Scleral ossicles: angiogenic scaffolds, a novel biomaterial for regenerative medicine applications†
Marta
Checchi
 *a,
Jessika
Bertacchini
*a,
Jessika
Bertacchini
 *a,
Francesco
Cavani
*a,
Francesco
Cavani
 a,
Maria Sara
Magarò
a,
Maria Sara
Magarò
 a,
Luca
Reggiani Bonetti
a,
Luca
Reggiani Bonetti
 b,
Geltrude Rita
Pugliese
b,
Geltrude Rita
Pugliese
 a,
Roberto
Tamma
a,
Roberto
Tamma
 c,
Domenico
Ribatti
c,
Domenico
Ribatti
 c,
Delphine B.
Maurel
c,
Delphine B.
Maurel
 d and
Carla
Palumbo
d and
Carla
Palumbo
 a
a
aDepartment of Biomedical, Metabolic Science and Neuroscience, University of Modena and Reggio Emilia, 41125 Modena, Italy. E-mail: marta.checchi@unimore.it; jbertacchini@unimore.it; Tel: +39 059.422.4858 Tel: +39 059.422.4842
bDepartment of Pathology, University of Modena and Reggio Emilia, 41125 Modena, Italy
cDepartment of Basic Medical Sciences, Neurosciences and Sensory Organs, Section of Human Anatomy and Histology, University of Bari Medical School, 70124 Bari, Italy
dPharmaceutical Sciences Department, University of Bordeaux, Bio-Tis, INSERM Unit 1026, 33076 Bordeaux, France
First published on 6th November 2019
Abstract
Given the current prolonged life expectancy, various pathologies affect increasingly the aging subjects. Regarding the musculoskeletal apparatus, bone fragility induces more susceptibility to fractures, often not accompanied by good ability of self-repairing, in particular when critical-size defects (CSD) occur. Currently orthopedic surgery makes use of allografting and autografting which, however, have limitations due to the scarce amount of tissue that can be taken from the donor, the possibility of disease transmission and donor site morbidity. The need to develop new solutions has pushed the field of tissue engineering (TE) research to study new scaffolds to be functionalized in order to obtain constructs capable of promoting tissue regeneration and achieve stable bone recovery over time. This investigation focuses on the most important aspect related to bone tissue regeneration: the angiogenic properties of the scaffold to be used. As an innovative solution, scleral ossicles (SOs), previously characterized as natural, biocompatible and spontaneously decellularized scaffolds used for bone repair, were tested for angiogenic potential and biocompatibility. To reach this purpose, in ovo Chorioallantoic Membrane Assay (CAM) was firstly used to test the angiogenic potential; secondly, in vivo subcutaneous implantation of SOs (in a rat model) was performed in order to assess the biocompatibility and the inflammatory response. Finally, thanks to the analysis of mass spectrometry (LCMSQE), the putative proteins responsible for the SO angiogenic properties were identified. Thus, a novel natural biomaterial is proposed, which is (i) able to induce an angiogenic response in vivo by subcutaneous implantation in a non-immunodeficient animal model, (ii) which does not induce any inflammatory response, and (iii) is useful for regenerative medicine application for the healing of bone CSD.
1. Introduction
Given the current prolonged life expectancy and the evermore aging world population, there is a rapid increase in musculoskeletal pathologies such as bone fragility inducing more susceptibility to fractures, often not accompanied by good ability of self-repairing and hence subjected to some complications like non-unions, as well as vertebral collapses, osteomyelitis, etc.2 As a result, costs and bone-related medical treatments increase constantly.3 Also, as a consequence of injuries, it is possible to incur large-sized fractures called critical-size defects (CSD) since the bone lesion is so severe that it prevents the skeletal segment from self-repairing.4 Recovery of significant skeletal defects could be partially abortive due to the perturbations that affect the regenerative process. Nowadays, surgical techniques include: bone grafting and joint arthroplasties, autografts (which are considered the gold standard in the field5,6) and allografts7–9 (that are often used to treat such bone defects). However, although all these strategies are commonly used in orthopedic surgery, these treatments have some limitations concerning their costs and their side effects such as potential infections, non-unions, donor site morbidity, difficulties in finding adequate donors and a limited amount of tissue to harvest. Moreover, severe fractures and replacements of injured skeletal segments are currently treated by means of metal implants that, instead, show some limitations such as post-transplant rejection and reduced bioactivity.10–12 Furthermore, implant mobility, inflammation and bone resorption are associated with failure of some metal implants. In many cases the biological system is not able to replace the bone loss that may be also complicated by the surrounding soft-tissue environment. Furthermore, the destruction of the local vascular network often occurs at the fracture site and for the bone healing process to be assured it is required to restore the vascularization previously. It is to be noted, in fact, that an essential prerequisite of fracture healing is the proper development of blood vessels and the complex interplay among blood vessel formation, oxygen supply, growth factors and cell proliferation influences the final healing outcome in large bone defects.13–17One of the most important aspects to take in consideration during the setting up of a new material for bone regeneration is its angiogenic potential.18 Angiogenesis is a complex process in which, during development, endothelial cells (ECs) switch from the “quiescent” to the “angiogenic phenotype” in response to the balance between anti- and pro-angiogenic factors.19 To do this, EC growth, migration, and tube formation are processes that occur under precise stimuli which regulate the cell–extracellular matrix (ECM) interactions in order to form new blood capillaries from the pre-existing ones.20,21
Angiogenesis and osteogenesis are tightly coupled: the angiogenesis process is essential and crucial for osteogenesis and bone regeneration, and osteogenesis is critical for the maintenance of a healthy and fully functional skeletal system.22 The blood vessels mediate the transport of circulating cells, oxygen, nutrients and waste products and provide angiogenic and angiocrine signals. Therefore, normal vascular formation plays a key role in both physiological and pathological processes of the skeletal system. The crucial role of the vessels has been already established during the various phases of bone osteogenesis, where two types of bone formation (i.e. static and dynamic) have been demonstrated to occur in close relationships with the vascular frameworks.23–26 In this sophisticated and finely modulated process, a variety of angiogenic factors are all widely expressed as primary inducers of vascular development.
In this regard in the last decade, regenerative medicine and TE have played a central role, proposing strategies for bone reconstitution with the ambition to overcome the complications associated with traditional techniques. In particular, many different scaffolds are developing with different properties, proposing new materials (i.e. 3D printable matrices/scaffolds) to be used as new strategies in order to trigger the onset of bone regeneration and optimize the cell growth.27–32 A variety of different exogenous chemical or physical stimuli were tested, such as soluble growth/differentiation factors, as well as the application of mechanical forces (i.e., loading).33–40 Finally, many types of cells have been used alone or in co-culture.41–48 An engineered scaffold-based strategy is the starting point of the recent innovative therapies, for which the choice of material properties, manufacturing methods and component treatments, is crucial. They are followed by pre-clinical in vitro studies and in vivo experimentation, which can be a prelude to a production of devices/constructs that meet the need/expectations of patients. Many of these materials display good mechanical properties but to the detriment of retaining a high porosity, so, scaffolds for TE which have demonstrated good outcomes in vitro have failed when implanted in vivo because of their insufficient capability of vascularization not permitting the cell infiltration and hence a proper vascularization.49
In this work it is proposed that the use of scleral ossicles (SOs) is an innovative solution.1 SOs are peculiar bony plates forming a ring at the scleral–corneal border of the eyeball of lower vertebrates. This proposed model is interesting because once SOs reach the definitive size, in the adult animal, they are subjected to mechanical stereotyped stress for their lifetime, solely with a protective function. Therefore, the bone remodeling processes (in response to metabolic need) must be avoided and, to do this, the osteocytes undergo massive apoptosis, making the ossicles like naturally decellularized bones.50–55 SOs have been suggested as natural biocompatible scaffolds for bone repair, inasmuch it has been reported that SOs are able to sustain cell adhesion, proliferation and differentiation and, moreover, they seem to have angiogenic potential in vitro.1 Definitely, the SOs act as a natural ECM-like platform to support angiogenesis and vascularization.
The aims of this study are: (I) to evaluate the angiogenic potential of SOs in ovo and in vivo, (II) to verify the absence of inflammatory potential upon in vivo SO subcutaneous implantation and finally (III) to investigate the factors responsible for the angiogenesis.
