm) at m/z 400) of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in MS and 30
000 in MS and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in MS2 (HR mode) or 15
000 in MS2 (HR mode) or 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (high-sensitivity mode). Automatic calibration was performed every five samples in positive ionization mode using an atmospheric-pressure chemical ionization (APCI) positive calibration solution (Sciex, Concord, Ontario, Canada) and every three samples in negative ionization mode using an APCI negative calibration solution (Sciex).
000 (high-sensitivity mode). Automatic calibration was performed every five samples in positive ionization mode using an atmospheric-pressure chemical ionization (APCI) positive calibration solution (Sciex, Concord, Ontario, Canada) and every three samples in negative ionization mode using an APCI negative calibration solution (Sciex).
TOF-MS scans were performed over a mass range of m/z 50 to m/z 1000, with an accumulation time of 100 ms and a collision energy (CE) of 5 eV. IDA scans were performed with an accumulation time of 100 ms and a CE of 35 eV with a CE spread of 15 eV over a mass range of m/z 50 to m/z 1000. Further criteria for IDA experiments were as follows: dynamic background subtraction on the four most intense ions; intensity threshold above 100 counts per second (cps); exclusion time of 5 s (half peak width) after two occurrences in high sensitivity mode. Data acquisition of MS parameters was controlled by Analyst TF software 1.7 (Sciex, Concord, Ontario, Canada).
At the sequence beginning, blanks and pooled QC samples were injected to allow instrument equilibration. Pooled QC samples as a measure for stability of the analytical system22 were injected every 5 samples. Additionally, to check for reproducibility of the data and retention time (RT) shifts, a system suitability test (SST) prepared according to Steuer et al.23 was measured every tenth sample. Analyte peak areas of SST compounds and potential identified markers were obtained by targeted peak area integration of precursor ions from TOF-MS data using MultiQuant V 2.1 (Sciex, Concord, Ontario, Canada).
2.6 Data processing, feature selection and statistical evaluation
For the evaluation of metabolites with constant concentrations (in the following referred to as “stable”) and altered metabolites between untreated and bleached hair samples, data-preprocessing, alignment, deconvolution, peak-picking, initial data normalization and filtering of TOF-MS data was performed in Progenesis QI (Waters Corp, Milford, USA). Data from IDA scans were incorporated for improved identification only. A pooled QC sample was chosen after automatic processing as an alignment reference. Peak-picking parameters were set as follows: automatic sensitivity method; sensitivity value 4; no minimum peak width; no retention time limits; ion species: [M + H]+, [M + 2H]2+, [M + H − H2O]+, [M + NH4]+, [M + Na]+, [M + 2Na]2+, [M − H]−, [M − 2H]−, [M − H2O − H]−, [M + Na − 2H]−, [M + FA − H]−. Data sets from HSST pos, HSST neg, HILIC pos and HILIC neg were filtered for features with an abundance >1000 cps in positive and >500 cps in negative ionization mode and available MS/MS spectra. All samples were finally normalized by the respective hair sample weight.Further feature selection and statistical analyses were performed in MetaboAnalyst 4.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24 and GraphPad Prism 7 (GraphPad Software, San Diego, CA. USA). Prior to statistical analyses in MetaboAnalyst, data was auto-scaled (mean-centered and divided by the standard deviation of each variable). Principal component analysis (PCA) was applied for first visualization of the data. To search for changed metabolites, volcano plots were applied with the following settings: Wilcoxon matched-pairs test (p < 0.01, false discovery rate (FDR) adjustment); fold-change analysis (threshold 2; significant paired count threshold 75%). The average loss of features in treated hair samples was calculated pairwise in percent with feature abundance in untreated hair samples set to 100%. Furthermore, GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA) was used to generate receiver operating characteristic (ROC) curves.
24 and GraphPad Prism 7 (GraphPad Software, San Diego, CA. USA). Prior to statistical analyses in MetaboAnalyst, data was auto-scaled (mean-centered and divided by the standard deviation of each variable). Principal component analysis (PCA) was applied for first visualization of the data. To search for changed metabolites, volcano plots were applied with the following settings: Wilcoxon matched-pairs test (p < 0.01, false discovery rate (FDR) adjustment); fold-change analysis (threshold 2; significant paired count threshold 75%). The average loss of features in treated hair samples was calculated pairwise in percent with feature abundance in untreated hair samples set to 100%. Furthermore, GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA) was used to generate receiver operating characteristic (ROC) curves.
2.7 Compound identification
Features of interest were searched against different databases on the MS and MS/MS level: the national institute of standards and technologies (NIST),25 the human metabolome database (HMDB),26 the online databases METLIN27 and Lipidblast28 and after calculation of the elemental composition in Progenesis QI as well as against an in-house database in PeakView V 2.2 (Sciex). Identity was confirmed by matching library search results, accurate precursor masses and fragment ions as well as RT to reference standards, if available. Identification results were classified to four identification confidence levels as suggested by the metabolomics standards initiative (MSI).293. Results and discussion
3.1 Bleaching procedure
To study the changes between cosmetically untreated and oxidatively treated hair samples, genuine hair samples were bleached in vitro. For this, we applied a concentration of 9% H2O2 during 30 min as this is to be expected to definitely produce an effect. In typical bleaching or hair dyeing products, H2O2 concentrations range from 2% to 12%.30,31 The H2O2 concentration chosen within our study therefore lies in the upper range. These bleaching conditions do not only lead to a visible brightening of the hair but also the hair structure carries away a severe damage as can already be seen by microscopic observation.19,32 This might also facilitate the leakage and the extraction from the hair matrix. Nevertheless, it is to be expected that individuals adulterating their hair on purpose with the goal of a negative hair testing result would accept and encourage a considerably high hair damage.3.2 Analytical procedure
Screening for potential differing metabolites due to oxidative hair treatment was performed with a previous evaluated LC-HRMS method.14,20 To avoid errors resulting from varying instrument performances over time, all samples were analyzed in randomized order and direct comparison of sample results was only performed within one batch. The analytical performance and the reproducibility of the batch analyses were monitored by deviation of RT and peak areas in SST and pooled QC samples. Mean coefficient of variations (CV) of peak areas were determined for all SST compounds and were 7.8 ± 2.9% (HSST pos), 11 ± 2.3% (HSST neg), 14 ± 5.3% (HILIC pos) and 21 ± 4.8% (HILIC neg). CV's of QC samples were <30% for the majority of the features (>75%), as is generally recommended for the assessment of the data quality.22 Hence, all experiments showed good stability of the data over the analytical runs and were therefore used for further data analysis.3.3 Feature selection
Progenesis QI was used for peak picking and data alignment. Compared to e.g. blood or urine samples, expected concentrations of exo- and endogenous compounds in hair samples are generally lower.1 To avoid missing compounds, the applied sensitivity value in Progenesis QI for peak-picking was optimized prior to further data analysis (data not shown). Increasing the sensitivity value lead to an increased number of detected compound ions. Even though this in turn also increased the background noise, it was accepted for this initial proof-of-concept study in order to detect as many potentially interesting candidates as possible. Processing in Progenesis QI initially lead to the detection of a maximum of 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compound ions. To allow for unambiguous identification, data was filtered for features (defined by RT and m/z) with MS/MS spectra only. Additionally, features with an abundance <1000 cps (in positive ionization) and <500 cps (in negative ionization) were removed due to their high CV's and low reproducibility.
000 compound ions. To allow for unambiguous identification, data was filtered for features (defined by RT and m/z) with MS/MS spectra only. Additionally, features with an abundance <1000 cps (in positive ionization) and <500 cps (in negative ionization) were removed due to their high CV's and low reproducibility.
