 Open Access Article
Open Access ArticleElectrospun nanofibers in cancer research: from engineering of in vitro 3D cancer models to therapy
Marta
Cavo†
a,
Francesca
Serio†
ab,
Narendra R.
Kale
c,
Eliana
D'Amone
a,
Giuseppe
Gigli
ab and
Loretta L.
del Mercato
 *a
*a
aInstitute of Nanotechnology, National Research Council (CNR-NANOTEC), c/o Campus Ecotekne, via Monteroni, 73100, Lecce, Italy. E-mail: loretta.delmercato@nanotec.cnr.it
bDepartment of Mathematics and Physics “Ennio De Giorgi”, University of Salento, via Arnesano, 73100, Lecce, Italy
cMIT-WPU, School of Pharmacy, Kothrud, Pune 411038, India
First published on 24th August 2020
Abstract
Electrospinning is historically related to tissue engineering due to its ability to produce nano-/microscale fibrous materials with mechanical and functional properties that are extremely similar to those of the extracellular matrix of living tissues. The general interest in electrospun fibrous matrices has recently expanded to cancer research both as scaffolds for in vitro cancer modelling and as patches for in vivo therapeutic delivery. In this review, we examine electrospinning by providing a brief description of the process and overview of most materials used in this process, discussing the effect of changing the process parameters on fiber conformations and assemblies. Then, we describe two different applications of electrospinning in service of cancer research: firstly, as three-dimensional (3D) fibrous materials for generating in vitro pre-clinical cancer models; and secondly, as patches encapsulating anticancer agents for in vivo delivery.
1. Introduction
Electrospinning (ES) is a worldwide recognized process to produce fibrous and porous two-dimensional (2D) and three-dimensional (3D) materials starting from a polymeric solution.1–4 The success of ES is mainly related to its versatility, despite being a simple and cost-effective technique: during the ES process, an electrified polymeric jet experiences bending instability and solidifies to produce long and continuous nanofibers with diameters ranging from a few tens of nanometers to a few micrometers.5ES has been significantly used for tissue engineering (TE) applications, since a huge number of different structures, morphologies and compositions can be obtained and adopted to make fibers suitable for vascular, bone, neural, tendon and ligament regeneration.6–14 In particular, a lot of studies have been published in the field of bone regeneration, where a nanofibrous substratum provides favourable conditions for bone-associated cell anchorage and growth;15,16 the random nature of fiber deposition produces the surface roughness suitable for cell attachment in bone scaffolds, as well as for osteoblastic differentiation and mineralization that are better regulated on nanofibrous surfaces than on more dense structures.17 ECM-like electrospun structures are also suitable for muscle cell adhesion, since myoblast cells require highly aligned (anisotropic) or unidirectionally oriented fibers to adhere and proliferate.18,19 The use of electrospun scaffolds as substrates for neural tissue engineering was also investigated; in this context, several groups have demonstrated how electrospun fibers show great efficacy in enhancing nerve regeneration.20,21 Last but not least, electrospinning has been used to produce thin tubes to be used as scaffolds to make blood vessels.22–26
In cancer treatment, nanofibers have many advantages such as the ability to obtain fibers with diameters ranging from nanometers to sub-micrometers, surface modification, alignment variation and drug encapsulation. Anticancer drug loaded ES nanofibers enable controlled and sustained drug release at the desired site of action with improved efficacy. ES nanofibers have also been used as an implant into a post-operative tumour cavity, to inhibit tumour recurrence and to prolong drug release at the tumour site.27 Furthermore, ES nanofibers provide a unique opportunity to build cell environments which can mimic the in vivo tumour micro-environment.28 Some of the recent studies towards 3D in vitro cancer model development are mentioned below.
Over the last few years, the research community has focused on the possibility to translate tissue engineering principles and methods into cancer research, with the aim to provide physiologically relevant 3D in vitro cancer models. This new branch is known as cancer tissue engineering and aims to explore cancer pathogenesis and evolution in a more realistic environment than 2D cultures or animal models.29–31 In this context, also the electrospinning technique has been repurposed to obtain 3D fibrous materials with properties similar to those found in native tumours.32 At the same time, in the last few years, electrospun matrices have gained attention also in clinical applications: thanks to their porous structure and ability to incorporate drugs within the fiber lumen, electrospun fiber matrices have been used as a transdermal drug delivery system (TDDS) to facilitate delivery through skin in a controlled way; this application is particularly relevant in the case of anticancer treatments that are often associated with several drawbacks, such as poor solubility and instability in the biological environment, adverse effects on healthy tissues and low concentration at tumour sites.33 We have listed some of the polymers used for anticancer drug delivery in vitro analysis using cancer cell lines (Table 1).
| Polymer | Fiber diameter | Drug | Cancer type | Ref. |
|---|---|---|---|---|
| Abbreviations: PLGA – poly lactic-co-glycolic acid, PLLA – poly(L-lactic acid), PEG–PLLA-poly(ethylene glycol)–poly(L-lactic acid), PCL – poly(ε-caprolactone), pNIPAM – poly(N-isopropylacrylamide), and PVA – poly(vinyl alcohol). | ||||
| PLGA | 430 nm | Doxorubicin hydrochloride and camptothecin | HepG-2 cells (human liver cancer) | 34 |
| PLLA | 300 nm | 5-Fluorouracil and oxaliplatin | Colorectal cancer | 35 |
| PEG–PLLA | 690 nm | Carmustine | Glioma C6 cells | 36 |
| PCL | 80–120 nm | 5-Fluorouracil and paclitaxel | TNBC cells (human triple negative breast cancer) | 37 |
| pNIPAM | 600–800 nm | Doxorubicin | Hela cells (human cervical cancer) | 38 |
| PVA | 260 nm | Doxorubicin | SKOV3 cells (ovary cancer) | 39 |
However, there are a few challenges in ES nanofiber applications such as achieving uniform drug distribution in ES fibers and drug compatibility in polymer solutions, avoiding initial burst release of drugs, and developing 3D scaffolds with desired porosity. Due to these critical parameters, all the electrospun nanofiber-based systems for cancer research are still in preclinical trials only. We discuss ahead the challenging parameters of ES fibers in cancer research.
All these facts highlight the multiple applications of electrospinning to solve the problems encountered in cancer research. The first part of this review will be dedicated to the summarization of the state of the art electrospinning-based 3D in vitro tumour models, subdivided according to the type of cancer studied or as membranes for intra- and extravasation studies; the second part will review the clinical applications of electrospun fibers for local therapy, giving overall a complete picture of electrospinning meeting cancer research, both in vitro and in vivo.