2. Experimental section
2.1. Extraction and preparation of the scleral ossicles (SOs)
Poultry processing waste, donated by a local butcher, was used as the source of SOs. Each eyeball of adult chickens (50–70 days old) was incised with a scalpel in order to extract the scleral ring. Under a stereomicroscope, the tissue membranes that cover the scleral ring were removed and each of the 13–14 SOs was disjoined. All bony plates were cleaned with PBS pH 7.4 (phosphate buffered saline) to eliminate the residues of eye liquid humor, dried and finally sterilized by exposure for 30 min under UV radiation.2.2. Chorioallantoic membrane (CAM) assay
Fertilized White Leghorn chicken eggs (Istituto Zooprofilattico di Puglia e Basilicata, Foggia, Italy) staged according to Hamburger and Hamilton (HH)56 were placed, at the onset, into an incubator and kept under humidity at 37 °C (day 0). At stage HH3 (day 3), a square window was opened into the eggshell after the removal of 2–3 ml of albumen so that the developing chorioallantoic membrane is detached from the shell itself and the underlying CAM vessels are observed. The window was sealed with a glass coverslip and the eggs were returned to the incubator. At stage HH8 (day 8 – when CAM is fully developed), the coverslips were removed and the top of the growing CAMs was visible; the CAM assay was performed as indicated by Ribatti et al. who explained the experimental procedure in order to evaluate the angiogenic potential either of a solid material or of a solution.57 Firstly, we evaluated the angiogenic potential of the sterile SO (1 mm-thick cross-section cut with scissors). Secondly, the angiogenic potential of different media has been tested (indicated in Fig. 1) for 48 h of culture. The coverslips were replaced after the different grafting, and the CAMs were examined daily until stage HH12 (day 12). Thereafter, the CAMs were fixed and photographed in ovo using a stereomicroscope equipped with a camera and an image analysis system (Olympus Italia, Opera/Milan, Italy). The angiogenic response was evaluated by counting the number of vessels appearing around the grafts (SO or media inoculation). For each condition 10 eggs per group were used.2.3. In vivo subcutaneous implantation of scleral ossicles
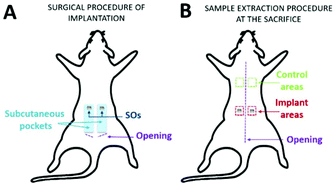
Thirteen 12-week-old male Wistar rats weighing 350–400 g were purchased from Janvier Labs (Saint Berthevin, France). The animal experiments were conducted at the Animal Facilities of the University of Bordeaux, France.All animal procedures were performed in accordance with the European Guidelines for Care and Use of Laboratory Animals, and approved by the Bordeaux Ethical Committee and by the French Ministry for Education, Research, and Innovation (agreement number APAFIS #4375-2016030408537165 v8). Animal housing and caretaking were provided by the animal facility in accordance with the national guidelines. All animals were kept under a 12 h–12 h light/dark cycle and the temperature was controlled.
After one week of adaptation, rats were shaved and the day after, one control rat was sacrificed (day 0). In the remaining twelve rats, sterile SO was placed subcutaneously. The rats were anesthetized with 4% isoflurane and maintained at 2% using a mask during the surgery. After scrubbing the skin with 0.2% sterile chlorhexidine, 2 cm dorsal incisions were made bilaterally on the sacral–lumbar area. Subcutaneous pockets (4–5 cm in length and 1.5 cm in width) were made bilaterally, parallel to the rostro-caudal axis in the direction of the head, and one SO per side was inserted (Fig. 2A). All the incisions were sutured with 3–4 sterile staples and the animals were followed throughout the post-implantation period up to euthanasia. After 1, 2, 4, and 12 weeks post-implantation, animals were euthanized using CO2 (TEM SEGA rodent system, Automate d'euthanasie v 4.5). The skin areas containing the SO were excised from each rat (implant areas) as well as skin control samples (control areas) from the dorsal thoracic region (Fig. 2B).
2.4. Histological staining and immunohistochemistry
Samples with/without SOs were decalcified for 4 days in 10% EDTA and then processed for paraffin embedding (Leica EG1150 H) following the procedure here described: 70% ethanol for 30 min, three times in 95% ethanol, three times in 100% ethanol, three times in toluene and two times in paraffin, 1 h for each. After paraffin embedding, twelve sections (5 μm) were obtained from each sample.Slides were deparaffinized by means of xylene (two times for 5 min each), hydrated through a graded series of alcohol (100%, 95%, 80% and 70%) for 5 min each and rinsed first with tap water and then with deionized water for 1 min each. After that, four slides (with two sections each) of both control (CTR) and implanted (IMPLANTED) samples, for each time point (weeks 1, 2, 4 and 12), were stained both with Hematoxylin & Eosin (H&E) or Masson's Trichrome (MT).
Briefly, for H&E staining, after the dehydration procedure, the slides were stained with Carazzi hematoxylin solution (DiaPath C0203), rinsed with tap water, stained with alcoholic 0.5% eosin Y (DiaPath C0353) and then rinsed with deionized water for 1 min each.
For Masson's trichrome staining, dehydrated slides were stained with Mayer's hemalum solution for 10 min, rinsed first with tap water for 10 min and then with deionized water for 1 min, before being exposed to 1% Fuchsin-Ponceau (1v/2v) for 1.5 min. After 1 min in 3% (w/v) phosphomolybdic acid, slides were run through acidified water (0.1% (v/v) glacial acetic acid) for 1 min, stained with 1% (w/v) light green for 1 min and rinsed with acidified water again for 1 min.
Immunohistochemistry (IHC) was performed using the automatic system Ventana Bench Mark XT according to the following protocol: samples were incubated for 15 min with buffer EZ prep (Ventana 950-102) at 72 °C; the antigen retrieval was performed by arranging slides in a Cell Conditioning 1 Buffer (Ventana 950-124) for 36 min; slides were incubated for 24 min with macrophage specific anti-CD68 clone PG-M1 (DakoCytomation M0876) at a concentration of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 in an antibody diluent (Ventana 251-018); samples were then treated with an ultraView Universal DAB Detection Kit (Ventana 760-500) and finally, slides were counterstained by immersion in hematoxylin II (Ventana 790-2208) for 12 min and mounted on slides. Stained sections were evaluated under a Nikon Eclipse Ni microscope (Nikon) equipped with a DS-Fi2 camera (Nikon) and processed by means of NIS-Elements D 5.11.00 software (Nikon).
300 in an antibody diluent (Ventana 251-018); samples were then treated with an ultraView Universal DAB Detection Kit (Ventana 760-500) and finally, slides were counterstained by immersion in hematoxylin II (Ventana 790-2208) for 12 min and mounted on slides. Stained sections were evaluated under a Nikon Eclipse Ni microscope (Nikon) equipped with a DS-Fi2 camera (Nikon) and processed by means of NIS-Elements D 5.11.00 software (Nikon).
2.5. Mass spectrometry, protein identification and annotation
Three replicates were prepared for mass spectrometry (LCMSQE) analysis according to the following protocol: four SOs were finely chopped and incubated with 50 μl of 50 ng μl−1 trypsin (PIERCE 90057S) at 37 °C for 16 h. Next, the trypsin was inactivated with 5% formic acid (with a volume equal to 10% of the final digestion volume) and subsequently dried in the Speed-vac for 1 h. Before performing the LCMSQE analysis, the samples were re-suspended in 40 μL of a mixture of water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile
acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) formic acid (97
formic acid (97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), sonicated for 10 min at room temperature and finally centrifuged at 12
2), sonicated for 10 min at room temperature and finally centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 r.c.f. for 10 min. Analyses were performed on an ESI Q Exactive Mass spectrometer (Thermo Scientific), controlled by an Xcalibur (v. 29 build 2926) and interfaced with an Ultimate 3000 UHPLC pump. The column (Zorbax SB-C18 RRHT, 2.1 × 50 mm, 1.8 μ particle size, Agilent Technologies), thermostated at 30 °C, was equilibrated with 0.3 ml min−1 of water and 0.1% formic acid (A) with 2% acetonitrile (B). Two different methods were used: 80 min for samples 1 and 2, and 3 h for sample 3. Briefly, in the first method after sample injection (20 μl), B% was kept at 2% for 2 min, then it was increased from 2 to 3% in 5 min, then linearly increased from 3 to 28% in 59 min; B% was then brought to 90% in 4 min and kept at 90% for 3 min, before the reconditioning step. The total runtime was 80 min. For the second method after sample injection (30 μl), B% was kept at 2% for 2 min, then it was linearly increased from 2 to 28% in 153 min; B% was then brought to 90% in 10 min and kept at 90% for 5 min, before the reconditioning step. The total run time was 3 h.
100 r.c.f. for 10 min. Analyses were performed on an ESI Q Exactive Mass spectrometer (Thermo Scientific), controlled by an Xcalibur (v. 29 build 2926) and interfaced with an Ultimate 3000 UHPLC pump. The column (Zorbax SB-C18 RRHT, 2.1 × 50 mm, 1.8 μ particle size, Agilent Technologies), thermostated at 30 °C, was equilibrated with 0.3 ml min−1 of water and 0.1% formic acid (A) with 2% acetonitrile (B). Two different methods were used: 80 min for samples 1 and 2, and 3 h for sample 3. Briefly, in the first method after sample injection (20 μl), B% was kept at 2% for 2 min, then it was increased from 2 to 3% in 5 min, then linearly increased from 3 to 28% in 59 min; B% was then brought to 90% in 4 min and kept at 90% for 3 min, before the reconditioning step. The total runtime was 80 min. For the second method after sample injection (30 μl), B% was kept at 2% for 2 min, then it was linearly increased from 2 to 28% in 153 min; B% was then brought to 90% in 10 min and kept at 90% for 5 min, before the reconditioning step. The total run time was 3 h.