For metabolites affected by cosmetic treatment, first visualization of the data by PCA analysis already showed clear separation of both groups in all four data sets as shown in Fig. 1. Moreover, volcano plots were applied to filter for features of significant interest. A significant paired count threshold of 75% was applied meaning that a minimum of 75% of the samples needed a consistent change above the given fold-change threshold. A total of 1053 (HSST pos), 903 (HSST neg), 1148 (HILIC pos) and 2005 (HILIC neg) features fulfilled the criteria and showed significant changes between untreated and bleached hair samples. The average loss of features across samples in percent can be found in Table 1.
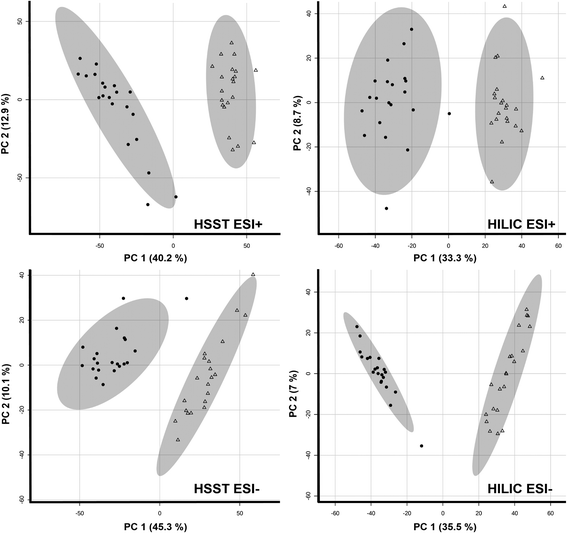 | ||
| Fig. 1 Principal component analyses (PCA) of untreated (triangle) and treated (9% H2O2, 30 min; radial form) hair samples (n = 21 each). | ||
| Feature name | Analytics | Adduct | Measured m/z | Delta [ppm] | RT [min] | FDR adjusted p-value | Fold change | Conc. change, mean loss [%] | Formula | (Tentative) identification | MSI level | Compound class | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ↑: increase after bleaching; ↓ decrease after bleaching. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | 0.90_104.1058 m/z | HSST pos | M + H | 104.1058 | −4.40 | 0.9 | 8.7 × 10−6 | 2.7 | ↓, −53.7% | C5H14NO | Choline | 1 | 1,2-Aminoalcohols | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 7.14_170.0102 m/z | HILIC pos | M + H | 170.0102 | −9.21 | 7.1 | 2.9 × 10−6 | 15.8 | ↑ | C3H7NO5S | Cysteic acid | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7.11_167.9968 m/z | HILIC neg | M − H | 167.9968 | −2.49 | 7.1 | 5.1 × 10−6 | 3.9 | ↑ | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | 1.90_139.0493 m/z | HSST pos | M + H | 139.0493 | −6.20 | 1.9 | 2.1 × 10−6 | 9.4 | ↓, −89.8% | C6H6N2O2 | Urocanic acid | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1.90_137.0359 m/z | HSST neg | M − H | 137.0359 | 1.53 | 1.9 | 5.8 × 10−5 | 12.8 | ↓, −50.6% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4.14_139.0491 m/z | HILIC pos | M + H | 139.0491 | −8.06 | 4.1 | 2.4 × 10−4 | 6.6 | ↓, −19.3% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | 0.87_174.0888 m/z | HSST neg | M − H | 174.0888 | 2.40 | 0.9 | 1.8 × 10−6 | 10.4 | ↓, −87.6% | C6H13N3O3 | Citrulline | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7.41_174.0882 m/z | HILIC neg | M − H | 174.0882 | −1.18 | 7.4 | 3.1 × 10−6 | 26.5 | ↓, −96.0% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | 0.82_189.1591 m/z | HSST pos | M + H | 189.1591 | −3.51 | 0.8 | 2.1 × 10−6 | 9.6 | ↓, −90.7% | C9H20N2O2 | Trimethyllysine | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10.89_189.1580 m/z | HILIC pos | M + H | 189.1580 | −9.43 | 10.9 | 2.9 × 10−6 | 9.0 | ↓, −87.1% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | 1.63_130.0490 m/z | HSST pos | M + H | 130.0490 | −6.60 | 1.6 | 2.1 × 10−6 | 2379.5 | ↓, −99.9% | C5H7NO3 | Pyroglutamic acid | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1.26_128.0356 m/z | HSST neg | M − H | 128.0356 | 2.27 | 1.3 | 1.8 × 10−6 | 97.8 | ↓, −98.4% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5.75_130.0486 m/z | HILIC pos | M + H | 130.0486 | −9.77 | 5.8 | 2.9 × 10−6 | 214.2 | ↓, −99.4% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5.73_129.0424n | HILIC neg | M − H, M + Cl | 128.0351 | −1.60 | 5.7 | 3.1 × 10−6 | 41.4 | ↓, −96.8% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | 0.80_199.9694 m/z | HSST neg | M − H | 199.9694 | 0.36 | 0.8 | 1.8 × 10−6 | 10.0 | ↑ | C3H7NO5S2 | Cysteine-sulfate | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7.04_201.9824 m/z | HILIC pos | M + H | 201.9824 | −7.21 | 7.0 | 2.9 × 10−6 | 11.0 | ↑ | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | 0.86_131.0825 m/z | HSST neg | M − H | 131.0825 | −1.14 | 0.9 | 1.8 × 10−6 | 4.9 | ↓, −75.7% | C5H12N2O2 | Ornithine | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9.67_131.0820 m/z | HILIC neg | M − H | 131.0820 | −4.61 | 9.7 | 3.1 × 10−6 | 62.3 | ↓, −97.8% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | 8.12_206.0804 m/z | HSST pos | M + H | 206.0804 | −3.90 | 8.1 | 2.1 × 10−6 | 4.1 | ↓, −76.1% | C11H11NO3 | DL-Indole-3-lactic acid | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3.51_204.0660 m/z | HILIC neg | M − H | 204.0660 | −2.86 | 3.5 | 3.1 × 10−6 | 7.7 | ↓, −87.3% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | 1.15_203.1491 m/z | HSST pos | M + H | 203.1491 | −5.56 | 1.2 | 2.1 × 10−6 | 6.1 | ↓, −82.5% | C8H18N4O2 | N,N-Dimethylarginine | 2 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9.23_203.1490 m/z | HILIC pos | M + H | 203.1490 | −6.43 | 9.2 | 2.9 × 10−6 | 21.2 | ↓, −94.3% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | 8.42_238.1068 m/z | HSST pos | M + H | 238.1068 | −2.57 | 8.4 | 2.1 × 10−6 | 3.8 | ↓, −71.2% | C12H15NO4 | N-lactoyl phenylalanine | 2 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | 6.59_189.1217 m/z | HILIC pos | M + H | 189.1217 | −8.61 | 6.6 | 7.0 × 10−6 | 3.0 | ↓, −52.9% | C8H16N2O3 | ε-Acetyllysine | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | 9.27_189.1329 m/z | HILIC pos | M + H | 189.1329 | −9.29 | 9.3 | 2.9 × 10−6 | 24.8 | ↓, −95.3% | C7H16N4O2 | Monomethylarginine | 2 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | 5.56_190.0539 m/z | HSST neg | M − H | 190.0539 | −2.13 | 5.6 | 1.8 × 10−6 | 676.1 | ↓, −100% | C7H13NO3S | Actetylmethionine | 2 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1.19_190.0534 m/z | HILIC neg | M − H | 190.0534 | −4.91 | 1.2 | 2.8 × 10−5 | 34.4 | ↓, −40.7% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | 0.84_152.0017 m/z | HSST neg | M − H | 152.0017 | −4.19 | 0.8 | 3.3 × 10−6 | 4.9 | ↑ | C3H7NO4S | Cysteinesulfinic acid | 2 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | 7.96_166.0625n | HSST neg | M − H, M − H2O − H | 165.0553 | −3.23 | 8.0 | 1.8 × 10−6 | 7.3 | ↓, −86.1% | C9H10O3 | Phenyllactic acid | 2 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | 5.65_182.0575n | HSST neg | M − H, M −H2O −H | 181.0504 | −2.45 | 5.7 | 1.8 × 10−6 | 8.1 | ↓, −87.2% | C9H10O4 | Hydroxyphenyllactic acid | 2 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3.82_181.0501 