2. Overview of the electrospinning technique
Electrospinning is basically an electro-hydrodynamic phenomenon: it begins with the exposure of a charged polymer to an electric field between a metal needle and a collector; this causes instability within the polymer solution that deforms the spherical droplet to a conical shape (Taylor cone); at this stage, ultrafine fibers are extruded from the conical polymer droplet, and are collected on a metallic collector kept at an optimized distance.6,40,41 The fiber formation process begins with jet initiation followed by jet whipping instability. A stable charged jet initiates when the applied voltage exceeds a critical value and the electrostatic force overcomes the fluid surface tension. Charge forces cause the whipping of the liquid jet towards the collector, allowing the polymer jet to stretch up to a diameter of a few nanometers. During this process, the jet shows non-axisymmetric (with reference to the jet center line) instabilities, i.e. bending instability and axisymmetric instability i.e. Rayleigh instability. Rayleigh instability is observed in the presence of two opposing forces on the surface area, i.e. the surface tension and electrostatic repulsion of charges in the jet.5,42 To successfully complete this process, ES apparatus has to be composed of a high voltage source (1–30 kV), a syringe pump to extrude the solution at a defined flow rate, a metallic needle and a conductive collector.41,43,44 The electrospinning process begins with sufficiently high electrostatic force generated by the increased applied voltage. The applied voltage is inversely proportional to the fiber diameter because of enhanced fiber stretching due to the higher voltage. In addition to the applied voltage, the viscosity of the solution plays a crucial role in the ES process. If low viscosity fluid is used, the solution cannot resist Rayleigh instability and break up into droplets. If the viscosity of the fluid is sufficiently high with long chain molecule content, the solution can withstand against Rayleigh instability and the fluid jet leads to nanofibers deposited on the collector.45,46Solvent evaporation plays a major role while the fluid jet flies towards the collector under the influence of electrostatic force. Solvent evaporation from the fluid jet occurs prior to obtaining a dry nanofiber deposited on the collector. There should be an optimum collecting distance, flow rate and viscosity of solution which are sufficient for solvent evaporation and fiber stretching. Decreased collecting distance and increased flow rate and viscosity result in non-evaporation of the solvent, which leads to bead like structures or an increased nanofiber diameter. Interplay among the applied voltage, solution viscosity, collecting distance, solvent evaporation and flow rate take place during the ES process and to obtain the desired nanofiber characteristics, it is necessary to optimize these parameters. Solvent selection is another critical parameter during the ES process. Highly volatile solvents may cause jet drying at the tip of the needle because of a low boiling point. Meanwhile, less volatile solvents can result in insufficient drying of the fluid jet and poor fiber formation. If the polymer dissolves in two solvents, one as the solvent and another as the non-solvent, ES leads to highly porous nanofibers.47–49
Although it appears at first as an easy method, the entire process depends on a multitude of parameters, such as environmental parameters (temperature and humidity), solution properties (viscosity, conductivity and surface tension) and governing variables (distance between the tip and the collector, electric potential, flow rate, selected polymers, and geometry of the collector). All these parameters can affect the capacity of the spinneret, or the fiber and pore diameters or the structure and morphology of the fibers. Notably, through the optimization of the cited parameters, fibers can be obtained randomly or in an ordered way, larger or thinner, with or without beads.
2.1. Configurations for electrospinning
The two main principles implemented to achieve ES construction are as follows:1. Manipulating the electric field via collector design (auxiliary electrodes):
Electrode arrangements and collector design had been varied to achieve better alignment of nanofibers. Parallel electrodes, multiple electrodes and charged pair electrodes had been tested to achieve the desired alignment of nanofibers. The use of a parallel electrode is one of the simplest ways to manipulate the electric field and drive the ES jet to swing back and forth between them, which leads to aligned fibers between the electrodes. The fiber should withhold its weight across the gap along with external electrostatic forces. A charged pair electrode has additional forces through alternate application of opposing charges. Alternate switching between charged electrodes causes the ES jet to bridge the gap between collector electrodes. For further improvement of the fiber alignment, steering electrodes were used simultaneously along with a parallel electrode collector.50,51
2. Moving the collector:
At the beginning, horizontal and vertical ES setups with flat collectors were proposed (Fig. 1a and b). Even though the gravitational force affecting the polymer is negligible with respect to the electric field forces, gravity has an effect on the shape of the polymer droplet and the Taylor cone; in particular, the shape of the droplet forming depends on the flatness of the needle tip; so in a vertical configuration the needle tip has to be straightened.52 Besides that, the real revolution in ES occurred when it was discovered that the geometry of the collector could influence the arrangement of the fibers and, in particular, could allow obtaining aligned fibers instead of random ones. This result can be achieved by substituting the traditional flat collector with a rotating collector (Fig. 1c).
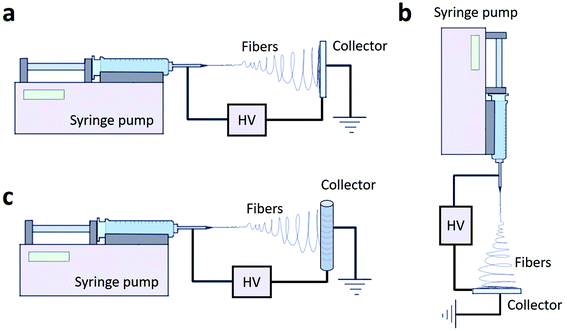 | ||
| Fig. 1 Different configurations of the ES set-up: horizontal (a), vertical (b) and with a rotating collector (c). | ||
The purpose of the rotating collector is to mechanically stretch the fibers helping them to align along the collector. Collector configurations include a solid cylinder which can rotate about its axis, a conducting wire wound on an insulated cylinder, a wired drum or a disc collector (Fig. 2a–d). With this last configuration, double- and triple-layer highly aligned crossbar structures were obtained.53
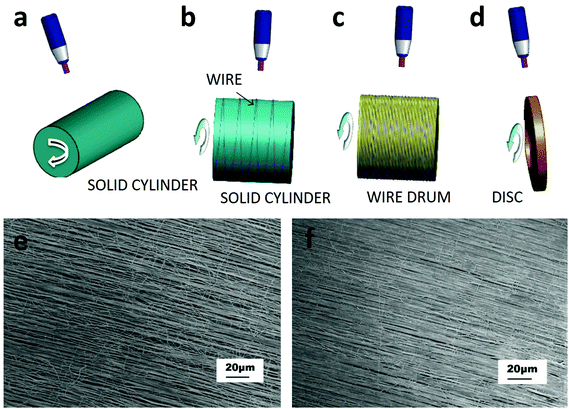 | ||
| Fig. 2 Category of ES set-ups based on the rotating device. These configurations include (a) a solid cylinder, (b) a wire wound on an insulated cylinder, (c) a wired drum, and (d) a disc collector.53 Well-aligned PVA fibers collected on a Teflon tube with different surface velocities: (e) about 1.3 m s−1 and (f) 2.3 m s−1.54 Reproduced from ref. 54 with permission from Elsevier, copyright 2006. | ||
Also in the case of rotating collectors, the alignment of fibers can be optimized by varying several parameters such as polymer viscosity and rotational speed, or improving the set-up with two oppositely placed needles as shown by Pan et al.54 In this work, fibers coming out of the two needles combined in a yarn, which was wound by a cylinder collector rotating at a high speed. Fibers manufactured by this method were continuous, well-aligned, and could be deposited over a large area; moreover, increasing the velocity improved the alignment of the fibers (Fig. 2e and f).
2.2. Materials
So far, more than fifty different polymers and solvents have been successfully electrospun into fibers with diameters in the range from 3 nm to 1 mm. The interactions between the solvent and polymer have been found to be crucial in ensuring a successful production of continuous nanofibers.55 In addition, the solvent selection is important for solution conductivity and may affect the fiber size distribution.56Substantially, the material choice depends on the application of the scaffold. For TE purposes, the choice is based on the properties of the tissue to be regenerated and biodegradable polymers are preferably used. The mostly synthetic materials used in TE are polyesters, such as poly(lactide) (PLA), poly(glycolide) (PGA) and poly(caprolactone) (PCL). Natural materials, such as cellulose, collagen, natural silk, fibrinogen, chitosan and hyaluronic acid, are of interest for use in regenerative medicine because the material becomes more recognizable for the cells.57 However, not all these natural polymers can be easily used with ES: for example, processing chitosan with ES is difficult because of its polycationic nature and its high viscosity in solution, and toxic or highly acidic solvent is needed for electrospinning;58 again, gelatin is not stable if electrospun as a single material; finally, hyaluronic acid solutions have very high viscosity and consequently producing uniform fibers from them can be difficult.59
The available literature in this field demonstrates the ability to electrospin natural polymers with the possibility to exploit their biocompatibility.60 However, when compared with synthetic materials, two main drawbacks such as poor mechanical performance and difficult electrospinnability are clear.