ESI (electrospray ionization) was performed in positive mode; the probe was heated at 290 °C, the capillary temperature was set at 270 °C; the following nitrogen flows (arbitrary units) were used to assist the ionization: Sheath Gas 40, Aux Gas 30, and Sweep Gas 3; capillary voltage was set to 3.5 kV and the S-Lens RF level was set at 55 (arbitrary units).
Centroided MS and MS2 spectra were recorded from 200 to 2000 m/z in FULL MS/dd-MS2 (TOP5) mode, at a resolution of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 17
000 and 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, respectively. The five most intense multi-charged ions were selected for MS2 nitrogen-promoted collision-induced dissociation (NCE = 28). A precursor active exclusion of 10 seconds was set; peptide-like isotope pattern ions were preferred. The mass spectrometer was calibrated before the start of the analyses.
500, respectively. The five most intense multi-charged ions were selected for MS2 nitrogen-promoted collision-induced dissociation (NCE = 28). A precursor active exclusion of 10 seconds was set; peptide-like isotope pattern ions were preferred. The mass spectrometer was calibrated before the start of the analyses.
MS raw data files were converted with MSConvert (version 3.0.19135-311725741 ProteoWizard) into mascot generic format and the peak lists were searched against Trembl (21902 sequences for Gallus gallus) for peptide sequences and C-Rap for contaminants with MASCOT (version 2.4, Matrix Science, London, UK).
Trypsin was set as a proteolytic enzyme, and deamidated-NQ and oxidized-M were set as variable modifications. One missed cleavage was allowed. Mass tolerances were set at 10 ppm for the precursor ions and 0.01 Da for the product ions. Automatic decoy database search was used to estimate the false discovery rate, which was adjusted to <1%.
All proteins identified from each experimental approach are listed in Table 1, ESI.† GO annotation was used to identify protein clusters, this was obtained using Protein ANalysis THrough Evolutionary Relationships58 (PANTHER version 14.1 (released 2019-03-12) contains 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 524 protein families, divided into 107
524 protein families, divided into 107![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 627 functionally distinct protein subfamilies), STRING database59 (version 11.0 since January 19, 2019 currently covers 9
627 functionally distinct protein subfamilies), STRING database59 (version 11.0 since January 19, 2019 currently covers 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 643
643![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 763 proteins from 2031 organisms) and the DAVID Bioinformatics Resources database60 (version 6.8).
763 proteins from 2031 organisms) and the DAVID Bioinformatics Resources database60 (version 6.8).
2.6. Statistical analysis
Experiments carried out in triplicate (as indicated in the Experimental section and in the respective paragraphs in the “Results and discussion” section) are defined respectively as two or three independent samples that were treated, isolated and analysed separately. All data are expressed as mean ± Standard Deviation (SD). Differences between the experimental conditions were evaluated by one-way analysis of variance (ANOVA) with Dunnett's corrections for multiple comparison or two-way analysis of variance (ANOVA) with Bonferroni corrections for multiple comparison; the type of analysis and significant p values are indicated in the figure legends. Statistical analyses and relative graphs were performed using GraphPad prism 6 (version 6.01).Differences are classified as significant with p < 0.05.
3. Results and discussion
3.1. In ovo angiogenic potential of scleral ossicles
In order to evaluate the angiogenic potential of the SOs in ovo, a peculiar analysis was conducted on the chick embryo chorioallantoic membrane (CAM). The CAM is an extra embryonic membrane which serves as a gas exchange surface and its function is supported by a dense capillary network.61 Some side considerations concern the ethical choice of the CAM model: the chick embryo that has not reached the 14th day of its gestation period does not feel pain and can therefore be used for experimentation without any ethical restrictions or prior protocol approval.62,63 Additionally, according to the “three Rs” rule, we have used a reduced number of animals for the successive in vivo experimentation (“reduction”), we minimized the pain and suffering from animals (“refinement”) and we have used an alternative (in ovo instead of in vivo) for the first experimental phase of the study (“replacement”).64,65 Because of its easy accessibility, the chicken CAM (frequently used as an experimental model, also as an alternative to the animal model, as just mentioned) has been used, in the present study, for the investigation of the angiogenic potential of SO.66–68 Moreover, in the development stages used in this experimentation (from day 8 to day 12) the immunocompetent chicken system is not fully developed and for this reason it is not responsive to an exogenous material.69The angiogenic potential of the SO was evaluated by the CAM assay, incubating firstly SO and subsequently media conditioned by SOs. Experiments were started at the stage HH8 of embryo development (when crucial steps as the development of the circulatory system occur) and carried out for four days, as described in the Experimental section.
In a preliminary experiment, SO was brought directly into contact with the growing CAM resulting in a strong vasculo-proliferative activity. Fig. 3 shows the appearance of CAM's vessels at stage HH12; the magnification of the dashed squared area shows a linear and orderly course of the vessels (Fig. 3A). The SO implant (removed because of its opacity and outlined with a dashed yellow line in Fig. 3B) induces a strong vasculo-proliferative reaction resulting in the formation of neo-formed vessels (yellow arrows) on the pre-existing ones. As an interesting aspect, it should be noted that the vascular course is extremely tortuous and irregular, representing the formation of new capillaries during tumour onset.
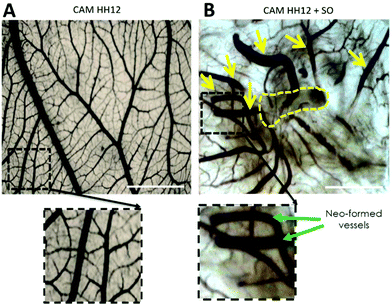 | ||
| Fig. 3 Appearance of CAM at stage HH12 with (B) or without SO (A). The yellow dashed line indicates the site of implantation of SO on the growing CAM (scale bar = 2 mm). | ||
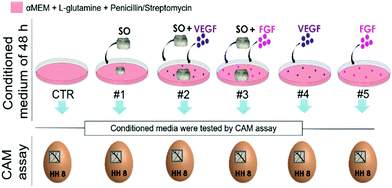
To better understand whether this vasculo-proliferative reaction is induced by factors released by the SO, six different conditioned media, obtained by culturing for 48 h a combination of serum free medium, SO and growth factors, were tested through the CAM assay (medium compositions are specified in Table 1).
| Media composition | |||
|---|---|---|---|
| αMEM + gluta + P/S | SO | GF | |
| CTR | + | − | − |
| #1 | + | + | − |
| #2 | + | + | + VEGF |
| #3 | + | + | + FGF |
| #4 | + | − | + VEGF |
| #5 | + | − | + FGF |
Both the medium conditioned by the presence of the SO-released-factors alone (#1) and the medium containing the combination of the two (#2 and #3) or the growth factors alone (#4 and #5) induce the formation of numerous allantoic vessels that develop radially with respect to the implant in a “spoked-wheel” pattern (white arrows in Fig. 4A). These observations are supported by the quantification of the number of vessels that moved away from the areas of medium inoculation under all conditions. The histogram in Fig. 4B is related to the quantification performed on the images of the CAM vessels at stage HH12 under all the conditions analyzed.
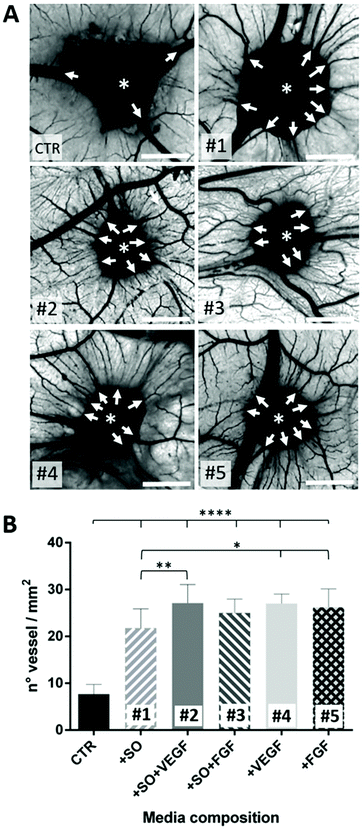 | ||
| Fig. 4 (A) Images of the CAM assay showing the inoculation site of the conditioned media (asterisks); medium composition is described in Table 1 (scale bar = 2 mm). The white arrows indicate the vessels developing radially around the inoculation site; (B) the number of vessels counted on the CAM for each conditioned medium. Statistical analysis was done using one-way ANOVA with Dunnett's corrections (*p < 0.05, **p < 0.01, ****p < 0.0001). | ||
In particular, the number of vessels is significantly higher under conditions #1 to #5 compared to the CTR (****p < 0.0001). It has been highlighted that the SO alone is able to induce an angiogenic response not so different (*p < 0.5) from conditions #4 and #5 (representing our positive controls). It is of note that the VEGF (#2) is able to potentiate the SO angiogenic response (1#) (**p < 0.01) suggesting an additive effect that is not observable with the FGF (#3). These findings suggest that the SO induces an important angiogenic reaction triggered by angiogenic factors released in the media where it is located. Furthermore, the VEGF acts as an enhancer of the SO angiogenic response while the FGF in combination with the SO effect does not affect the angiogenic potential of the SOs.70–72 The last evidence probably means that the FGF angiogenic response is not potentiated because of a “saturation” effect (viz. every CAM, despite the sum of favorable conditions, is not able to develop vessels beyond a certain number73). As the final consideration, concerning the data so far obtained on this aspect, it is correct to assert that the SOs have a good angiogenic potential in ovo.