m/z | HILIC neg | M −H | 181.0501 | −2.66 | 3.8 | 7.2 × 10−6 | 20.5 | ↓, −82.6% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 18 | 7.13_188.0559 m/z | HILIC neg | M −H | 188.0559 | −2.65 | 7.1 | 3.1 × 10−6 | 27.6 | ↓, −96.3% | C7H11NO5 | Acetylglutamic acid | 2 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 19 | 6.77_124.0069 m/z | HILIC neg | M −H | 124.0069 | −4.13 | 6.8 | 3.1 × 10−6 | 9.2 | ↓, −89.2% | C2H7NO3S | Taurine | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 20 | 7.26_104.0343 m/z | HILIC neg | M −H | 104.0343 | −9.53 | 7.3 | 3.1 × 10−6 | 21.1 | ↓, −92.6% | C3H7NO3 | Serine | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 21 | 0.84_118.0499 m/z | HSST neg | M −H | 118.0499 | −8.70 | 0.8 | 5.7 × 10−6 | 2.9 | ↓, −59.1% | C4H9NO3 | Threonine | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 22 | 2.37_132.1007 m/z | HSST pos | M + H | 132.1007 | −9.49 | 2.4 | 2.1 × 10−6 | 4.9 | ↓, −74.6% | C6H13NO2 | Isoleucine/leucine | 2 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5.40_130.0870 m/z | HILIC neg | M − H | 130.0870 | −2.43 | 5.4 | 3.1 × 10−6 | 4.8 | ↓, −73.6% | Isoleucine | 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 23 | 2.21_132.1006 m/z | HSST pos | M + H | 132.1006 | −9.66 | 2.2 | 2.1 × 10−6 | 5.3 | ↓, −77.5% | C6H13NO2 | Leucine/isoleucine | 2 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2.23_130.0874 m/z | HSST neg | M −H | 130.0874 | 0.37 | 2.2 | 1.8 × 10−6 | 3.8 | ↓, −67.4% | Leucine/isoleucine | 2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5.57_130.0870 m/z | HILIC neg | M −H | 130.0870 | −2.71 | 5.6 | 3.1 × 10−6 | 4.3 | ↓, −72.6% | Leucine | 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 24 | 0.81_175.1182 m/z | HSST pos | M + H | 175.1182 | −4.47 | 0.8 | 2.1 × 10−6 | 6.4 | ↓, −83.2% | C6H14N4O2 | Arginine | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 0.82_173.1043 m/z | HSST neg | M − H | 173.1043 | −0.5 | 0.8 | 1.8 × 10−6 | 5.9 | ↓, −82.4% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9.68_173.1037 m/z | HILIC neg | M − H | 173.1037 | −3.99 | 9.7 | 7.2 × 10−6 | 68.8 | ↓, −89.1% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 25 | 9.84_145.0976 m/z | HILIC neg | M − H | 145.0976 | −4.39 | 9.8 | 3.1 × 10−6 | 11.5 | ↓, −87.7% | C6H14N2O2 | Lysine | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 26 | 4.30_166.0856 m/z | HSST pos | M + H | 166.0856 | −4.20 | 4.3 | 2.1 × 10−6 | 3.2 | ↓, −65.6% | C9H11NO2 | Phenylalanine | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 27 | 2.36_182.0803 m/z | HSST pos | M + H | 182.0803 | −4.54 | 2.4 | 2.1 × 10−6 | 4.3 | ↓, −75.3% | C9H11NO3 | Tyrosine | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2.33_180.0667 m/z | HSST neg | M − H | 180.0667 | 0.64 | 2.3 | 3.3 × 10−6 | 3.5 | ↓, −64.9% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6.08_180.0666 m/z | HILIC neg | M − H | 180.0666 | −0.14 | 6.1 | 3.1 × 10−6 | 4.9 | ↓, −75.1% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 28 | 5.45_227.0778 m/z | HSST pos | M + Na, M + H | 227.0778 | −6.36 | 5.5 | 2.1 × 10−6 | 7.3 | ↓, −85.0% | C11H12N2O2 | Tryptophan | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5.48_203.0823 m/z | HILIC neg | M −H | 203.0823 | −1.46 | 5.5 | 3.1 × 10−6 | 3.6 | ↓, −68.0% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 29 | 0.79_156.0756 m/z | HSST pos | M + H | 156.0756 | −7.27 | 0.8 | 8.7 × 10−6 | 6.8 | ↓, −76.0% | C6H9N3O2 | Histidine | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 30 | 0.83_147.0519n | HSST pos | M + H, M + H − H2O | 148.0594 | −8.55 | 0.8 | 2.1 × 10−6 | 6.3 | ↓, −82.8% | C5H9NO4 | Glutamic acid | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 0.84_147.0531n | HSST neg | M −H, M − H2O −H | 146.0460 | −0.18 | 0.8 | 1.8 × 10−6 | 7.0 | ↓, −82.3% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7.55_147.0518n | HILIC pos | M + H, M + H − H2O | 148.0595 | −9.46 | 7.6 | 2.9 × 10−6 | 7.4 | ↓, −79.8% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 31 | 1.17_116.0714 m/z | HSST neg | M −H | 116.0714 | −2.23 | 1.2 | 1.8 × 10−06 | 4.5 | ↓, −70.0% | C5H11NO2 | Valine | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6.07_116.0712 m/z | HILIC neg | M −H | 116.0712 | −4.07 | 6.1 | 7.2 × 10−6 | 4.7 | ↓, −69.4% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 32 | 0.80_133.0376n | HSST neg | M − H, M + Na −2H | 132.0303 | 0.70 | 0.8 | 1.8 × 10−06 | 15.9 | ↓, 92.2% | C4H7NO4 | Aspartic acid | 1 | Amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7.58_132.0299 m/z | HILIC neg | M − H | 132.0299 | −2.45 | 7.6 | 3.1 × 10−6 | 18.5 | ↓, −91.4% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 33 | 0.86_310.1125 m/z | HSST pos | M + H | 310.1125 | −2.33 | 0.9 | 2.1 × 10−6 | 89.5 | ↓, −99.9% | C11H19NO9 | N-Acetylneuraminic acid | 1 | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 0.90_308.0984 m/z | HSST neg | M − H | 308.0984 | −1.13 | 0.9 | 1.8 × 10−6 | 137.6 | ↓, −99.1% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6.80_309.1055n | HILIC pos | M + H | 310.1128 | −1.52 | 6.8 | 2.9 × 10−6 | 14.0 | ↓, −92.4% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6.78_308.0984 m/z | HILIC neg | M −H | 308.0984 | −1.02 | 6.8 | 3.1 × 10−6 | 247.1 | ↓, −99.6% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 34 | 7.78_162.1103 m/z | HILIC pos | M + H | 162.1103 | −6.20 | 7.8 | 5.0 × 10−6 | 11.5 | ↓, −83.0% | C7H15NO3 | Carnitine (C0) | 1 | Carnitines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 35 | 6.46_218.1367 m/z | HILIC pos | M + H | 218.1367 | −9.31 | 6.5 | 4.8 × 10−5 | 7.8 | ↓, −56.4% | C10H19NO4 | Propionylcarnitine (C3) | 1 | Carnitines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 36 | 6.07_232.1525 m/z | HILIC pos | M + H | 232.1525 | −7.79 | 6.1 | 2.9 × 10−6 | 21.9 | ↓, −91.2% | C11H21NO4 | Butyrylcarnitine (C4) | 1 | Carnitines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 37 | 6.11_246.1697 m/z | HSST pos | M + H | 246.1697 | −1.22 | 6.1 | 2.1 × 10−6 | 9.0 | ↓, −89.9% | C12H23NO4 | 2-Methylbutyroylcarnitine | 2 | Carnitines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 38 | 7.89_260.1844 m/z | HSST pos | M + H | 260.1844 | −4.71 | 7.9 | 2.1 × 10−6 | 203.0 | ↓, −99.4% | C13H25NO4 | Hexanoylcarnitine (C6) | 1 | Carnitines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5.57_260.1834 m/z | HILIC pos | M + H | 260.1834 | −8.49 | 5.6 | 2.9 × 10−6 | 45.4 | ↓, −97.5% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 39 | 9.33_274.2001 m/z | HSST pos | M + H | 274.2001 | −4.36 | 9.3 | 2.1 × 10−6 | 362.6 | ↓, −99.2% | C14H27NO4 | Heptanoylcarnitine (C7) | 2 | Carnitines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 40 | 10.55_288.2173 m/z | HSST pos | M + H | 288.2173 | 1.14 | 10.6 | 2.1 × 10−6 | 38.6 | ↓, −97.0% | C15H29NO4 | Octanoylcarnitine (C8) | 1 | Carnitines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5.33_288.2157 m/z | HILIC pos | M + H | 288.2157 | −4.44 | 5.3 | 2.9 × 10−6 | 73.0 | ↓, −98.7% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 41 | 5.42_286.1983 m/z | HILIC pos | M + H − H2O, M + H | 286.1983 | −9.81 | 5.4 | 2.9 × 10−6 | 1290.1 | ↓, −99.9% | C15H29NO5 | Hydroxyoctanoylcarnitine | 2 | Carnitines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 42 | 11.54_302.2303 m/z | HSST pos | M + H | 302.2303 | −7.59 | 11.5 | 2.1 × 10−6 | 12.0 | ↓, −89.5% | C16H31NO4 | Nonanoylcarnitine (C9) | 2 | Carnitines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5.26_302.2311 m/z | HILIC pos | M + H | 302.2311 | −5.00 | 5.3 | 2.9 × 10−6 | 13.0 | ↓, −92.6% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 43 | 12.36_316.2464 m/z | HSST pos | M + H | 316.2464 | −5.83 | 12.4 | 2.1 × 10−6 | 14.2 | ↓, −91.2% | C17H33NO4 | Decanoylcarnitine (C10) | 1 | Carnitines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5.08_315.2392n | HILIC pos | M + H | 316.2465 | −5.65 | 5.1 | 2.9 × 10−6 | 3.8 | ↓, −72.6% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 44 | 5.48_332.2400 m/z | HILIC pos | M + H | 332.2400 | −9.43 | 5.5 | 2.9 × 10−6 | 1836.5 | ↓, −100% | C17H33NO5 | Hydroxydecanoylcarnitine | 2 | Carnitines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 45 | 7.76_377.1486 m/z | HSST pos | M + H | 377.1486 | 8.13 | 7.8 | 3.7 × 10−6 | 2.1 | ↓, −49.5% | C17H20N4O6 | Riboflavin | 1 | Cofactors | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 46 | 1.15_123.0544 m/z | HILIC pos | M + H | 123.0544 | −7.31 | 1.2 | 4.9 × 10−5 | 3.4 | ↓, −53.1% | C6H6N2O | Nicotinamid | 1 | Cofactors | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 47 | 7.13_665.1229n | HILIC pos | M + 2H, M + H | 666.1280 | −2.75 | 7.1 | 2.9 × 10−6 | 75.7 | ↑ | C21H29N7O14P2 | NADH | 2 | Cofactors | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 48 | 8.54_174.0879n | HSST pos | M + H − H2O, M + H, M + NH4 | 192.1160 | −7.63 | 8.5 | 3.4 × 10−9 | 2.0 | ↓, −48.0% | C8H14O4 | Suberic acid | 1 | Fatty acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 49 | 9.94_188.1048n | HSST neg | M − H, M + Na − 2H | 187.0975 | −0.41 | 9.9 | 1.7 × 10−3 | 2.6 | ↓, −28.9% | C9H16O4 | Azelaic acid | 1 | Fatty acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 50 | 3.73_89.0243 m/z | HILIC neg | M − H | 89.0243 | −1.56 | 3.7 | 3.1 × 10−6 | 189.5 | ↓, −98.9% | C3H6O3 | Lactic acid | 2 | Hydroxy acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 51 | 5.42_157.0359 m/z | HILIC neg | M −H | 157.0359 | −5.26 | 5.4 | 3.1 × 10−6 | 53.7 | ↓, −98.3% | C4H6N4O3 | Allantoin | 2 | Imidazoles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 52 | 5.49_146.0592 m/z | HILIC pos | M + H | 146.0592 | −6.03 | 5.5 | 2.9 × 10−6 | 3.6 | ↓, −66.4% | C9H7NO | 1H-Indole-4-carboxaldehyde | 2 | Indoles and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 53 | 3.90_198.0046 m/z | HSST neg | M − H | 198.0046 | 1.09 | 3.9 | 1.8 × 10−6 | 83.8 | ↑ | C7H5NO6 | PTCA | 1 | Melanin degradation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 54 | 5.34_198.0045 m/z | HSST neg | M − H | 198.0045 | 0.52 | 5.3 | 1.8 × 10−6 | 18.7 | ↑ | C7H5NO6 | IsoPTCA | 2 | Melanin degradation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 55 | 6.82_241.9944 m/z | HSST neg | M − H | 241.9944 | 0.53 | 6.8 | 1.8 × 10−6 | 126.8 | ↑ | C8H5NO8 | PTeCA | 1 | Melanin degradation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 56 | 5.06_220.1171 m/z | HSST pos | M + H | 220.1171 | −4.07 | 5.1 | 2.1 × 10−6 | 6.5 | ↓, −82.3% | C9H17NO5 | Pantothenic acid | 2 | Monocarboxylic acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3.41_218.1030 m/z | HILIC neg | M − H | 218.1030 | −1.69 | 3.4 | 3.1 × 10−6 | 21.1 | ↓, −95.8% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 57 | 4.00_268.0840n | HSST pos | M + H, M + Na | 269.0913 | 12.12 | 4.0 | 2.1 × 10−6 | 3.7 | ↓, −71.3% | C10H12N4O5 | Inosine | 1 | Nucleosides and nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4.00_267.0730 m/z | HSST neg | M − H | 267.0730 | −2.02 | 4.0 | 3.3 × 10−6 | 3.8 | ↓, −68.0% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4.82_269.0873 m/z | HILIC pos | M + H | 269.0873 | −2.95 | 4.8 | 5.0 × 10−6 | 3.2 | ↓, −63.9% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4.81_267.0724 m/z | HILIC neg | M − H | 267.0724 | −3.92 | 4.8 | 3.1 × 10−6 | 4.4 | ↓, −71.6% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 58 | 4.56_268.1036 m/z | HSST pos | M + H | 268.1036 | −1.49 | 4.6 | 5.1 × 10−4 | 2.3 | ↑ | C10H13N5O4 | Adenosine | 1 | Nucleosides and nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 59 | 5.48_284.0967 m/z | HILIC pos | M + H | 284.0967 | −7.90 | 5.5 | 2.9 × 10−6 | 3.4 | ↓, −71.3% | C10H13N5O5 | Guanosine | 2 | Nucleosides and nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5.48_282.0832 m/z | HILIC neg | M − H | 282.0832 | −4.19 | 5.5 | 3.1 × 10−6 | 5.4 | ↓, −80.4% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 60 | 1.23_348.0700 m/z | HSST pos | M + H | 348.0700 | −0.97 | 1.2 | 2.1 × 10−6 | 3.5 | ↑ | C10H14N5O7P | Adenosine monophosphate (AMP) | 2 | Nucleosides and nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1.26_346.0553 m/z | HSST neg | M − H | 346.0553 | −1.54 | 1.3 | 1.8 × 10−6 | 3.1 | ↑ | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6.99_348.0696 m/z | HILIC pos | M + H | 348.0696 | −2.31 | 7.0 | 2.9 × 10−6 | 3.4 | ↑ | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 61 | 1.19_560.0779 m/z | HSST pos | M + H | 560.0779 | −1.81 | 1.2 | 5.2 × 10−6 | 2.6 | ↑ | C15H23N5O14P2 | Adenosine diphosphate ribose (ADP-ribose) | 2 | Nucleosides and nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1.21_558.0639 m/z | HSST neg | M −H | 558.0639 | −0.94 | 1.2 | 7.5 × 10−7 | 2.8 | ↑ | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7.21_560.0752 m/z | HILIC pos | M + H | 560.0752 | −6.71 | 7.2 | 2.9 × 10−6 | 3.5 | ↑ | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 62 | 3.28_282.1194 m/z | HSST pos | M + H | 282.1194 | −1.14 | 3.3 | 2.1 × 10−6 | 51.2 | ↓, −98.3% | C11H15N5O4 | Methyladenosine | 2 | Nucleosides and nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6.70_282.1183 m/z | HILIC pos | M + H | 282.1183 | −4.83 | 6.7 | 2.9 × 10−6 | 28.4 | ↓, −92.5% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 63 | 5.47_312.1291 m/z | HSST pos | M + H | 312.1291 | −3.77 | 5.5 | 2.1 × 10−6 | 2.3 | ↓, −56.7% | C12H17N5O5 | N,N-Dimethylguanosine | 2 | Nucleosides and nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4.02_312.1275 m/z | HILIC pos | M + H | 312.1275 | −8.91 | 4.0 | 9.7 × 10−4 | 2.4 | ↓, −52.1% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 64 | 1.23_426.0220 m/z | HSST neg | M − H | 426.0220 | −0.21 | 1.2 | 1.8 × 10−6 | 10.7 | ↑ | C10H15N5O10P2 | Adenosine diphosphate (ADP) | 2 | Nucleosides and nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7.95_428.0334 m/z | HILIC pos | M + H | 428.0334 | −7.77 | 8.0 | 2.9 × 10−6 | 11.5 | ↑ | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 65 | 3.52_166.0709 m/z | HSST pos | M + H | 166.0709 | −8.51 | 3.5 | 2.1 × 10−6 | 8.2 | ↓, −91.0% | C6H7N5O | Methylguanine | 2 | Purines and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 66 | 5.34_181.0711 m/z | HSST pos | M + H | 181.0711 | −5.17 | 5.3 | 2.1 × 10−6 | 9.2 | ↓, −87.4% | C7H8N4O2 | Theobromine | 1 | Purines and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1.13_181.0715 m/z | HILIC pos | M + H | 181.0715 | −3.03 | 1.1 | 2.9 × 10−6 | 6.7 | ↓, −86.5% | Theobromine/Theophylline | 2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 67 | 5.95_181.0716 m/z | HSST pos | M + H | 181.0716 | −2.36 | 6.0 | 2.1 × 10−6 | 6.1 | ↓, −73.2% | C7H8N4O2 | Theophylline | 1 | Purines and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 68 | 4.00_137.0457 m/z | HSST pos | M + H | 137.0457 | 2.59 | 4.0 | 2.1 × 10−6 | 4.3 | ↓, −76.9% | C5H4N4O | Hypoxanthine | 1 | Purines and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 69 | 1.23_169.0347 m/z | HSST pos | M + H | 169.0347 | −5.49 | 1.2 | 2.1 × 10−6 | 632.1 | ↓, −99.9% | C5H4N4O3 | Uric acid | 1 | Purines and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1.26_167.0210 m/z | HSST neg | M − H | 167.0210 | −0.23 | 1.3 | 1.5 × 10−5 | 225.8 | ↓, −80.5% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5.76_167.0207 m/z | HILIC neg | M − H | 167.0207 | −2.01 | 5.8 | 3.1 × 10−6 | 110.9 | ↓, −98.2% | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
To screen for metabolites remaining stable after treatment and between subjects, further filtering for features with a CV <30% only was conducted. Additionally, a fold-change threshold ≤1.2 and significance level with p ≥ 0.05 had to be fulfilled. In total, 126 (HSST pos), 61 (HSST neg), 220 (HILIC pos) and 35 (HILIC neg) stable features were detected. These are low numbers compared to the number of total compound ions detected, assuming that the vast majority of hair metabolites is affected through the quite aggressive oxidative bleaching procedure. To avoid missing of important stable features through the applied MS/MS filter, the same processing has been performed including also features without MS/MS data. The number of stable features however only increased minimally (e.g. from 126 to 144 features in HSST pos mode).
3.4 Feature identification
Hit lists of features were manually checked for positive hits. In total, 69 altered metabolites could be identified, thereof 43 metabolites with an identification level 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 29 after comparison of MS/MS spectra and RT to a reference standard as summarized in Table 1. For the purpose of the study, only metabolites with an identification level of 2 and better were considered in this study as unambiguous identification is necessary for their implementation into routine analysis. No stable metabolites could be identified with sufficient confidence (level 2 or better). Compounds resulting clearly from exogenous origin (e.g. caffeine, dexpanthenol) were not further investigated as inter-individual differences are unpredictable and marker suitability is considered as low. Some compounds were detected on both columns and/or different ionization modes and are listed as a single compound in Table 1. If insufficient chromatographic separation hindered an exact identification, the MSI level was assigned to level 2. From the hit lists of significantly altered features, a high number of features could be assigned to the compound class of peptides (see also Table S1†). As the protein content of hair is very high,33 this supports the hypothesis of an enhanced degradation and break-down of proteins from the hair matrix, thus extending the feature hit list.
29 after comparison of MS/MS spectra and RT to a reference standard as summarized in Table 1. For the purpose of the study, only metabolites with an identification level of 2 and better were considered in this study as unambiguous identification is necessary for their implementation into routine analysis. No stable metabolites could be identified with sufficient confidence (level 2 or better). Compounds resulting clearly from exogenous origin (e.g. caffeine, dexpanthenol) were not further investigated as inter-individual differences are unpredictable and marker suitability is considered as low. Some compounds were detected on both columns and/or different ionization modes and are listed as a single compound in Table 1. If insufficient chromatographic separation hindered an exact identification, the MSI level was assigned to level 2. From the hit lists of significantly altered features, a high number of features could be assigned to the compound class of peptides (see also Table S1†). As the protein content of hair is very high,33 this supports the hypothesis of an enhanced degradation and break-down of proteins from the hair matrix, thus extending the feature hit list.