3. Electrospinning for 3D in vitro cancer models
Traditionally, cancer biology research has involved in vitro analysis of cell behaviour predominantly using two-dimensional (2D) cell cultures and in vivo animal models:61 in detail, 2D models are routinely used as initial systems for evaluating the effectiveness of molecules as potential therapeutic drugs; this initial screening precedes animal studies before advancing to human clinical trials.61 The dissimilarities in cell behaviour between 2D cultures and real tumours are well known and they mainly derive from changes in gene expression originating from the different interactions to which the cells are subjected within a 2D microenvironment if compared to a more natural 3D microenvironment.62–70 A striking example of this is represented by the unequal nutrient concentration to which cells are exposed: in 2D cultures, cells are uniformly exposed to nutrients, while in vivo the concentration of soluble factors influencing cell proliferation is characterized by spatial gradients that play a vital role in biological differentiation, organ development and countless other biological processes.71–74 Cancer cells would alter their behaviour in 2D culture from their in vivo behaviour. However, 3D scaffolds will mimic in vivo conditions which will help in precise analysis of the cellular response against anticancer drugs. It is therefore not surprising that many aspects of tumorigenesis are still not fully understood.75 Below, we have listed noted differences of cell line behaviour on 3D scaffolds from that on 2D culture of the same material (Table 2).| 3D nanofiber scaffolds | Fiber diameter | Cell line | Noted difference from 2D culture | Ref. |
|---|---|---|---|---|
| Abbreviations: PCL – poly(ε-caprolactone), PEG – poly(ethylene glycol), PCL – poly(ε-caprolactone), PVA – poly(vinyl alcohol), PEOT – poly(ethylene oxide terephthalate), PDMS – poly(dimethyl siloxane), and PMMA – poly(methylmethacrylate). | ||||
| PCL | 400 nm to 2 μm | CT26 colon cancer cells and bone marrow-derived dendritic cells (BM-DC) | Lipopolysaccharide (LPS)-activated BM-DCs showed increased expression of CD86 and major histocompatibility complex class II | 76 |
| PEG | <600 nm | NIH 3T3 cells | Five times higher cell proliferation | 77 |
| PCL | 295–701 nm | Triple negative breast cancer (TNBC) cells | Higher mammosphere forming capacity and aldehyde dehydrogenase activity, higher cell proliferation and elongation | 78 |
| — | Ewing sarcoma cells | More resistant to traditional cytotoxic drugs, also exhibited remarkable differences in the expression pattern of the insulin-like growth factor-1 receptor/mammalian target of the rapamycin pathway | 79 | |
| Silk | 497 nm | HN12 cells | Increased paclitaxel drug concentration in order to achieve a cytotoxic effect | 80 |
| PVA and PEOT | 5–10 μm | Pancreatic ductal adenocarcinoma cells (PDAC) | Primary PDAC cells showed good viability and synthesized tumour-specific metalloproteinases (MMPs) such as MMP-2, and MMP-9 | 81 |
| PDMS and PMMA | 1.15–3.6 μm | Human lung cancer epithelial cells (A549) | Leads to the formation of cell spheroids from single cells | 82 |
On the other side, there is a growing awareness of the limitations of animal research and its inability to make reliable predictions for human clinical trials. Indeed, animal studies seem to overestimate by about 30% the likelihood that a treatment will be effective because negative results are often unpublished.83 For all these reasons there is growing interest in developing new 3D in vitro cancer models to be adopted as pre-screening models and to recapitulate the 3D microenvironment where cancer cells live.
3.1. Electrospun fibers as extracellular matrix (ECM)-like materials to mimic the cancer microenvironment
Cancer cells within a solid tumour are in close contact with the ECM, which is mainly composed of collagen, fibronectin, elastin, laminin and proteoglycan84,85 with additional components for specific tissues.86 In a pathological environment such as the cancer one, the ECM cannot be considered just as a passive environment surrounding cells, but as an active component with a critical role in regulating tumour cell properties and behaviour.87 Specifically, in breast cancer, the ECM stiffness affects cancer development and migration;73,88,89 in ovarian cancer patients, the expression of collagen VI correlates with the tumour grade and it has been shown to be upregulated in drug-resistant cancer cells, which can subsequently remodel the ECM to further promote chemoresistance;90 again, in small cell lung cancer (SCLC) the adhesion of cancer cells to the ECM enhances tumorigenicity and confers resistance to chemotherapeutic agents.91 Ideally, a 3D tumour model should be able to recapitulate this cell–ECM crosstalk. In order to study the ECM–cancer cell interactions in a 3D environment, biomimetic substrates have been produced.92 A number of strategies exist, and above all Matrigel, a commercial basement membrane preparation, is used.93–95 Besides that, a lot of 3D scaffolds have been used to culture cancer cells in vitro, allowing for their growth and proliferation in a context closer to reality than 2D traditional cultures. Electrospun fiber scaffolds provide an effective artificial 3D culture matrix due to their biomimicry of the architecture of the ECM96,97 and high versatility in biochemical stimuli, including growth factors, adhesion molecules, and drugs.60,98 In the following paragraphs, we will analyse the state of the art in vitro 3D tumour models with a focus on electrospun micro- and nanofibres, citing some of the most relevant works in the field. The paragraphs have been subdivided according to the cancer type.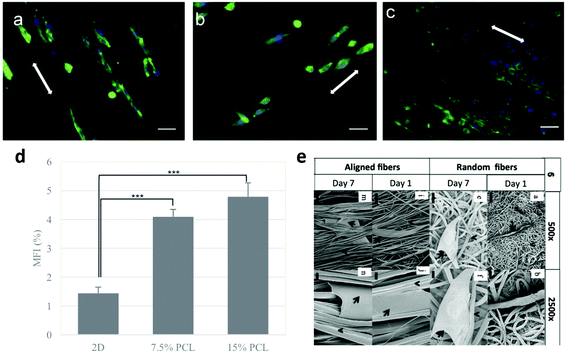 | ||
| Fig. 3 Response of different breast cancer cell lines on aligned PCL fibers: (a) 4T1, (b) MTCL, and (c) MDA-MB-231. Cells were cultured on PCL fibers for 3 days. Green represents the actin cytoskeleton and blue the nucleus of a cell, scale bar = 40 μm.98 (d) Mammosphere forming index (MFI) of MCF-7 cells after 2D and 3D culture on electrospun scaffolds. Statistically significant level was p < 0.001.102 (e) SEM images of MDA-MB-231 cells on fibrous scaffolds after day 1 and day 7 of culture. The arrows depict the cell body and the arrowheads depict the fibers.101 Reproduced from ref. 98 with permission from John Wiley and Sons, copyright 2017, and ref. 102 with permission from Elsevier, copyright 2011. | ||
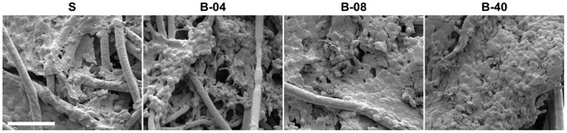 | ||
| Fig. 4 SEM micrographs of Ewing sarcoma cell culture under static and flow perfusion conditions (S, static; B-04, 0.04 mL min−1; B-08, 0.08 mL min−1; and B-40, 0.40 mL min−1). After 10 days of culture, scaffold surfaces for the different experimental groups were examined by SEM (scale bar, 50 μm).110 | ||
3.2. Electrospun fibers as membranes to study intra- and extravasation
All the 3D models presented in the previous section are promising in bridging the gap between in vitro and in vivo testing; however, they fail to mimic some specific and crucial steps of tumour evolution, including a cell motility and metastasis cascade. Considering the lethal impact of these mechanisms, there is a strong need to provide not only realistic in vitro models of primary tumours, but also reliable new models for intra- and extravasation, the two main phases of metastasis. The National Cancer Institute described extravasation as “the movement of cells out of a blood vessel into tissue during inflammation or metastasis (the spread of cancer)” and intravasation as “the movement of a cell or a foreign substance through the wall of a blood or lymph vessel into the vessel itself”. Cancer cells invade the stroma and metastasize to blood vessels as singlets or clusters. This stromal invasion is followed by extravasation into and intravasation from the blood vessels and subsequent invasion of neighbouring tissues. Invasive cells then re-establish distal colonies, causing metastasis.116Thanks to their fibril structures, electrospun membranes have been proposed to mimic the tumour–blood vessel interface, the physical zones where intra- and extravasation take place, or specific zones where metastasis can occur, such as the bone. Some examples of hybrid systems are already present in the literature. For instance, Cavo and colleagues combined an alginate-Matrigel based gel with embedded breast cancer cells with electrospun membranes to examine the cell invasion capability; the results showed that the cells were able to migrate through the gels and attach to the membrane mimicking the vascular walls hosted within a bioreactor.72 In a different work, Ali et al. successfully grew human bone-derived osteoblast cells onto electrospun PCL scaffolds to mimic the bone niche and integrated it into a bioengineered 3D in vitro model recapitulating prostate cancer metastasis to the bone.117 Again, Rabionet et al. developed and used PCL scaffolds for 3D breast cancer cell culture by using ES nanofibers.78
Despite the little supporting literature, combining 3D tumour models with cellular infiltration throughout nanofiber scaffolds seems to be crucial for realizing completely engineered cancer models and to better recapitulate the microenvironment of cancer cells.