3.2. In vivo subcutaneous implantation of scleral ossicles
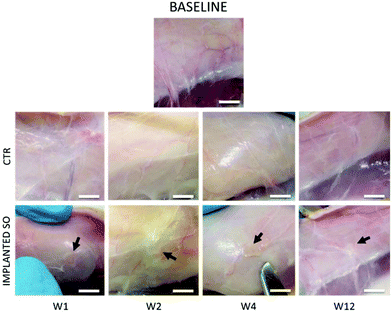
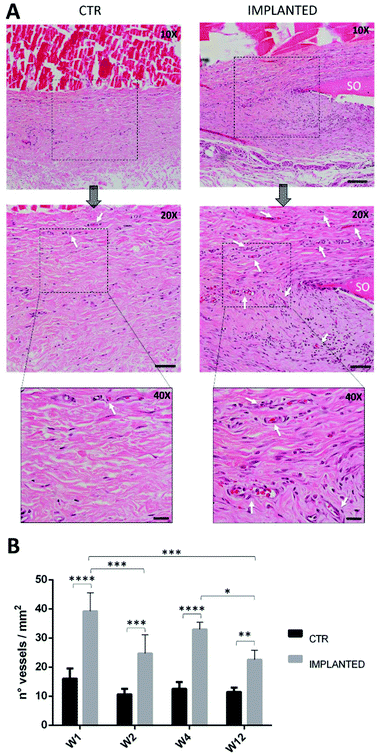
Based on the previous CAM results, indicating a strong angiogenic potential of SOs, it was decided to test the biocompatibility and inflammation response in a rat model through the subcutaneous implantation in the dorsal region for 1, 2, 4 and 12 weeks.No loss of animals was observed throughout the procedure, not even after surgery. All rats had good recovery and no animals showed pain after surgery. Following euthanasia, a longitudinal incision was made in the dorsal region above the spine; then, both sides were opened to (i) retrieve the samples of skin (sample area of 1 × 1 cm) including the SO (implant area in Fig. 2B) and (ii) take the CTR samples (control areas in Fig. 2B). All implanted SOs were visible by gross inspections at all the time points and no sepsis or evidence of infection was seen (Fig. 5).
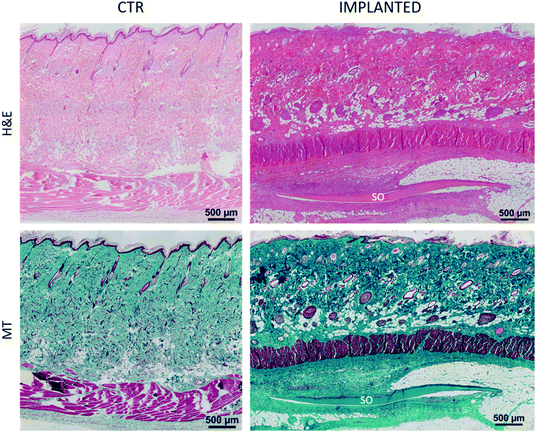
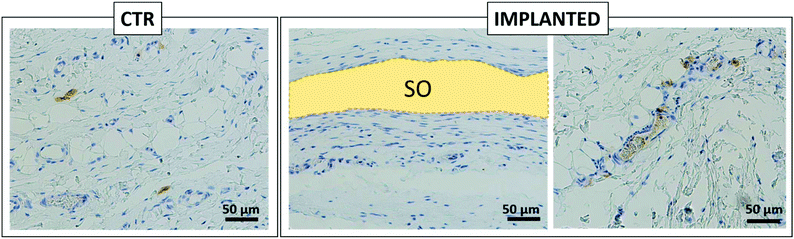
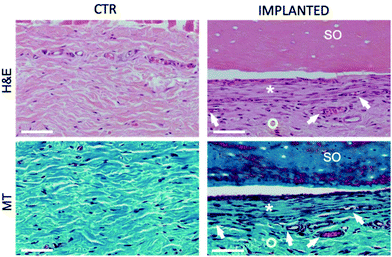
Furthermore, histological analysis showed interesting aspects. The SOs, under both the macroscopic (Fig. 5) and histological views (Fig. 6), neither show induction of any inflammatory/edema signs nor macrophage migration and infiltration of new fibroblasts or necrosis at all the time points (1, 2, 4 and 12 weeks).74 This is also in line with the IHC analysis: as expected, the presence of CD68 positive cells was observed both in the control rat tissue (Fig. 7-CTR) and in the implanted animals, in proximity to blood vessels at a distance from the implant site (right panels in Fig. 7-IMPLANTED). Interestingly, in the area close to the SO implant, CD68 positive cells were not detected. On this basis, we can deduce that the presence of CD68 positive elements is not related to the implant site/procedure. Hence, we can conclude that SOs are biocompatible and well tolerated from the host organism which does not show any reaction due to foreign bodies75 and also SOs were not subjected to rejection.76,77
We were positively surprised to see no inflammatory reaction, considering that SOs come from chickens and were put in a non-immunodeficient rat model.78,79 This is probably due to the fact that the scaffold proposed is already naturally decellularized. In fact, reduced/absent immune responses have been observed in autologous, allogenic, and xenogenic decellularized scaffolds, as demonstrated for other artificially decellularized scaffolds by other authors.80,81 Interestingly, in the area close to the SO, the connective tissue displays a well-organized and compact collagen texture (asterisks in Fig. 8) compared to the adjacent collagen that has a looser organization and presents a greater quantity of vessels (circles in Fig. 8). This different organization of the connective tissue is visible immediately adjacent to the SO at all the time points tested.
Quantification of blood vessels is showed in Fig. 9: the histogram clearly shows that the amount of blood vessels found in the IMPLANTED samples is statistically significant compared to the CTR. Furthermore, significant differences in the quantity of vessels in the IMPLANTED samples are visible especially between weeks 1–2, 1–12 and 4–12, but decrease with time. The authors speculate that these data suggest the hypothesis that at the onset, the early vascularization process depends on factors issued by SOs, and overtime the consolidation of the vascular environment is strongly aimed at the osteogenesis induction. As an element of discussion, it is worth emphasizing that neovascularization is a positive response to the introduction of a foreign body in an organism, that ensures a proper supply of biochemical signals, oxygen and nutrient to the cells of the tissue surrounding the implant.82 The extensive blood vessel formation suggests the onset of a series of events able to speed up the vascularization, thus enhancing the efficacy of tissue formation. These features are very important for a biomaterial able to stimulate angiogenesis without amplifying inflammation, underlining that, in this case, angiogenesis is an essential prelude to osteogenesis processes.83,84
3.3. Mass spectrometry analysis
Following in ovo and in vivo observations, demonstrating that SO has a very strong ability to induce angiogenesis, we conducted further investigation by mass spectrometry in order to determine which factors were responsible for these effects.Since both the SOs and the conditioned media were inducers of angiogenesis, the protein content of SOs was analyzed by mass spectrometry analysis (LCMSQE). All proteins were categorized in GO annotations analyzing molecular functions, biological processes and protein classes.
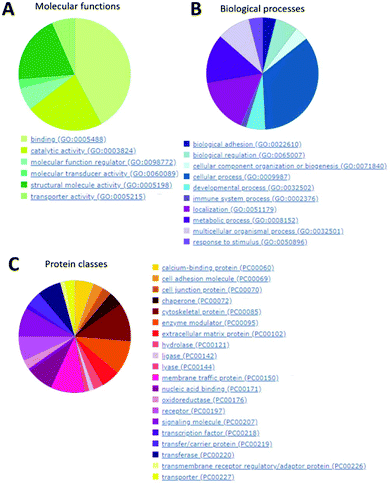
In Fig. 10, it is clearly visible that the majority of proteins found inside the SOs (panel A) are involved in binding activity, catalytic activity and structural molecular activity (such as extracellular matrix structural constituent, etc.).