As discussed in detail in a previous publication, the “hair metabolome” is still insufficiently described.20 Thus, many metabolites which might be hair specific might not be represented within the different databases and are therefore unamenable to identification. The majority of identified metabolites was found to be decreased in concentration or even totally degraded through the bleaching process. Higher levels due to bleaching were only observed for adenosine and its derivatives. Neo-formation or the detection of direct oxidation products was only observed for melanin degradation products and oxidation products of cysteine and cystine. The difficulty to identify new oxidation products is based on the fact that spectral databases used for metabolomics deal with endogenous compounds altered under physiological conditions only. The bleaching process however is an “artificial” influence on the endogenous metabolites which produces oxidation products that might not be present under physiological conditions and have therefore not (yet) been described and studied. Hence, they might rarely be present in currently available databases. In addition, follow-up reactions of oxidation products or multiple oxidations are difficult to predict and to identify in a complex sample. Even after identification, such oxidation products might rarely be available as reference material for routine analysis. Therefore, an intensified search for further oxidation products was omitted and focus applied on the (high) number of identified compounds.
3.5 Marker interpretation
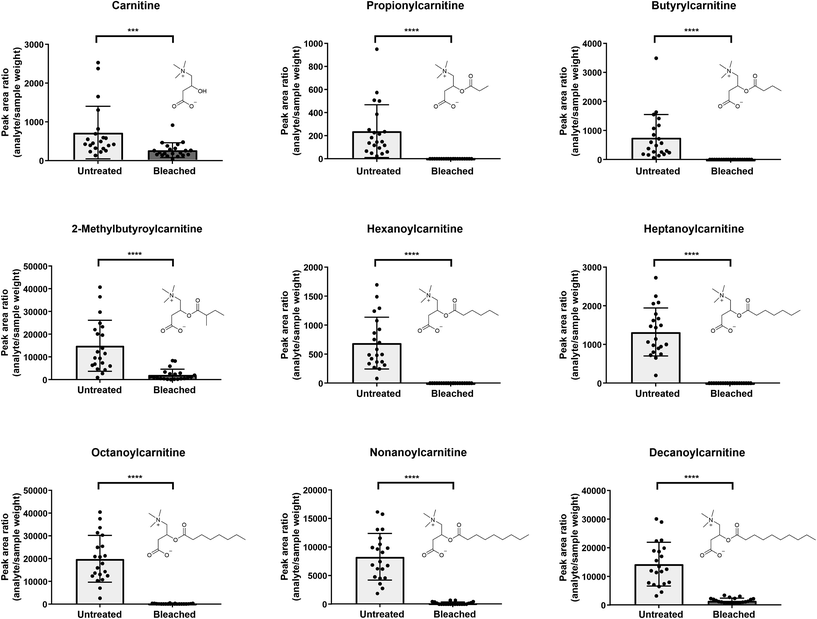
In general, a high number of compounds was found to be significantly altered between untreated and bleached hair samples. Altogether, with this untargeted workflow, we found compound changes in all directions: increase, decrease, total degradation and neo-formation (see Table 1). To act as a potential biomarker, a compound should ideally be usable in qualitative screening procedures without the time-intensive and complex establishment of a cut-off value. This is only true for newly formed (oxidation) products that are missing in authentic hair samples. Total degradation of endogenous compounds could be useful, but might also depend on the sensitivity of the method. The used Q-TOF method was selected for its broad screening and identification properties, but lacks sensitivity compared to typical quantification instruments such as triple quadrupols in multiple reaction monitoring mode. Compounds that are undetectable in the current setting might still be measurable in low amounts when more sensitive methods are applied. For metabolites that are not solely formed through adulteration but only altered in abundance, high inter-individual concentration differences in the metabolome due to lifestyle habits or diseases (‘endogenous variances’) and/or due to the complex nature of hair samples (‘exogenous variances’) are expected, even without oxidative treatments. Therefore, at best, markers from different metabolic pathways should be screened for. Furthermore, additional strategies should be considered, such as formation of metabolite ratios or normalization to endogenous metabolites unaffected by treatment and homogenously distributed among the population. Compound classes predominantly identified as significantly altered after bleaching include melanin degradation products, carnitines, amino acids and derivatives, as well as purines, nucleosides and derivatives and will be discussed in the following subchapters.As depicted in Fig. 3, five out of 9 detected carnitines were degraded after bleaching. Mainly low abundant acylcarnitines were undetectable, assuming that complete analyte degradation after bleaching might be dependent on initial metabolite concentrations and/or instrument sensitivity. If increased lipophilicity causes higher incorporation into the hair matrix,1 increasing chain length of the acyl moiety should result in higher levels in bleached hair samples. Yet, this correlation could not be confirmed in our study.
Selecting various metabolites over one single marker can increase confidence of the analytical findings. Kluge et al. investigated the suitability of endogenous biomolecules as validity parameters for the detection of manipulated urine samples.41 Samples were considered as manipulated if less than six out of 10 endogenous biomolecules (therefrom six acylcarnitines: C3, butyrylcarnitine (C4), isovalerylcarnitine, hexanoylcarnitine (C6), heptanoylcarnitine (C7) and octanoylcarnitine (C8)) were identified by an untargeted LC-MS/MS screening method.41 In line, we propose, that undetectability of highly abundant carnitines such as e.g. C8 in hair would highlight a sample as suspicious. C4 and C8 especially showed high prevalences with detection rates >98% in authentic urine samples.41 However, similar comprehensive data on expected prevalence of carnitines in hair samples is still missing.