4. Electrospun nanofibrous devices for cancer therapy
Cancer treatment represents one of the most crucial issues in clinical management. Uncontrolled growth, immortality and the ability to metastasize are significant characteristics of cancer cells.118 This paragraph aims to explore the existing applications of electrospun nanofibers in cancer therapy, mostly focusing on drug/gene delivery and cancer cell detection/sensing and highlighting the challenges and future directions for applications of electrospun nanofibers in cancer research.4.1. Drug delivery
Conventional surgery, radiotherapy and chemotherapy are currently the most adapted treatment modalities for cancer.119 However, these options are often limited and inadequate.120 Actually, the clinical use of the most engaged chemotherapeutics against several solid and hematopoietic cancers, including breast cancer, osteosarcomas, aggressive lymphomas, and leukaemia, is usually limited by the severe and harmful side effects at therapeutic concentrations.63 Moreover, in order to maximize the therapeutic effects, many patients need to take excess amounts of drugs, orally or via systematic injection, which might trigger side effects in healthy tissues.121In order to limit the common systemic side effects and toxicity, researchers are currently focusing their work particularly on postoperative chemotherapy applications for developing innovative and sophisticated treatments to specifically target cancer cells.
Targeted microcarrier technologies such as micro- and nanoparticles, liposomes and nanofibrous materials are designed for drug delivery in a controlled manner to a specific site. The release of drugs in a controlled way is a complex challenge in which systems provide an optimal amount of the drug to a specific target at a predetermined rate and for a definite time period (ranging from days to years).122
Controlled release systems offer advantages when compared to conventional drug therapies. Firstly, they maintain the drug in the desired therapeutic range in the blood by a single administration and avoid oscillating drug levels attributed to the standard trend of injected drugs in the blood (the blood level of the drug rises, reaches a peak plasma level and then declines) which cause alternating periods of ineffectiveness and toxicity. Other advantages comprise the localized drug delivery to a specific target of the body. In this manner, one can guarantee an improvement of the pharmacological properties of free drugs and an increment of the patient compliance with a reduced amount of the needed drug and a decreased frequency of drug administration.122–124 Moreover, in contrast to sustained release formulations, such as suspensions and emulsions, which are influenced by the environmental conditions and therefore are subjected to patient variations,122 controlled release systems enable the drug to be preserved in a polymeric material. The release of the drug through the polymeric system network is controlled by two main mechanisms: (1) drug diffusion, which is the most common release mechanism, whereby the drug moves in a concentration gradient from the inner part to the system's outer side up to the body; and (2) chemical mechanisms, including the degradation of the polymer or the cleavage of the drug from a polymer linkage to allow the drug release.122
Electrospun fibers represent an ideal platform for drug delivery since they provide a high surface area to volume ratio and may favour abundant drug release.125 Various delivery systems have been developed by loading antibiotics, anticancer drugs, proteins and also nucleic acids. The application of electrospun fibers for chemotherapy has become popular more recently, in particular as a local drug delivery system after a surgical operation for removing solid tumours.121
The active molecules can be (i) physically or chemically attached to the surface of the fibers or (ii) mixed with the polymers. In the following, the different loading methods will be presented.
The ES process with two or more constituents is one major approach to produce nanofibers encapsulating pharmaceuticals or other bioactive components with a significant drug loading ability. This configuration requires the application of the conventional ES device (a single nozzle with a single capillary) in which both drugs and polymers are mixed together in the same solvent and then electrospun into drug-loaded fibers.130–133
Ranganath et al. reported the successful production of electrospun poly-(D,L-lactide-co-glycolide) fibers to deliver paclitaxel for postsurgical chemotherapy against malignant brain tumours. The device showed sustained paclitaxel release over 80 days in vitro and demonstrated tumour growth inhibition in an animal study.134 Xu and co-workers developed an implantable biodegradable system for controlled release of a 1,3-bis(2-chloroethyl)-1-nitrosourea anticancer drug by ES a copolymer solution. The drug well dispersed through the polymer matrix and its release appeared directly proportional to the loaded drug amount.36 Liu et al. designed biodegradable drug carriers for in situ treatment of liver cancer based on electrospun PLLA nanofibers loaded with doxorubicin hydrochloride in order to treat locally an unresectable liver cancer or to prevent a post-surgery tumour recurrence avoiding the systemic chemotherapy. The amount of doxorubicin hydrochloride loaded was completely released by diffusion from the nanofibers.135
Following this approach, Zong et al. developed an electrospun cisplatin-loaded delivery system to treat cervix cancer in mice via vaginal implantation. It showed a good mucoadhesive property and high anti-tumour power thanks to the cisplatin release from the nanofibers.136
Ramachandran et al. developed a localized nanoimplant for controlled delivery of the anti-cancer drug, temozolomide, in orthotopic brain-tumour. Specifically, a library of drug loaded (20 wt%) electrospun nanofibers of PLGA–PLA–PCL blends was produced with distinct in vivo brain-release kinetics (hours to months). Orthotopic rat glioma implanted wafers showed constant drug release (116.6 μg day−1) with negligible leakage into peripheral blood (<100 ng) showing an ∼1000-fold differential drug dosage in the tumour versus peripheral blood. Most importantly, an implant with a one-month release profile resulted in long-term (>4 month) survival in 85.7% animals, whereas a 7 day releasing implant showed tumour recurrence in 54.6% animals, exhibiting a median survival of only 74 days.137
However, ES is not immune to a few drawbacks. Above all, this method of synthesis leads to an initial burst release of the drug.130,138 This is due to fast diffusional effects and probably it is predisposed by the low physical interactions between the drug and the polymer, so that, during the process, most of the drug molecules are likely to be localized around the fiber surface.130,139,140 As a result, most of the drug amount came in contact with the surrounding water solution and this caused a subsequent large burst release at short times.139 In addition, the burst release is often affected by the biodegradable feature of the chosen carrier.130–132,140 Thus, a special caution is required to tailor both the release and the degradation rate, for example by varying the polymer blend composition and the ratio of amorphous to crystalline segments in order to control whether drug release occurs via diffusion alone or by diffusion and scaffold degradation.141,142
In contrast to ES, researchers have developed an original approach to limit the initial burst and, in this sense, to extend the drug release over a long period of time.