They are also mainly involved in cellular processes, localization and metabolic processes (Fig. 10B). Finally, the most abundant protein classes are cytoskeletal proteins (mainly actin and microtubule families), receptors and signaling molecules (Fig. 10C).
These pieces of evidence could suggest that several proteins belonging to these categories are also involved in the angiogenesis process.85–87
Next, in order to validate the previous observations concerning the SO angiogenic effects, we analyzed all the hits found by LCMSQE analysis by means of proteomic and interatomic databases. A series of proteins were identified as belonging to the Gene Ontology (GO) annotations: vasculature development (GO0001944), cardiovascular system development (GO0072358), regulation of vasculature development (GO1901342) and negative regulation of vasculature development (GO1901343).
All the proteins defined in Table 2 are involved in the pathways of angiogenesis or vascular development and act as positive or negative regulators of these processes. In the literature, it is reported that the angiogenic processes of endothelial cell growth, migration and tube formation are finely regulated by the balance between pro- and anti-angiogenic factors, cell–extracellular matrix interactions and matrix-degrading proteases.128
| ID Uniprot | Protein name | Protein sequence coverage % | MASCOT protein score | emPAI % |
|---|---|---|---|---|
| DCN | Decorin88–91 | 45.9 | 1818 | 31.9 |
| MYH10 | Myosin, heavy chain 1092–94 | 41.3 | 1240 | 11.7 |
| MYH9 | Myosin, heavy chain 995–97 | 42.1 | 1087 | 10.2 |
| COL1A2 | Collagen α-2(I) chain98–100 | 19.7 | 656 | 6.0 |
| SERPINF1 | Serpin peptidase inhibitor101–103 | 34.4 | 239 | 5.3 |
| TMED2 | Transmembrane p24 trafficking protein 2104 | 13.9 | 67 | 4.2 |
| FN1 | Fibronectin105,106 | 12.2 | 64 | 0.6 |
| THBS1 | Thrombospondin1107–109 | 5.9 | 57 | 0.3 |
| COL1A1 | Collagen α-1(I) chain105,110,111 | 34.2 | 54 | 21.5 |
| MYLK | Myosin light chain kinase112 | 3.5 | 51 | 0.5 |
| NCL | Nucleolin95,97,113 | 8.5 | 50 | 0.6 |
| RAP1A | Member of RAS oncogene114–119 | 17.6 | 46 | 2.1 |
| THBS4 | Thrombospondin 4120–122 | 9.0 | 44 | 0.5 |
| THY1 | Thy-1 membrane glycoprotein123–125 | 27.3 | 36 | 2.4 |
| MYL3 | Myosin, light chain 3126,127 | 11.9 | 35 | 2.0 |
Therefore, to complete the previous investigations on the proteins found in SOs, the LCMSQE analysis confirmed the presence of proteins with angiogenic properties capable of inducing angiogenic reactions in the surrounding tissue.
In particular, the first protein with the highest MASCOT protein score is decorin (DCN), a chondroitin/dermatan sulphate proteoglycan, belonging to the small leucine-rich proteoglycan family, which is involved in the collagen fibril assembly to control fibril formation and regulating their mechanical properties. It also plays an important role in the induction of a specific set of metalloproteinases and the concomitant formation of cord-like structures and cell-lined cavities. Furthermore, decorin has also been found to be expressed in a significant amount around neovessels after varicose vein surgery in patients,129 and it has been shown in animal models that the injured cornea deficient for decorin has compromised angiogenesis.88 Moreover, it is worth noting that decorin activates angiogenesis through the IGF1R/AKT/VEGF pathway,130 one of the most important signal transduction mechanisms involved in speeding the cell proliferation in normal and cancer cells.131 In parallel decorin seems to be involved in anti-angiogenic processes; fragments of decorin seem to depress VEGF-induced focal adhesion kinase phosphorylation and assembly of focal adhesions.89 A retardation in cornea neovascularization has also been observed via a downregulation of proangiogenic molecules including VEGF, linked with an overexpression of decorin.132
Therefore, decorin's role in angiogenesis (pro- or anti-angiogenic) seems to be dependent on the molecular microenvironment (where angiogenesis is induced). Decorin can either promote or inhibit angiogenesis and thus, impacts the fate (life and death) of endothelial cells.90
Other relatively abundant proteins found on SOs are myosin heavy chain 9 and 10. There are two non-muscle myosin II also namely NMHC IIA and NMHC IIB, respectively.96 In particular myosin-9, identified as a nucleolin-binding protein, is the physical linker between nucleolin and the cytoskeleton. This actin-based motor protein provides an essential function, together with those of the VEGF and ECM, in linking nucleolin and the cytoskeleton in order to mediate nucleolin's function in angiogenic processes.97 Nucleolin has also been described as a nucleolar protein involved in cell proliferation, cytokinesis, replication, embryogenesis and nucleogenesis. Moreover, cell surface nucleolin is essential for migration and tube formation of endothelial cells. During angiogenesis, the upregulation of cell–surface nucleolin is attributed to the shuttle of nucleolin from the nucleus to the cell membrane, mediated by myosin-9.95
In summary, the LCMSQE analysis supports the previous in vitro and in vivo analysis showing that the proteins present in SOs could induce angiogenic effects.
4. Conclusions
In this research, we demonstrated the capability of the previously characterized SOs to release bioactive molecules able to induce a clear angiogenic response. A variety of techniques were used to investigate, firstly, the angiogenic response in ovo and in vivo and, secondly, the biocompatibility in vivo. After observing both the angiogenic potential and the biocompatibility, the putative actors of these effects were also sought by highlighting which angiogenic factors could be released from this peculiar material. In conclusion, the scaffold investigated in the present study might be considered a good candidate as a bone substitute or a TE scaffold for future clinical applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Filippo Genovese for helping with performing LCMSQE and in the protein identification phase, Mathilde Fénélon for her help in surgeries and Sylvie Rey for her support with the histological staining procedures. This work was supported by the UIMORE FAR funds (ID No. JJKH20190073KJ) and a collaboration with BioTis of Bordeaux. The authors acknowledge the Fondazione Cassa di Risparmio di Modena for funding the UHPLC-ESI-Q Exactive system at the Centro Interdipartimentale Grandi Strumenti (CIGS). The study was also supported by funds “Department of Excellence 2018-2021” (Department of Biomedical, Metabolic and Neural Sciences).Notes and references
- M. Checchi, J. Bertacchini, G. Grisendi, A. Smargiassi, A. Sola, M. Messori and C. Palumbo, Biomedicines, 2018, 6, 1–14 CrossRef.
- R. Agarwal and A. J. García, Adv. Drug Delivery Rev., 2015, 94, 53–62 CrossRef CAS.
- N. Maffulli, R. Papalia, B. Zampogna, G. Torre, E. Albo and V. Denaro, Surgeon, 2016, 14, 345–360 CrossRef CAS.
- E. H. Schemitsch, J. Orthop. Trauma, 2017, 31, S20–S22 CrossRef.
- T. C. Fitzgibbons, M. A. Hawks, S. T. McMullen and D. J. Inda, J. Am. Acad. Orthop. Surg., 2011, 19, 112–120 CrossRef.
- I. G. Winson and A. Higgs, Foot Ankle Clin., 2010, 15, 553–558 CrossRef PubMed.
- S. M. Graham, A. Leonidou, N. Aslam-Pervez, A. Hamza, P. Panteliadis, M. Heliotis, A. Mantalaris and E. Tsiridis, Expert Opin. Biol. Ther., 2010, 10, 885–901 CrossRef CAS.
- C. M. Soardi, S. Spinato, D. Zaffe and H. L. Wang, Clin. Oral Implants Res., 2011, 22, 560–566 CrossRef.
- P. Kumar, G. Fathima and B. Vinitha, J. Pharm. BioAllied Sci., 2013, 5, 125–127 CrossRef.
- M. Windrich, M. Grimmer, O. Christ, S. Rinderknecht and P. Beckerle, Biomed. Eng. Online, 2016, 15, 5–19 CrossRef.
- J. C. Van Egmond, H. Verburg and N. M. C. Mathijssen, Acta Orthop., 2015, 86, 708–713 CrossRef.
- D. Abi-Hanna, J. Kerferd, K. Phan, P. Rao and R. Mobbs, World Neurosurg., 2018, 109, 188–196 CrossRef.
- C. J. Percival and J. T. Richtsmeier, Dev. Dyn., 2013, 242, 909–922 CrossRef.
- H. Schell, G. N. Duda, A. Peters, S. Tsitsilonis, K. A. Johnson and K. Schmidt-Bleek, J. Exp. Orthop., 2017, 4, 1–11 CrossRef.
- A. Carlier, N. van Gastel, L. Geris, G. Carmeliet and H. Van Oosterwyck, PLoS Comput. Biol., 2014, 10, 1–21 CrossRef.
- M. Richard and E. Thomas, Injury, 2011, 42, 551–555 CrossRef.