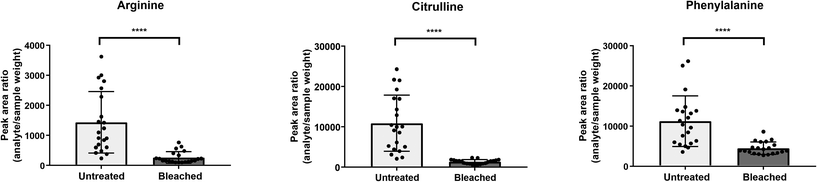
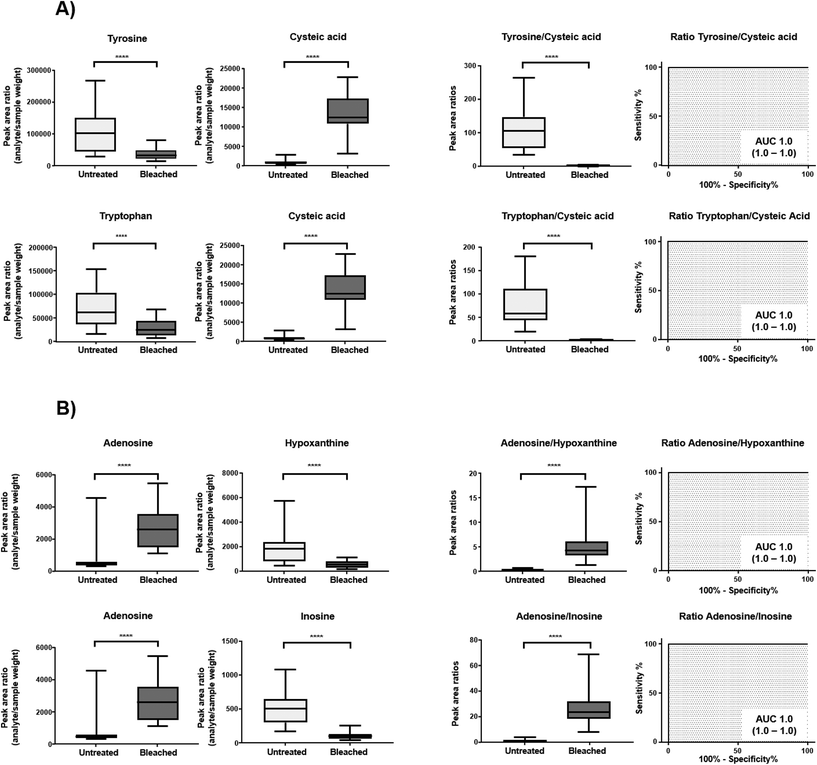
Cysteine is predominantly present in the keratin structure. Sulphur groups of cysteine are oxidized to cystine forming disulphide bonds in keratin and giving strength to the hair structure. The sulphur atoms of cysteine and cystine are sensitive to oxidative processes which leads to the formation of cysteic acid. As also shown by other authors, cysteic acid is significantly increased in bleached hair samples and is widely monitored as a marker for bleach-induced hair damage.18 Moreover, the untargeted workflow allowed for the additional identification of cysteinesulfate and cysteinesulfinic acid as oxidation products of cystine and cysteine respectively. Even though all three cysteine derivatives were formed during bleaching, they were also detectable in untreated hair samples. On the contrary, despite the intense bleaching, amino acids and derivatives that decreased in treated samples still remained detectable in bleached hair samples, as exemplarily depicted for a selection of amino acids in Fig. 4. We therefore tested different biomarker ratios of elevated and lowered amino acids for their ability to increase the discrimination power and to avoid the need for defined cut-off values. Ratios of tyrosine/cysteic acid and tryptophan/cysteic acid in untreated and treated hair samples are given in Fig. 5A. ROC curves were applied to illustrate the discrimination power of biomarker ratios. With calculated area under the curve (AUC) values approaching one, the prediction power of a biomarker or biomarker ratio in terms of specificity and sensitivity increases. In contrast to the single amino acids, complete discrimination of untreated and treated hair samples was possible by biomarker ratios. That means, if the ratio of tyrosine/cysteic acid falls below a certain threshold, this would denote an authentic hair sample as suspicious. Higher numbers of hair samples followed by amino acid (semi-) quantification will be necessary to confirm these initial findings prior to routine applications.
Overall, best markers identified within this subgroup were uric acid and N-acetylneuraminic acid, as they were totally degraded. These results are comparable to studies by Steuer et al. who investigated the detection and marker suitability of endogenous biomolecules in urine samples adulterated with potassium nitrite.23 The authors were additionally able to identify corresponding oxidation products of uric acid as 5-hydroxy-isourate and 5-hydroxy-2-oxo-4-ureido-2,5-dihydro-1H-imidazole-5-carboxylate, albeit in very low abundance. These oxidation products could not be identified in our study even after targeted data evaluation. Most likely, concentrations of these markers in hair are far below the sensitivity limit of the method.
Finally, the herein used HRMS method is a highly complex, sophisticated and time-intensive method. Typical routine drug screenings are usually performed on triple quad instruments with only one chromatographic column and in positive ionization mode only. Therefore, our study focused on the identification of a broad range of potential biomarkers that could then be implemented individually into routine methods. Useful combinations of markers and/or marker ratios in terms of metabolite coverage and detection rates in general and sufficient analytical confidence in a routine setting (sensitive detection, analytical validity) allowing for reliable detection of adulteration attempts then have to be evaluated in routine methods. Altogether, the applicability of identified markers or marker ratios to real case samples from routine case work as well as their suitability to detect other adulteration attempts and hair treatments like e.g. permanent hair dyeing or thermal hair straightening should be investigated in future studies.
4. Conclusions
Using an untargeted hair metabolomics workflow, numerous potential markers and marker ratios for indirect detection of oxidative hair manipulation were identified. The variety of identified markers now offers the possibility to individually implement a biomarker selection into routine methods of different laboratories. Further studies with higher numbers of hair samples are needed to test the routine applicability in more sensitive targeted approaches and to determine general detection rates of endogenous biomolecules, selectivity and specificity. Furthermore, the applicability of identified markers to other cosmetic hair treatments should be studied to further improve the interpretation of hair testing results.5. Compliance with ethical standards
All samples were analyzed in full conformance with Swiss laws (statement of Cantonal Ethics Board of the Canton of Zurich: BASEC-no. Req-2017-00946). According to Swissethics (Humanforschungsgesetz), no further ethical approval from the cantonal ethic commission is necessary if the research is not aiming to investigate diseases or functions of the human body as is the case in the current study.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
The authors would like to express their gratitude to Emma Louise Kessler, MD for her generous legacy she donated to the Institute of Forensic Medicine at the University of Zurich, Switzerland for research purposes.References
- F. Pragst and M. A. Balikova, Clin. Chim. Acta, 2006, 370, 17–49 CrossRef CAS PubMed.
- C. Jurado, P. Kintz, M. Menendez and M. Repetto, Int. J. Leg. Med., 1997, 110, 159–163 CrossRef CAS.
- G. Skopp, L. Potsch and M. R. Moeller, Forensic Sci. Int., 1997, 84, 43–52 CrossRef CAS.
- I. Kerekes and M. Yegles, Ther. Drug Monit., 2013, 35, 527–529 CrossRef CAS PubMed.
- J. Ettlinger, L. Kirchen and M. Yegles, Drug Test. Anal., 2014, 6(Suppl 1), 74–77 CrossRef CAS PubMed.
- T. M. Binz, M. R. Baumgartner and T. Kraemer, Forensic Sci. Int., 2014, 244, 20–24 CrossRef CAS PubMed.
- M. del Mar Ramirez Fernandez, S. M. R. Wille, V. Di Fazio and N. Samyn, Forensic Sci. Int., 2019, 297, 277–283 CrossRef CAS PubMed.
- L. Morini, A. Zucchella, A. Polettini, L. Politi and A. Groppi, Forensic Sci. Int., 2010, 198, 23–27 CrossRef CAS PubMed.
- N. Van Elsue and M. Yegles, Forensic Sci. Int., 2019, 297, 270–276 CrossRef CAS PubMed.
- S. Petzel-Witt, W. Pogoda, C. Wunder, A. Paulke, M. Schubert-Zsilavecz and S. W. Toennes, Drug Test. Anal., 2018, 10, 177–183 CrossRef CAS PubMed.
- S. Witt, C. Wunder, A. Paulke, M. A. Verhoff, M. Schubert-Zsilavecz and S. W. Toennes, Drug Test. Anal., 2016, 8, 826–831 CrossRef CAS PubMed.
- D. Ammann, R. Becker, A. Kohl, J. Hanisch and I. Nehls, Forensic Sci. Int., 2014, 244, 30–35 CrossRef CAS PubMed.
- S. Petzel-Witt, S. I. Meier, M. Schubert-Zsilavecz and S. W. Toennes, Drug Test. Anal., 2018, 10, 768–773 CrossRef CAS PubMed.
- L. Eisenbeiss, T. M. Binz, M. R. Baumgartner, A. E. Steuer and T. Kraemer, Drug Test. Anal., 2020, 12, 230–238 CrossRef CAS PubMed.
- S. Luong, R. Shimmon, J. Hook and S. Fu, Anal. Bioanal. Chem., 2012, 403, 2057–2063 CrossRef CAS PubMed.
- S. Luong and S. Fu, Drug Test. Anal., 2014, 6, 277–287 CrossRef CAS PubMed.