A good approach to prolong the therapeutic release of the drug from electrospun fibers is to add an inorganic nanocarrier into the fibers, such as silica based nanoparticles143–145 and hydroxyapatite.146
For example, Zhou et al. implemented the conventional ES setup by firstly loading the anticancer doxorubicin drug into SiO2 nanoparticles and then mixing the resulting solution with the polymer solution.143 In the study of Zheng et al., amoxicillin, a model drug, was adsorbed to the hydroxyapatite surface particles before to disperse them into PLGA solution ready to be electrospun. The model showed a sustained release profile and a limited initial burst release.146 Moreover, novel drug carriers with a higher affinity for both drug molecules and polymer solution were then engaged. An example is the star polymer ad hoc synthesized in the work of Balakrishnana and co-workers which is optimal for further applications. The star polymer was blended with the more traditional PCL and the system was used for sustained release of doxorubicin.147
Another big issue in mixing the drug and polymer together is the exposition of drugs to harsh solvents, often necessary to dissolve the most used polymers. A range of biomolecules, including anti-cancer drugs, can be easily immobilized on the surface of electrospun fibers via a chemical or physical method for sustained drug delivery. This way of surface functionalization has at least two main advantages when compared to conventional ES. Firstly, physical and chemical drug immobilization after fiber synthesis can avoid the denaturation or inactivation of the drug generally caused by a high voltage or harsh organic solvents; secondly, the possibility to add additional materials after the ES process leads to avoiding of unexpected but usual trouble in terms of electrospinnability, for instance the difficulties in dissolving such biomolecules in an organic solvent because of their high molecular weight and the charges present on their surfaces.
Physical non-specific random adsorption is governed by electrostatic interactions, hydrophobic interactions, hydrogen bonding, and van der Waals interactions between drugs and the fiber surface.148 However, non-specific adsorption allows the formation of weak links and it may favour a significant burst release. This is the reason why it is rarely used for anticancer therapy applications.
A more applied strategy is immobilization after chemical treatment of the fiber surface, by which it is possible to tailor fiber adhesion properties by adding appropriate functional groups such as amine, carboxyl, hydroxyl or thiol groups suitable for further immobilization.149 Chemical conjugation methods are better for finely controlling the amount of the incorporated drug on the nanofiber mesh than physically trapping the drug on the surface, and they exhibit slow drug release kinetics with reduced initial burst release.150 For instance, Volpato and co-workers reported a chemical immobilization of heparin-containing polyelectrolyte complex nanoparticles and a basic fibroblast growth factor onto chitosan electrospun fibers. They prevented the burst release effect by post-fabrication modification of the fiber surface by adding a single bilayer of a polyelectrolyte multilayer composed of N,N,N-trimethyl chitosan and heparin.151 Im et al. limited the initial burst by modifying cross-linked hydrogel fibers through fluorination before the addition of the model drug.152 A versatile surface modification method is the approach performed by Ma et al. that worked on a porous nanofiber device made of a chitosan/polyethylene oxide mixture. After fabrication, the nanofibers were soaked in water to remove polyethylene oxide and then immersed in a solution with the anticancer drug to load it. Finally, the porous drug-loaded device was dipped in hyaluronic acid solution with the aim to control drug release thanks to the chitosan matrix and hyaluronic acid affinity.153 The best promising strategy to limit the initial burst release is the use of core–shell fibers with coaxial needles.154–156 The Co-axial ES technique has high potential in biomedical applications for core/sheath nanofibers since the polymer shell generally protects the core compounds from direct exposure to the external environment and, consequently, contributes to the prolonged drug release.157 Coaxial ES is very useful mainly for the encapsulation of bioactive compounds which are susceptible to harsh organic solvents (drugs, proteins, cells and nucleic acids).155,156 For instance, Zhang and collaborators demonstrated the successful loading of fluorescein isothiocyanate-conjugated bovine serum albumin, a model protein, in a water-soluble core of poly(ethylene glycol) within a PCL shell. These core–shell fibers showed a reduction of the initial burst release compared to fibers electrospun from the mixture of the poly(ethylene glycol) and PCL obtained with a conventional setup.156 Mickova et al. reported the development of a nanofiber–liposome system for drug delivery. They tested the incorporation of liposomes containing the horseradish peroxidase drug with co-electrospinning and with coaxial electrospinning and demonstrated that the traditional setup does not preserve the integrity of the liposome which, in contrast, is well preserved. In this second case, the therapeutic activity of the encapsulated drug was demonstrated.158
Another approach to slow down the kinetics of release is the development of multilayer stacked nanofibers by the subsequent ES of two or more different solutions. Generally, these systems combine an initial fast release with a sustained release thanks to layers with different diffusion pathways and degradation behaviours. The versatility of layer by layer fiber deposition leads to a range of modifications and implementation to make it suitable also for multidrug delivery systems. Combining multiple treatments in different times plays a key role in chemotherapy for cancer treatment. Okuda and co-workers created tetra-layered drug-loaded nanofiber meshes for dual release. The construction was performed with a first drug-loaded layer, a second barrier mesh, a third drug-loaded layer and a final barrier mesh. An in vitro release assay demonstrated that the tetra-layered device can provide timed dual release of the respective drugs by tailoring the morphological features of each component mesh like the barrier mesh thickness.148–150,159
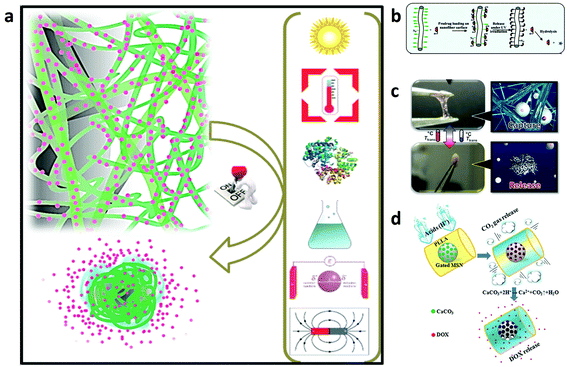 | ||
| Fig. 5 (a) Illustration of the mechanisms of on-demand drug release under biological, chemical, light, temperature, magnetic, and electric field stimuli.163 (b) Schematic of the photocontrolled drug delivery based on the host–guest linkage on the photoresponsive nanofiber surface.161 (c) A nanofiber web that captures and releases cells by switching to a hydrogel-like structure in response to temperature changes.164 (d) A drug-loaded intelligent electrospun fiber mat which reacts only in pathological acidic environments by producing CO2 gas with corresponding water penetration into the core of the fibers and rapid drug release.165 Reproduced from ref. 161 with permission from American Chemical Society, copyright 2009; ref. 163 with permission from John Wiley and Sons, copyright 2014; ref. 164 with permission from John Wiley and Sons, copyright 2012; and ref. 165 with permission from John Wiley and Sons, copyright 2015. | ||
In these systems, the drug is localized at a specific targeted area by internal or external forces and then activated. Recently, magnetic particles carrying drug molecules have been developed to target the drugs to specific sites in the body using external magnetic fields. Shortly after concentration in the targeted region, the drug molecules were gradually released, thus improving their therapeutic efficiency, while lowering the collateral toxic side effects on healthy cells or tissues.