- J. M. Kanczler and R. O. C. Oreffo, Eur. Cells Mater., 2008, 15, 100–114 CrossRef CAS.
- L.-J. Chen and H. Kaji, Lab Chip, 2017, 17, 4186–4219 RSC.
- A. Neve, F. P. Cantatore, N. Maruotti, A. Corrado and D. Ribatti, Biomed. Res. Int., 2014, 2014, 1–10 CrossRef.
- J. Sottile, Biochim. Biophys. Acta, Rev. Cancer, 2004, 1654, 13–22 CrossRef CAS.
- Y. Du, S. C. B. Herath, Q. G. Wang, D. A. Wang, H. H. Asada and P. C. Y. Chen, Sci. Rep., 2016, 6, 1–14 CrossRef CAS.
- K. K. Sivaraj and R. H. Adams, Development, 2016, 143, 2706–2715 CrossRef CAS.
- G. Marotti, M. Ferretti, M. A. Muglia, C. Palumbo and S. Palazzini, Bone, 1992, 13, 363–368 CrossRef CAS.
- G. Marotti, Ital. J. Anat. Embryol., 2010, 115, 123–126 Search PubMed.
- M. Ferretti, C. Palumbo, M. Contri and G. Marotti, Anat. Embryol., 2002, 206, 21–29 CrossRef PubMed.
- M. Ferretti, C. Palumbo, L. Bertoni, F. Cavani and G. Marotti, Anat. Rec., Part A, 2006, 288, 1158–1162 CrossRef.
- Z. Wang, Z. Li, Z. Li, B. Wu, Y. Liu and W. Wu, Acta Biomater., 2018, 81, 129–145 CrossRef CAS.
- A. Noori, S. J. Ashrafi, R. Vaez-Ghaemi, A. Hatamian-Zaremi and T. J. Webster, Int. J. Nanomed., 2017, 12, 4937–4961 CrossRef CAS.
- A. E. Jakus, A. L. Rutz, S. W. Jordan, A. Kannan, S. M. Mitchell, C. Yun, K. D. Koube, S. C. Yoo, H. E. Whiteley, C. P. Richter, R. D. Galiano, W. K. Hsu, S. R. Stock, E. L. Hsu and R. N. Shah, Sci. Transl. Med., 2016, 8, 1–16 Search PubMed.
- M. Kim, J. Son, H. Lee, H. Hwang, C. H. Choi and G. Kim, Curr. Appl. Phys., 2014, 14, 1–7 CrossRef.
- B. Zhang, P. Zhang, Z. Wang, Z. Lyu and H. Wu, J. Zhejiang Univ., Sci., B, 2017, 18, 963–976 CrossRef CAS.
- G. Turnbull, J. Clarke, F. Picard, P. Riches, L. Jia, F. Han, B. Li and W. Shu, Bioact. Mater., 2018, 3, 278–314 CrossRef.
- H. Zigdon-Giladi, A. Khutaba, R. Elimelech, E. E. Machtei and S. Srouji, J. Biomed. Mater. Res., Part A, 2017, 105, 2712–2721 CrossRef CAS.
- N. Bloise, L. Petecchia, G. Ceccarelli, L. Fassina, C. Usai, F. Bertoglio, M. Balli, M. Vassalli, G. C. De Angelis, P. Gavazzo, M. Imbriani and L. Visai, PLoS One, 2018, 13, 1–19 CrossRef.
- J. Yin, S. Qiu, B. Shi, X. Xu, Y. Zhao, J. Gao, S. Zhao and S. Min, Biomed. Mater., 2018, 13, 025001 CrossRef.
- K. Hu and B. R. Olsen, Bone, 2016, 91, 30–38 CrossRef CAS.
- D. Lopes, C. Martins-Cruz, M. B. Oliveira and J. F. Mano, Biomaterials, 2018, 185, 240–275 CrossRef CAS.
- B. P. Hung, D. L. Hutton and W. L. Grayson, Stem Cell Res. Ther., 2013, 4, 1–7 CrossRef.
- C. O. Urrutia, M. V. Dominguez-García, J. Flores-Estrada, A. Laguna-Camacho, J. Castillo-Cadena and M. V. Flores-Merino, in Scaffolds in Tissue Engineering - Materials, Technologies and Clinical Applications, 2017, pp. 147–161 Search PubMed.
- F. Zhao, T. J. Vaughan and L. M. McNamara, Biomech. Model. Mechanobiol., 2016, 15, 561–577 CrossRef.
- Y.-C. Li, K. Zhu and T.-H. Young, J. Thorac. Dis., 2017, 9, 455–459 CrossRef.
- L. Zhang, Y. Morsi, Y. Wang, Y. Li and S. Ramakrishna, Jpn. Dent. Sci. Rev., 2013, 49, 14–26 CrossRef.
- D. Fauza, Best Pract. Res. Clin. Obstet. Gynaecol., 2004, 18, 877–891 CrossRef PubMed.
- U. G. Longo, M. Loppini, A. Berton, L. La Verde, W. S. Khan and V. Denaro, Curr. Stem Cell Res. Ther., 2012, 7, 272–281 CrossRef CAS.
- X. Sun, Y. Kang, J. Bao, Y. Zhang, Y. Yang and X. Zhou, Biomaterials, 2013, 34, 4971–4981 CrossRef CAS.
- B. S. Kim, J. S. Kim, S. S. Yang, H. W. Kim, H. J. Lim and J. Lee, Biomater. Res., 2015, 19, 1–9 CrossRef.
- M. Tanaka, Y. Sato, H. Haniu, H. Nomura, S. Kobayashi, S. Takanashi, M. Okamoto, T. Takizawa, K. Aoki, Y. Usui, A. Oishi, H. Kato and N. Saito, PLoS One, 2017, 12, 1–18 CAS.
- S. Almubarak, H. Nethercott, M. Freeberg, C. Beaudon, A. Jha, W. Jackson, R. Marcucio, T. Miclau, K. Healy and C. Bahney, Bone, 2016, 83, 197–209 CrossRef CAS.
- F. J. O’ Brien, Mater. Today, 2011, 14, 88–95 CrossRef.
- T. A. Franz-Odendaal and B. K. Hall, J. Morphol., 2006, 267, 1326–1337 CrossRef.
- K. Jourdeuil and T. A. Franz-Odendaal, Anat. Rec., 2012, 295, 691–698 CrossRef.
- T. A. Franz-Odendaal, B. K. Hall and P. E. Witten, Dev. Dyn., 2006, 235, 176–190 CrossRef CAS.
- F. C. Lima, L. G. Vieira, A. L. Q. Santos, S. B. S. De Simone, L. Q. R. Hirano, J. M. M. Silva and M. F. Romao, Braz. J. Morphol. Sci., 2009, 26, 165–169 Search PubMed.
- C. Palumbo, L. Presutti, E. Genovese, F. Cavani, P. Sena, M. Benincasa and M. Ferretti, Ital. J. Anat. Embryol., 2012, 117, 2012 Search PubMed.
- C. Palumbo, F. Cavani, P. Sena, M. Benincasa and M. Ferretti, Calcif. Tissue Int., 2012, 90, 211–218 CrossRef CAS.
- V. Hamburger and H. L. Hamilton, J. Morphol., 1951, 88, 49–92 CrossRef CAS.
- D. Ribatti, A. Gualandris, M. Bastaki, A. Vacca, M. Iurlaro, L. Roncali and M. Presta, J. Vasc. Res., 1997, 34, 455–463 CrossRef CAS.