- W. B. Dunn, D. Broadhurst, P. Begley, E. Zelena, S. Francis-McIntyre, N. Anderson, M. Brown, J. D. Knowles, A. Halsall, J. N. Haselden, A. W. Nicholls, I. D. Wilson, D. B. Kell, R. Goodacre and Human Serum Metabolome Consortium, Nat. Protoc., 2011, 6, 1060–1083 CrossRef CAS PubMed.
- K. M. Joo, A. R. Kim, S. N. Kim, B. M. Kim, H. K. Lee, S. Bae, J. H. Lee and K. M. Lim, Exp. Dermatol., 2016, 25, 729–731 CrossRef CAS PubMed.
- A. J. Grosvenor, S. Deb-Choudhury, P. G. Middlewood, A. Thomas, E. Lee, J. A. Vernon, J. L. Woods, C. Taylor, F. I. Bell and S. Clerens, Int. J. Cosmet. Sci., 2018, 40, 536–548 CrossRef CAS PubMed.
- L. Eisenbeiss, A. E. Steuer, T. M. Binz, M. R. Baumgartner and T. Kraemer, Anal. Bioanal. Chem., 2019, 411, 3963–3977 CrossRef CAS PubMed.
- M. I. Boxler, T. D. Schneider, T. Kraemer and A. E. Steuer, Drug Test. Anal., 2019, 11, 678–696 CrossRef CAS PubMed.
- H. G. Gika, G. A. Theodoridis, R. S. Plumb and I. D. Wilson, J. Pharm. Biomed. Anal., 2014, 87, 12–25 CrossRef CAS PubMed.
- A. E. Steuer, K. Arnold, T. D. Schneider, M. Poetzsch and T. Kraemer, Anal. Bioanal. Chem., 2017, 409, 6235–6244 CrossRef CAS PubMed.
- J. Chong, O. Soufan, C. Li, I. Caraus, S. Li, G. Bourque, D. S. Wishart and J. Xia, Nucleic Acids Res., 2018, 46, W486–W494 CrossRef CAS PubMed.
- E. P. J. Linstrom and W. G. Mallard, NIST Chemistry WebBook, NIST Standard Reference Database Number 69, National Institute of Standards and Technology, Gaithersburg MD, 2001, p. 20899, DOI:10.18434/T4D303, (retrieved November 8, 2018).
- D. S. Wishart, D. Tzur, C. Knox, R. Eisner, A. C. Guo, N. Young, D. Cheng, K. Jewell, D. Arndt, S. Sawhney, C. Fung, L. Nikolai, M. Lewis, M. A. Coutouly, I. Forsythe, P. Tang, S. Shrivastava, K. Jeroncic, P. Stothard, G. Amegbey, D. Block, D. D. Hau, J. Wagner, J. Miniaci, M. Clements, M. Gebremedhin, N. Guo, Y. Zhang, G. E. Duggan, G. D. Macinnis, A. M. Weljie, R. Dowlatabadi, F. Bamforth, D. Clive, R. Greiner, L. Li, T. Marrie, B. D. Sykes, H. J. Vogel and L. Querengesser, Nucleic Acids Res., 2007, 35, D521–D526 CrossRef CAS PubMed.
- C. Guijas, J. R. Montenegro-Burke, X. Domingo-Almenara, A. Palermo, B. Warth, G. Hermann, G. Koellensperger, T. Huan, W. Uritboonthai, A. E. Aisporna, D. W. Wolan, M. E. Spilker, H. P. Benton and G. Siuzdak, Anal. Chem., 2018, 90, 3156–3164 CrossRef CAS PubMed.
- T. Kind, K. H. Liu, D. Y. Lee, B. DeFelice, J. K. Meissen and O. Fiehn, Nat. Methods, 2013, 10, 755–758 CrossRef CAS PubMed.
- L. W. Sumner, A. Amberg, D. Barrett, M. H. Beale, R. Beger, C. A. Daykin, T. W. Fan, O. Fiehn, R. Goodacre, J. L. Griffin, T. Hankemeier, N. Hardy, J. Harnly, R. Higashi, J. Kopka, A. N. Lane, J. C. Lindon, P. Marriott, A. W. Nicholls, M. D. Reily, J. J. Thaden and M. R. Viant, Metabolomics, 2007, 3, 211–221 CrossRef CAS PubMed.
- A. Guerra-Tapia and E. Gonzalez-Guerra, Actas Dermosifiliogr., 2014, 105, 833–839 CrossRef CAS PubMed.
- S. Harrison and R. Sinclair, J. Cosmet. Dermatol., 2003, 2, 180–185 CrossRef CAS PubMed.
- R. Dawber, Clin. Dermatol., 1996, 14, 105–112 CrossRef CAS PubMed.
- S. S. Adav, R. S. Subbaiaih, S. K. Kerk, A. Y. Lee, H. Y. Lai, K. W. Ng, S. K. Sze and A. Schmidtchen, Sci. Rep., 2018, 8, 1599 CrossRef PubMed.
- S. Ito, Y. Nakanishi, R. K. Valenzuela, M. H. Brilliant, L. Kolbe and K. Wakamatsu, Pigm. Cell Melanoma Res., 2011, 24, 605–613 CrossRef CAS PubMed.
- S. Ito, K. Wakamatsu and T. Sarna, Photochem. Photobiol., 2018, 94, 409–420 CrossRef CAS PubMed.
- K. Wakamatsu, Y. Nakanishi, N. Miyazaki, L. Kolbe and S. Ito, Pigm. Cell Melanoma Res., 2012, 25, 434–445 CrossRef CAS PubMed.
- S. Ito, A. Pilat, W. Gerwat, C. M. Skumatz, M. Ito, A. Kiyono, A. Zadlo, Y. Nakanishi, L. Kolbe, J. M. Burke, T. Sarna and K. Wakamatsu, Pigm. Cell Melanoma Res., 2013, 26, 357–366 CrossRef CAS PubMed.
- S. Li, D. Gao and Y. Jiang, Metabolites, 2019, 9, 36 CrossRef CAS PubMed.
- K. Foitzik, E. Hoting, T. Forster, P. Pertile and R. Paus, Exp. Dermatol., 2007, 16, 936–945 CrossRef CAS PubMed.
- G. Brotzu, A. M. Fadda, M. L. Manca, T. Manca, F. Marongiu, M. Campisi and F. Consolaro, Dermatol. Ther., 2019, 32, e12778 CrossRef PubMed.
- J. Kluge, L. Rentzsch, D. Remane, F. T. Peters and D. K. Wissenbach, Drug Test. Anal., 2018, 10, 1536–1542 CrossRef CAS PubMed.
- A. H. Rashaid, P. B. Harrington and G. P. Jackson, Anal. Chem., 2015, 87, 7078–7084 CrossRef CAS PubMed.
- C. R. Robbins, in Chemical and Physical Behavior of Human Hair, 2012, ch. 2, pp. 105–176, DOI:10.1007/978-3-642-25611-0_2.
- T. Iwabuchi, R. Ideta, R. Ehama, H. Yamanishi, M. Iino, Y. Nakazawa, T. Kobayashi, M. Ohyama and J. Kishimoto, J. Dermatol., 2016, 43, 567–570 CrossRef CAS PubMed.
- Y. Watanabe, T. Nagashima, N. Hanzawa, A. Ishino, Y. Nakazawa, M. Ogo, T. Iwabuchi and M. Tajima, Int. J. Cosmet. Sci., 2015, 37, 579–587 CrossRef CAS PubMed.
- M. Giorgio, G. I. Dellino, V. Gambino, N. Roda and P. G. Pelicci, Redox Biol., 2020, 29, 101398 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0an01265c |
| This journal is © The Royal Society of Chemistry 2020 |