For instance, Kim and co-workers reported temperature-responsive electrospun nanofibers for ‘on–off’ switchable release of dextran as a drug model.166 Afterwards, Kim et al. designed smart nanofibers with bi-functional action by ES a chemically crosslinkable temperature-responsive polymer with magnetic nanoparticles and the anticancer drug (doxorubicin) for induction of skin cancer apoptosis. The nanofibers showed simultaneous heat generation and drug release in response to ‘on–off’ switching during an alternating magnetic field. The alternating magnetic field triggers self-generated heat from the incorporated nanoparticles and induces the deswelling of polymer networks in the nanofiber with consequent drug release. The double effects of the drug and heat resulted in successful treatment of human melanoma cells with about 70% of cancer cell mortality and this demonstrated their high potential as a manipulative hyperthermia material and a switchable drug release platform.160
Since cancer cells are known to secrete acids which reduce the pH to below 6.8, Zhao et al. reported in their work the production of a smart drug release system upon stimulation via pH changes. The system was realized by developing polymer fibers embedding drug loaded-mesoporous silica nanoparticles functionalized with CaCO3 as an inorganic cap to control the opening of the pore entrances of the mesoporous silica nanoparticles inside the fibers. CaCO3 starts to dissolve in Ca2+ and CO2 gas just after a reduction of the physiological pH in response to an acidic environment. During the release assay, the system demonstrated an improvement of the drug release amount as the pH decreased. Moreover, the in vitro tests with HeLa confirmed its cytotoxic ability due to the release of the drug in an acidic environment.165
4.2. Electrospun fibers for gene delivery
RNA interference (RNAi) is a promising and powerful approach for cancer treatment. It is a mechanism by which double stranded RNAs mediate sequence-specific gene silencing, providing a new tool in the fight against cancer. On the other hand, uncontrolled transgene expression in non-target organs/sites/cells is problematic due to the high biological activities of transgene products. Furthermore, undesirable biodistribution of vectors leads to their loss and vector-dependent side effects. Thus, gene delivery systems that are targeted to specific organs/sites/cells are important for not only efficacy but also safety. Since the initial study of Fang and Reneker, demonstrating the possibility of ES pure DNA nanofibers, the use of this technique for a variety of applications has truly increased to a significant extent.167Electrospun nanofiber mediated siRNA delivery may serve as a potential alternative since the nanofiber can provide sustained long-term delivery at the tumour site (Fig. 6).168
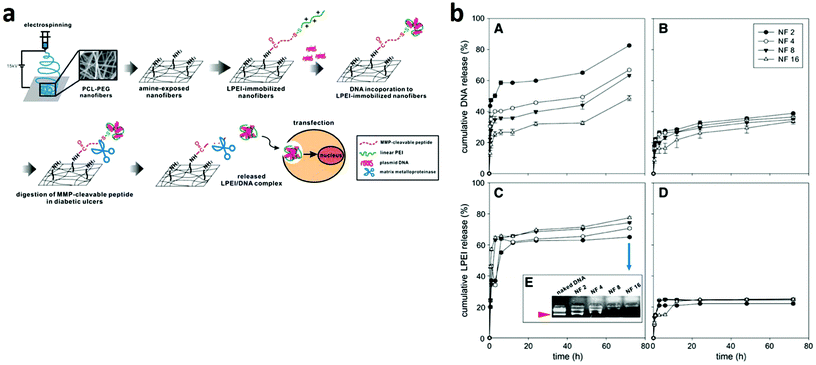 | ||
| Fig. 6 (a) Schematic diagram of an MMP-responsive electrospun nanofibrous matrix for gene delivery. (b) Release profiles of DNA (A and B) and LPEI (C and D) from the nanofibrous matrix in the presence of MMP-2.168 Reproduced from ref. 168 with permission from Elsevier, copyright 2010. | ||
In this regard, Achille and co-workers demonstrated the feasibility of loading into biodegradable fibers a bioactive plasmid encoding for short hairpin (sh) RNA against the cell cycle specific protein, Cdk2. The system led to the release of plasmid DNA during the fiber degradation, for over 21 days which suppressed the proliferation of MCF-7 breast cancer cells and affected their viability.169 Lei and co-workers incorporated a RNAi plasmid designed to specifically suppress MMP-2, an essential proteinase regulating brain tumour invasion and angiogenesis in tumour cells, and a cytotoxic drug paclitaxel into PLGA based fibers to achieve a sustained release of both agents. It was found that DNA nanoparticles complexed with the gene carrier polyethyleneimine and embedded in microfibers were able to inhibit MMP-2 mRNA and protein expression and, particularly, through an in vivo test on an intracranial xenograft tumour model in BALB/c nude mice it was proved that the gene/drug dual delivery microfibers were able to impose significant tumour regression compared with single drug delivery microfibers and commercial drug treatment.170 We have listed some of the ES nanofiber materials used for gene delivery in cancer cell lines (Table 3).
| Material | Gene particle | Cell line | Ref. |
|---|---|---|---|
| Abbreviations: PCL – poly(ε-caprolactone), PEG – poly(ethylene glycol), PLLA – poly(L-lactic acid), and PEI – poly(ethylenimine). | |||
| PCL | pCMVb-GFP | 3T3 (murine fibroblast cells) | 171 |
| miRNA-145 | HuH-7 (hepatocyte derived cellular carcinoma cells) | 172 | |
| PCL–PEG | pEGFP-N1 | NIH3T3 cells | 168 |
| PLLA | pGL3 | COS-7 cells | 14 |
| PEI–PEG | pMSCV | HEK293 cells | 173 |
4.3. Electrospun fibers for immunosensors/cancer screening
Evidence suggests that the detection and treatment of early stage cancer are essential in improving survival. However, early detection is difficult as it is usually asymptomatic until the disease has spread.174 Results of previous studies highlighted that circulating antibodies to cancer associated antigens will likely be present in the early stages of disease and their detection might be the best way to identify early stage malignancies.175Recent advances in nanoscience and nanotechnology have opened up new horizons for the development of biosensors of enhanced sensitivity, specificity, detection time, and low cost. Sensor miniaturization provides great versatility for incorporation into multiplexed, portable, wearable, and even implantable medical devices. The attractiveness of such nanomaterials relies not only on their ability to act as efficient and stabilizing platforms for the biosensing elements, but also on their small size, large surface area, high reactivity, controlled morphology and structure, biocompatibility, and in some cases electrocatalytic properties. Structuring the transducer at the nanoscale contributes towards enlarging the overall surface available for bioreceptor immobilization. Incorporation of nanomaterials into the sensing layers is also often associated with higher mass transfer rates, acceleration and magnification of the transduction process, contributing to signal amplification and a faster biosensor response. In particular, fibers produced by ES are very interesting in this area since they can be chemically and physically decorated with bioactive molecules such as cell-recognizable ligands and provide a large surface area.176 Sensing elements are principally enzymes, antibodies, and more scarcely DNA strands or aptamers and the mechanism of action is the electrochemical transduction.