- J. C. Venter, M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, J. D. Gocayne, P. Amanatides, R. M. Ballew, D. H. Huson, J. R. Wortman, Q. Zhang, C. D. Kodira, X. H. Zheng, L. Chen, M. Skupski, G. Subramanian, P. D. Thomas, J. Zhang, G. L. G. Miklos, C. Nelson, S. Broder, A. G. Clark, J. Nadeau, V. A. Mckusick, N. Zinder, A. J. Levine, R. J. Roberts, M. Simon, C. Slayman, M. Hunkapiller, R. Bolanos, A. Delcher, I. Dew, D. Fasulo, M. Flanigan, L. Florea, A. Halpern, S. Hannenhalli, S. Kravitz, S. Levy, C. Mobarry, K. Reinert, K. Remington, J. Abu-threideh, E. Beasley, K. Biddick, V. Bonazzi, R. Brandon, M. Cargill, I. Chandramouliswaran, R. Charlab, K. Chaturvedi, Z. Deng, V. Di Francesco, P. Dunn, K. Eilbeck, C. Evangelista, A. E. Gabrielian, W. Gan, W. Ge, F. Gong, Z. Gu, P. Guan, T. J. Heiman, M. E. Higgins, R. Ji, Z. Ke, K. A. Ketchum, Z. Lai, Y. Lei, Z. Li, J. Li, Y. Liang, X. Lin, F. Lu, G. V. Merkulov, N. Milshina, H. M. Moore, A. K. Naik, V. A. Narayan, B. Neelam, D. Nusskern, D. B. Rusch, S. Salzberg, W. Shao, B. Shue, J. Sun, Z. Y. Wang, A. Wang, X. Wang, J. Wang, M. Wei, R. Wides, C. Xiao, C. Yan, A. Yao, J. Ye, M. Zhan, W. Zhang, H. Zhang, Q. Zhao, L. Zheng, F. Zhong, W. Zhong, S. C. Zhu, S. Zhao, D. Gilbert, S. Baumhueter, G. Spier, C. Carter, A. Cravchik, T. Woodage, F. Ali, H. An, A. Awe, D. Baldwin, H. Baden, M. Barnstead, I. Barrow, K. Beeson, D. Busam, A. Carver, M. L. Cheng, L. Curry, S. Danaher, L. Davenport, R. Desilets, S. Dietz, K. Dodson, L. Doup, S. Ferriera, N. Garg, A. Gluecksmann, B. Hart, J. Haynes, C. Haynes, C. Heiner, S. Hladun, D. Hostin, J. Houck, T. Howland, C. Ibegwam, J. Johnson, F. Kalush, L. Kline, S. Koduru, A. Love, F. Mann, D. May, S. Mccawley, T. Mcintosh, I. Mcmullen, M. Moy, L. Moy, B. Murphy, K. Nelson, C. Pfannkoch, E. Pratts, V. Puri, H. Qureshi, M. Reardon, R. Rodriguez, Y. Rogers, D. Romblad, B. Ruhfel, R. Scott, C. Sitter, M. Smallwood, E. Stewart, R. Strong, E. Suh, R. Thomas, N. N. Tint, S. Tse, C. Vech, G. Wang, J. Wetter, S. Williams, M. Williams, S. Windsor, E. Winn-deen, K. Wolfe, J. Zaveri, K. Zaveri, J. F. Abril, R. Guigo, A. Kejariwal, H. Mi, B. Lazareva, T. Hatton, A. Narechania, K. Diemer, A. Muruganujan, N. Guo, S. Sato, V. Bafna, S. Istrail, R. Lippert, R. Schwartz, B. Walenz, S. Yooseph, D. Allen, A. Basu, J. Baxendale, L. Blick, M. Caminha, J. Carnes-stine, P. Caulk, Y. Chiang, M. Coyne, C. Dahlke, A. D. Mays, M. Dombroski, M. Donnelly, D. Ely, S. Esparham, C. Fosler, H. Gire, S. Glanowski, K. Glasser, A. Glodek, M. Gorokhov, K. Graham, B. Gropman, M. Harris, J. Heil, S. Henderson, J. Hoover, D. Jennings, C. Jordan, J. Jordan, J. Kasha, L. Kagan, C. Kraft, A. Levitsky, M. Lewis, X. Liu, J. Lopez, D. Ma, W. Majoros, J. Mcdaniel, S. Murphy, M. Newman, T. Nguyen, N. Nguyen, M. Nodell, S. Pan, J. Peck, M. Peterson, W. Rowe, R. Sanders, J. Scott, M. Simpson, T. Smith, A. Sprague, T. Stockwell, R. Turner, E. Venter, M. Wang, M. Wen, D. Wu, M. Wu, A. Xia, A. Zandieh and X. Zhu, Science, 2001, 291, 1304–1351 CrossRef CAS.
- M. Gomez de Agüero, S. C. Ganal-Vonarburg, T. Fuhrer, S. Rupp, U. Yasuhiro, H. Li, A. Steinert, M. Heikenwalder, S. Hapfelmeier, U. Sauer, K. D. McCoy and A. J. Macpherson, Science, 2016, 351, 1296–1302 CrossRef.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Nat. Protoc., 2009, 4, 44–57 CrossRef CAS.
- D. Ribatti, Reprod. Toxicol., 2017, 70, 97–101 CrossRef CAS.
- Institutional Animal Care and Use Committee (IACUC), Policy on protocol for Use Use of Avian Embryos and Eggs. An association of New England Medical Center and Tufts, 2016 Search PubMed.
- ILAR news, 1991, 33.4: 68–70.
- W. M. S. Russell and R. L. Burch, The principles of humane experimental technique, 1959 Search PubMed.
- C. S. Kue, K. Y. Tan, M. L. Lam and H. B. Lee, Exp. Anim., 2015, 64, 129–138 CrossRef CAS.
- O. Baum, F. Suter, B. Gerber, S. A. Tschanz, R. Buergy, F. Blank, R. Hlushchuk and V. Djonov, Microcirculation, 2010, 17, 447–457 CAS.
- U. Montecorboli, T. Annese, C. Marinaccio and D. Ribatti, Int. J. Dev. Biol., 2015, 59, 461–464 CrossRef CAS.
- P. Nowak-sliwinska, T. Segura and M. L. Iruela-arispe, Angiogenesis, 2014, 17, 779–804 CrossRef CAS PubMed.
- D. Ribatti, in Cardiovascular Development. Methods in Molecular Biology (Methods and Protocols), Humana Press, Totowa, NJ, 2012, vol. 843, pp. 47–57 Search PubMed.
- S. Singh, B. M. Wu and J. C. Y. Dunn, J. Biomed. Mater. Res., Part A, 2012, 100, 720–727 CrossRef.
- D. Ribatti, C. Urbinati, B. Nico, M. Rusnati, L. Roncali and M. Presta, Dev. Biol., 1995, 170, 89–49 CrossRef.
- X. Xin, S. Yang, G. Ingle, C. Zlot, L. Rangell, J. Kowalski, R. Schwall, N. Ferrara and M. E. Gerritsen, Am. J. Pathol., 2001, 158, 1111–1120 CrossRef CAS.
- D. O. DeFouw, V. J. Rizzo, R. Steinfeld and R. N. Feinberg, Microvasc. Res., 1989, 38, 136–147 CrossRef CAS.
- Z. Sheikh, P. J. Brooks, O. Barzilay, N. Fine and M. Glogauer, Materials, 2015, 8, 5671–5701 CrossRef CAS.
- J. M. Anderson, Semin. Immunol., 2008, 20, 86–100 CrossRef CAS.
- Y. Xin, Y. Yuan, F. Chi, J. Wang and J. Yang, Chin. Med. J., 2015, 128, 2124–2125 CrossRef.
- U. Ezomike, M. Ituen and S. Ekpemo, J. West Afr. Coll. Surg., 2011, 1, 53–59 Search PubMed.
- A. Prudente, W. J. Fávaro, P. L. Filho and C. L. Z. Riccetto, Int. Braz. J. Urol., 2016, 42, 585–593 CrossRef PubMed.
- B. A. Imhof and D. Dunon, Horm. Metab. Res., 1997, 29, 614–621 CrossRef CAS.
- S. Sangkert, J. Meesane, S. Kamonmattayakul and W. Lin, Mater. Sci. Eng., C, 2016, 58, 1138–1149 CrossRef CAS PubMed.
- K. Wilson, A. Terlouw, K. Roberts, J. C. Wolchok and M. B. Program, J. Mater. Sci.: Mater. Med., 2016, 27, 1–29 CrossRef CAS.
- S. Ghanaati, M. J. Webber, R. E. Unger, C. Orth, J. F. Hulvat, S. E. Kiehna, M. Barbeck, A. Rasic, S. I. Stupp and C. J. Kirkpatrick, Biomaterials, 2009, 30, 6202–6212 CrossRef CAS.
- C. Fredriksson, M. Hedhammar, R. Feinstein, K. Nordling, G. Kratz, J. Johansson, F. Huss and A. Rising, Materials, 2009, 2, 1908–1922 CrossRef CAS.
- L. Chu, G. Jiang, X. Le Hu, T. D. James, X. P. He, Y. Li and T. Tang, J. Mater. Chem. B, 2018, 6, 4197–4204 RSC.
- A. M. Goodwin, Microvasc. Res., 2007, 74, 172–183 CrossRef CAS.
- D. Guidolin, A. Vacca, G. G. Nussdorfer and D. Ribatti, Microvasc. Res., 2004, 67, 117–124 CrossRef CAS.
- B. Vailhé, D. Vittet and J. J. Feige, Lab. Invest., 2001, 81, 439–452 CrossRef.
- E. Schönherr, C. Sunderkötter, L. Schaefer, S. Thanos, S. Grässel, Å. Oldberg, R. V. Iozzo, M. F. Young and H. Kresse, J. Vasc. Res., 2004, 41, 499–508 CrossRef.
- K. N. Sulochana, H. Fan, S. Jois, V. Subramanian, F. Sun, R. M. Kini and R. Ge, J. Biol. Chem., 2005, 280, 27935–27948 CrossRef CAS.
- H. Järveläinen, A. Sainio and T. N. Wight, Matrix Biol., 2015, 43, 15–26 CrossRef PubMed.