Ali et al. reported the fabrication of an efficient, label-free, selective and highly reproducible immunosensor with unprecedented sensitivity (femto-molar) to detect a breast cancer biomarker for early diagnostics. Mesoporous zinc oxide nanofibers are synthesized by the ES technique and their fragments are electrophoretically deposited on an indium tin oxide glass substrate and conjugated via covalent or electrostatic interactions with a biomarker, the epidermal growth factor receptor 2. Label-free detection of the breast cancer biomarker using this point-of-care device is achieved by an electrochemical impedance technique that has high sensitivity (7.76 kΩ μM−1) and can detect a 1 fM (4.34 × 10−5 ng mL−1) concentration. The excellent impedimetric response of this immunosensor enables fast detection (128 s) in a wide detection test range (1.0 fM–0.5 μM).177
Paul et al. reported a novel biosensor platform based on multiwalled carbon nanotube embedded zinc oxide nanowires for the ultrasensitive detection of carcinoma antigen-125. The system was synthesized by simple ES. The label-free detection was performed by the differential voltammetry technique and it demonstrated excellent sensitivity (90.14 μA (U mL−1)−1 cm−2) with a detection limit of 0.00113 U mL−1 concentration.178 Soares et al. reported the fabrication of immunosensors based on nanostructured mats of electrospun nanofibers of polyamide 6 and poly(allylamine hydrochloride) coated with either multiwalled carbon nanotubes or gold nanoparticles, whose three-dimensional structure was suitable for the immobilization of anti-CA19-9 antibodies to detect the pancreatic cancer biomarker CA19-9. By using impedance spectroscopy, the sensing platform was able to detect CA19-9 with a sensitivity and detection limit of 1.84 and 1.57 U mL−1 for the nanostructured architectures containing multiwalled carbon nanotubes and gold nanoparticles, respectively. The high sensitivity and selectivity of the immunosensors were also explored in tests with blood serum from patients with distinct concentrations of CA19-9, for which the impedance spectral data were processed with a multidimensional projection technique. The robustness of the immunosensors in dealing with patient samples without suffering interference from analytes present in biological fluids is promising for a simple, effective diagnosis of pancreatic cancer at early stages.179 More recently, a novel class of pH-sensing ES fiber scaffolds has been reported by embedding ratiometric pH sensing capsules directly within the lumen of electrospun organic fibers.180 Upon proton-induced switching, the hybrid ratiometric organic fibers undergo optical changes that could be recorded with fluorescence detectors and correlated with the local proton concentration with micrometer-scale spatial resolution. Biocompatible ES fiber mats with pH sensing properties could be used for in vivo spatio-temporal measurements of the extracellular acidity of live cells, finding potential applications in drug screening and drug discovery for evaluating the efficacy of glycolysis inhibiting anti-cancer drugs in real time (Fig. 7).181,182
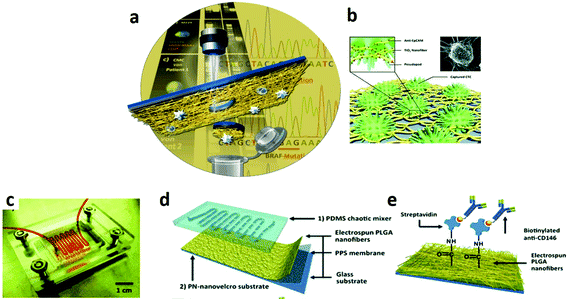 | ||
| Fig. 7 (a) A method to detect and isolate single circulating melanoma cells from normal white blood cells with a laser microdissection technique.181 (b) A nanofiber platform TiO2-based and biological cell-capture agent to capture circulating tumour cells.182 (c) Picture of a nanostructured chip for melanoma cell isolation composed of (d) an overlaid PDMS chaotic mixer and a transparent nanovelcro substrate; (e) NHS chemistry is used to covalently anchor streptavidin for conjugation of biotinylated anti-CD146, a melanoma-specific antibody.181 Reproduced from ref. 181 with permission from John Wiley and Sons, copyright 2013 and ref. 182 with permission from John Wiley and Sons, copyright 2012. | ||
5. Conclusions
The high versatility, cost effectiveness and scalability of ES have been inspiring the design of novel strategies in cancer research. The use of electrospun scaffolds, such as polymer nanofibers that recapitulate the fibrous architecture of the native ECM environment, could help in the manufacture of xeno-free and highly controlled 3D in vitro tumour models for cell-based compound screening, mechanistic studies and pre-clinical drug discovery. Various approaches propose multifunctional and stimuli-responsive electrospun patches for topical release of therapeutics in a controlled and sustained way with improved therapeutic efficacy, reduced side effects and prolonged life expectancy of patients. Electrospun nanostructured mats also represent attractive sensor platforms to be used for fast diagnosis of cancers at early stages. Although numerous improvements have been achieved by employing electrospun scaffolds in cancer diagnosis and therapy, in depth in vivo studies are indispensable before use in clinical trials and medical device products.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the ERC Starting Grant INTERCELLMED (project number 759959), the My First AIRC Grant (MFAG-2019, project number 22902), the FISR-C.N.R., Tecnopolo di Nanotecnologia e Fotonica per la medicina di precisione (project number: B83B17000010001) and Tecnopolo per la medicina di precisione – Regione Puglia (project number: B84I18000540002).References
- Y. M. Shin, M. M. Hohman, M. P. Brenner and G. C. Rutledge, Polymer, 2001, 42, 09955–09967 CrossRef CAS.
- J. M. Deitzel, J. D. Kleinmeyer, J. K. Hirvonen and N. C. Beck Tan, Polymer, 2001, 42, 8163–8170 CrossRef CAS.
- K. Y. Lee, L. Jeong, Y. O. Kang, S. J. Lee and W. H. Park, Adv. Drug Delivery Rev., 2009, 61, 1020–1032 CrossRef CAS PubMed.
- H. Homayoni, S. A. H. Ravandi and M. Valizadeh, Carbohydr. Polym., 2009, 77, 656–661 CrossRef CAS.
- D. H. Reneker and A. L. Yarin, Polymer, 2008, 49, 2387–2425 CrossRef CAS.
- D. Deshawar and P. Chokshi, Polymer, 2017, 131, 34–49 CrossRef CAS.
- T. J. Sill and H. A. von Recum, Biomaterials, 2008, 20, 1989–2006 CrossRef PubMed.
- E. D. Boland, J. A. Matthews, K. J. Pawlowski, D. G. Simpson, G. E. Wnek and G. L. Bowlin, Front. Biosci., 2004, 9, 1422–1432 CrossRef CAS PubMed.
- J. A. Matthews, G. E. Wnek, D. G. Simpson and G. L. Bowlin, Biomacromolecules, 2002, 3, 32–238 CrossRef PubMed.
- S. Eap, A. Ferrand, C. Mendoza-Palomares, A. Hébraud, J. F. Stoltz, D. Mainard, G. Schlatter and N. Benkirane-Jessel, in Bio-Medical Materials and Engineering, 2012, pp. 137–141 Search PubMed.
- N. J. Schaub, C. D. Johnson, B. Cooper and R. J. Gilbert, J. Neurotrauma, 2016, 33, 1405–1415 CrossRef PubMed.
- F. Ajalloueian, H. Tavanai, J. Hilborn, O. Donzel-Gargand, K. Leifer, A. Wickham and A. Arpanaei, BioMed Res. Int., 2014, 475280 Search PubMed.
- A. Nandakumar, H. Fernandes, J. De Boer, L. Moroni, P. Habibovic and C. A. Van Blitterswijk, Macromol. Biosci., 2010, 10, 1365–1373 CrossRef CAS PubMed.
- W. Chang, X. Mu, X. Zhu, G. Ma, C. Li, F. Xu and J. Nie, Mater. Sci. Eng., C, 2013, 33, 4369–4376 CrossRef CAS PubMed.
- J. Erben, K. Pilarova, F. Sanetrnik, J. Chvojka, V. Jencova, L. Blazkova, J. Havlicek, O. Novak, P. Mikes, E. Prosecka, D. Lukas and E. Kuzelova Kostakova, Mater. Lett., 2015, 143, 172–176 CrossRef CAS.
- F. Sharifi, S. M. Atyabi, D. Norouzian, M. Zandi, S. Irani and H. Bakhshi, Int. J. Biol. Macromol., 2018, 115, 243–248 CrossRef CAS PubMed.
- A. Wubneh, E. K. Tsekoura, C. Ayranci and H. Uludağ, Acta Biomater., 2018, 80, 1–30 CrossRef CAS PubMed.
- B. M. Baker, A. O. Gee, R. B. Metter, A. S. Nathan, R. A. Marklein, J. A. Burdick and R. L. Mauck, Biomaterials, 2008, 29, 2348–2358 CrossRef CAS PubMed.
- M. T. Wolf, C. L. Dearth, S. B. Sonnenberg, E. G. Loboa and S. F. Badylak, Adv. Drug Delivery Rev., 2015, 84, 208–221 CrossRef CAS PubMed.
- O. Suwantong, S. Waleetorncheepsawat, N. Sanchavanakit, P. Pavasant, P. Cheepsunthorn, T. Bunaprasert and P. Supaphol, Int. J. Biol. Macromol., 2007, 40, 217–223 CrossRef CAS PubMed.
- H. Cao, T. Liu and S. Y. Chew, Adv. Drug Delivery Rev., 2009, 61, 1055–1064 CrossRef CAS PubMed.
- Y. M. Ju, J. S. Choi, A. Atala, J. J. Yoo and S. J. Lee, Biomaterials, 2010, 31, 4313–4321 CrossRef CAS PubMed.
- H. Ahn, Y. M. Ju, H. Takahashi, D. F. Williams, J. J. Yoo, S. J. Lee, T. Okano and A. Atala, Acta Biomater., 2015, 16, 14–22 CrossRef CAS PubMed.