- J. Lai, F. Chen, J. Chen, G. Ruan, M. He, C. Chen, J. Tang and D. W. Wang, Sci. Rep., 2017, 7, 1–11 CrossRef.
- C. A. Franco, J. Blanc, A. Parlakian, R. Blanco, I. M. Aspalter, N. Kazakova, N. Diguet, E. Mylonas, J. Gao-Li, A. Vaahtokari, V. Penard-Lacronique, M. Fruttiger, I. Rosewell, M. Mericskay, H. Gerhardt and Z. Li, J. Cell Sci., 2013, 126, e1 CrossRef.
- L. A. Ridge, K. Mitchell, A. Al-Anbaki, W. M. Shaikh Qureshi, L. A. Stephen, G. Tenin, Y. Lu, I. E. Lupu, C. Clowes, A. Robertson, E. Barnes, J. A. Wright, B. Keavney, E. Ehler, S. C. Lovell, K. E. Kadler and K. E. Hentges, PLoS Genet., 2017, 13, 1–40 Search PubMed.
- A. Wang, X. Ma, M. Anne, C. Liu, S. Kawamoto and R. S. Adelstein, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14656–14650 Search PubMed.
- Y. Huang, H. Shi, H. Zhou, X. Song, S. Yuan and Y. Luo, Blood, 2006, 107, 3564–3571 CrossRef CAS.
- J. England and S. Loughna, Cell. Mol. Life Sci., 2013, 70, 1221–1239 CrossRef CAS.
- Y. Ding, N. Song, C. Liu, T. He, W. Zhuo, X. He, Y. Chen, X. Song, Y. Fu and Y. Luo, Arterioscler. Thromb. Vasc. Biol., 2012, 32, 126–134 Search PubMed.
- S. Tamilzhalagan, D. Rathinam and K. Ganesan, Mol. Carcinog., 2017, 56, 1590–1602 CrossRef.
- A. M. Buga, C. Margaritescu, C. J. Scholz, E. Radu, C. Zelenak and A. Popa-Wagner, Front. Aging Neurosci., 2014, 6, 1–20 Search PubMed.
- A. Caporali and C. Emanueli, Trends Cardiovasc. Med., 2011, 21, 162–166 CrossRef CAS.
- J. Beckers, F. Herrmann, S. Rieger, A. L. Drobyshev, M. Horsch, M. H. De Angelis and B. Seliger, Int. J. Cancer, 2005, 114, 590–597 CrossRef CAS.
- J. G. Ren, C. Jie and C. Talbot, Med. Hypotheses, 2005, 64, 74–78 CrossRef CAS.
- X. He, R. Cheng, S. Benyajati and J. Ma, Clin. Sci., 2015, 128, 805–823 CrossRef.
- J. A. Pérez-Valencia, F. Prosdocimi, I. M. Cesari, I. R. Da Costa, C. Furtado, M. Agostini and F. D. Rumjanek, Sci. Rep., 2018, 8, 1–18 CrossRef.
- M. Mongiat, E. Andreuzzi, G. Tarticchio and A. Paulitti, Int. J. Mol. Sci., 2016, 17, 2–26 Search PubMed.
- P. A. Murphy, S. Begum and R. O. Hynes, PLoS One, 2015, 10, 1–16 Search PubMed.
- J. Lawler, J. Cell. Mol. Med., 2002, 6, 1–12 CrossRef CAS.
- P. R. Lawler and J. Lawler, Cold Spring Harbor Perspect. Med., 2012, 2, 1–13 Search PubMed.
- I. Ligi, S. Simoncini, E. Tellier, P. F. Vassallo, F. Sabatier, B. Guillet, E. Lamy, G. Sarlon, C. Quemener, A. Bikfalvi, M. Marcelli, A. Pascal, B. Dizier, U. Simeoni, F. Dignat-George and F. Anfosso, Vasc. Biol., 2011, 118, 1699–1709 CAS.
- L. C. van Kempen, J. Rijntjes, I. Maor-Cornelissen, S. Vincent-Naulleau, M. J. Gerritsen, D. J. Ruiter, M. C. R. van Dijk, C. Geffrotin and G. N. P. van Muijen, Int. J. Cancer, 2008, 122, 1019–1029 CrossRef CAS.
- T. Twardowski, A. Fertala, J. P. Orgel and J. D. San Antonio, Curr. Pharm. Des., 2007, 130, 3608–3621 CrossRef.
- Q. Shen, R. R. Rigor, C. D. Pivetti, M. H. Wu and S. Y. Yuan, Cardiovasc. Res., 2010, 87, 272–280 CrossRef CAS.
- I. Ugrinova, M. Petrova, M. Chalabi-Dchar and P. Bouvet, Adv. Protein Chem. Struct. Biol., 2018, 111, 133–164 Search PubMed.
- J. Yan, F. Li, D. A. Ingram and L. A. Quilliam, Mol. Cell. Biol., 2008, 28, 5803–5810 CrossRef CAS.
- G. Carmona, A. Orlandi, A. M. Zeiher, S. Dimmeler, E. Chavakis, S. Göttig, R. Henschler, J. Scheele, T. Bäuerle, M. Jugold and F. Kiessling, Blood, 2009, 113, 488–497 CrossRef CAS.
- S. Lakshmikanthan, M. Sobczak, C. Chun, A. Henschel, J. Dargatz, R. Ramchandran and M. Chrzanowska-Wodnicka, Blood, 2011, 118, 2015–2026 CrossRef CAS.
- J. Menon, R. C. Doebele, S. Gomes, E. Bevilacqua, K. M. Reindl and M. R. Rosner, PLoS One, 2012, 7, 1–10 Search PubMed.
- H. Yamamoto, M. Ehling, K. Kato, K. Kanai, M. Van Lessen, M. Frye, D. Zeuschner, M. Nakayama, D. Vestweber and R. H. Adams, Nat. Commun., 2015, 6, 1–14 Search PubMed.
- J. L. Arbiser, Blood, 2011, 118, 1719–1720 CrossRef CAS PubMed.
- S. Muppala, E. Frolova, R. Xiao, I. Krukovets, S. Yoon, G. Hoppe, A. Vasanji, E. Plow and O. Stenina-Adognravi, Arterioscler., Thromb., Vasc. Biol., 2015, 35, 1975–1986 CrossRef CAS.
- S. Muppala, R. Xiao, I. Krukovets, D. Verbovetsky, R. Yendamuri, N. Habib, P. Raman, E. Plow and O. Stenina-Adognravi, Oncogene, 2017, 36, 5189–5198 CrossRef CAS.
- J. Liu, G. Cheng, H. Yang, X. Deng, C. Qin, L. Hua and C. Yin, Mol. Med. Rep., 2016, 14, 1451–1458 CrossRef CAS.
- C. Sauzay, K. Voutetakis, A. Chatziioannou and E. Chevet, Front. Cell Dev. Biol., 2019, 7, 1–11 CrossRef.
- G. Jurisic, M. Iolyeva, S. T. Proulx, C. Halin and M. Detmar, Exp. Cell Res., 2010, 316, 2982–2992 CrossRef CAS.
- J. A. Bradley, G. Ramirez and J. S. Hagood, BioFactors, 2009, 35, 258–265 CrossRef CAS.
- G. M. Polachini, L. M. Sobral, A. M. Mercante, A. F. Paes-leme, F. C. Xavier, T. Henrique, D. M. Guimarães, A. Vidotto, E. E. Fukuyama, J. F. Gòis-Filho, P. M. Cury, O. A. Curioni, P. J. Michaluart, A. M. Silva, V. Wünsch-Filho, F. D. Nunes, A. M. Leopoldino and E. H. Tajara, PLoS One, 2012, 7, 1–13 CrossRef.
- F. Z. Marques, A. E. Campain, Y. H. J. Yang and B. J. Morris, Hypertension, 2010, 56, 319–324 CrossRef CAS.
- D. H. Kong, M. R. Kim, J. H. Jang, H. J. Na and S. Lee, Int. J. Mol. Sci., 2017, 18, 1–25 Search PubMed.
- S. Reich-Schupke, A. Mumme, P. Altmeyer and M. Stuecker, Dermatol. Surg., 2011, 37, 480–485 CrossRef CAS.
- J. Lai, F. Chen, J. Chen, G. Ruan, M. He, C. Chen, J. Tang and D. W. Wang, Sci. Rep., 2017, 7, 1–11 CrossRef.
- J. Bertacchini, M. Guida, B. Accordi, L. Mediani, A. M. Martelli, P. Barozzi, E. P. Iii, L. Liotta, G. Milani, M. Giordan and M. Luppi, Leukemia, 2014, 28, 2197–2205 CrossRef CAS.
- R. R. Mohan, J. C. K. Tovey, A. Sharma, G. S. Schultz, J. W. Cowden and A. Tandon, PLoS One, 2011, 6, 1–11 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9bm01234f |
| This journal is © The Royal Society of Chemistry 2020 |