- P. Uttayarat, A. Perets, M. Li, P. Pimton, S. J. Stachelek, I. Alferiev, R. J. Composto, R. J. Levy and P. I. Lelkes, Acta Biomater., 2010, 6, 4229–4237 CrossRef CAS PubMed.
- M. B. Browning, D. Dempsey, V. Guiza, S. Becerra, J. Rivera, B. Russell, M. Höök, F. Clubb, M. Miller, T. Fossum, J. F. Dong, A. L. Bergeron, M. Hahn and E. Cosgriff-Hernandez, Acta Biomater., 2012, 8, 1010–1021 CrossRef CAS PubMed.
- S. Rayatpisheh, D. E. Heath, A. Shakouri, P. O. Rujitanaroj, S. Y. Chew and M. B. Chan-Park, Biomaterials, 2014, 35, 2713–2719 CrossRef CAS PubMed.
- L. Poláková, J. Širc, R. Hobzová, A.-I. Cocârţă and E. Heřmánková, Int. J. Pharm., 2019, 558, 268–283 CrossRef PubMed.
- K. M. Kennedy, A. Bhaw-Luximon and D. Jhurry, Acta Biomater., 2017, 50, 41–55 CrossRef CAS PubMed.
- G. Jensen, C. Morrill and Y. Huang, Acta Pharm. Sin. B, 2018, 8, 756–766 CrossRef PubMed.
- E. Burdett, F. K. Kasper, A. G. Mikos and J. A. Ludwig, Tissue Eng., Part B, 2010, 63, 351–359 CrossRef PubMed.
- L. G. Griffith and G. Naughton, Science, 2002, 295, 1009–1014 CrossRef CAS PubMed.
- O. Hartman, C. Zhang, E. L. Adams, M. C. Farach-Carson, N. J. Petrelli, B. D. Chase and J. F. Rabolt, Biomacromolecules, 2009, 10, 019–2032 CrossRef PubMed.
- V. Agrahari, V. Agrahari, J. Meng and A. K. Mitra, in Emerging Nanotechnologies for Diagnostics, Drug Delivery and Medical Devices, 2017, pp. 189–215 Search PubMed.
- J. Wei, J. Hu, M. Li, Y. Chen and Y. Chen, RSC Adv., 2014, 4, 28011–28019 RSC.
- J. Zhang, X. Wang, T. Liu, S. Liu and X. Jing, Drug Delivery, 2016, 23, 784–790 CrossRef CAS PubMed.
- X. Xu, X. Chen, X. Xu, T. Lu, X. Wang, L. Yang and X. Jing, J. Controlled Release, 2006, 114, 307–316 CrossRef CAS PubMed.
- S. Iqbal, M. H. Rashid, A. S. Arbab and M. Khan, J. Biomed. Nanotechnol., 2017, 13, 355–366 CrossRef CAS PubMed.
- P. Slemming-Adamsen, J. Song, M. Dong, F. Besenbacher and M. Chen, Macromol. Mater. Eng., 2015, 300, 1226–1231 CrossRef CAS.
- E. Yan, Y. Fan, Z. Sun, J. Gao, X. Hao, S. Pei, C. Wang, L. Sun and D. Zhang, Mater. Sci. Eng., C, 2014, 41, 217–223 CrossRef CAS PubMed.
- J. Doshi and D. H. Reneker, J. Electrostat., 1995, 35, 151–160 CrossRef CAS.
- H. Zong, X. Xia, Y. Liang, S. Dai, A. Alsaedi, T. Hayat, F. Kong and J. H. Pan, Mater. Sci. Eng., C, 2018, 92, 1075–1091 CrossRef CAS PubMed.
- Z.-M. Huang, Y.-Z. Zhang, M. Kotaki and S. Ramakrishna, Compos. Sci. Technol., 2003, 63, 2223–2253 CrossRef CAS.
- L. J. Chen, J. Der Liao, S. J. Lin, Y. J. Chuang and Y. S. Fu, Polymer, 2009, 50, 3516–3521 CrossRef CAS.
- C. Su, C. Lu, H. Cao, F. Gao, J. Chang, Y. Li and C. He, Mater. Lett., 2017, 204, 8–11 CrossRef CAS.
- H. Y. Jian, S. V. Fridrikh and G. C. Rutledge, Polymer, 2006, 47, 4789–4797 CrossRef.
- N. Okutan, P. Terzi and F. Altay, Food Hydrocolloids, 2014, 39, 19–26 CrossRef CAS.
- X.-F. Wu, Y. Salkovskiy and Y. A. Dzenis, Appl. Phys. Lett., 2011, 98, 223108 CrossRef.
- O. S. Yördem, M. Papila and Y. Z. Menceloğlu, Mater. Des., 2008, 29, 34–44 CrossRef.
- J. Zeng, H. Haoqing, A. Schaper, J. H. Wendorff and A. Greiner, e-Polymers, 2003, 3, 102–110 Search PubMed.
- Y. Wu, L. A. Carnell and R. L. Clark, Polymer, 2007, 48, 5653–5661 CrossRef CAS.
- D. Li, G. Ouyang, J. T. McCann and Y. Xia, Nano Lett., 2005, 5, 913–916 CrossRef CAS PubMed.
- D. Rodoplu and M. Mutlu, J. Eng. Fibers Fabr., 2012, 7, 118–123 CAS.
- R. Sahay, V. Thavasi and S. Ramakrishna, J. Nanomater., 2011, 1–17 Search PubMed.
- H. Pan, L. Li, L. Hu and X. Cui, Polymer, 2006, 47, 4901–4904 CrossRef CAS.
- L. Wannatong, A. Sirivat and P. Supaphol, Polym. Int., 2004, 53, 1851–1859 CrossRef CAS.
- I. Hayati, A. I. Bailey and T. F. Tadros, J. Colloid Interface Sci., 1987, 117, 205–221 CrossRef CAS.
- H. Shin, S. Jo and A. G. Mikos, Biomaterials, 2003, 24, 4353–4364 CrossRef CAS PubMed.
- F. Croisier and C. Jérôme, Eur. Polym. J., 2013, 49, 780–792 CrossRef CAS.
- E. K. Brenner, J. D. Schiffman, E. A. Thompson, L. J. Toth and C. L. Schauer, Carbohydr. Polym., 2012, 87, 926–929 CrossRef CAS.
- O. Hartman, C. Zhang, E. L. Adams, M. C. Farach-Carson, N. J. Petrelli, B. D. Chase and J. F. Rabolt, Biomaterials, 2010, 31, 5700–5718 CrossRef CAS PubMed.
- C. J. Lovitt, T. B. Shelper and V. M. Avery, Biology, 2014, 3, 345–367 CrossRef PubMed.
- B. K. Jong, Semin. Cancer Biol., 2005, 15, 365–377 CrossRef PubMed.
- H. Liu, J. Lin and K. Roy, Biomaterials, 2006, 27, 5978–5989 CrossRef CAS PubMed.
- M. Turetta, F. Del Ben, G. Brisotto, E. Biscontin, M. Bulfoni, D. Cesselli, A. Colombatti, G. Scoles, G. Gigli and L. L. del Mercato, Curr. Med. Chem., 2018, 25, 4616–4637 CrossRef CAS PubMed.
- A. Polini, L. L. del Mercato, A. Barra, Y. S. Zhang, F. Calabi and G. Gigli, Drug Discovery Today, 2019, 24, 517–525 CrossRef CAS PubMed.
- M. R. Carvalho, D. Barata, L. M. Teixeira, S. Giselbrecht, R. L. Reis, J. M. Oliveira, R. Truckenmüller and P. Habibovic, Sci. Adv., 2019, 5, eaaw1317 CrossRef CAS PubMed.
- D. Barata, C. Van Blitterswijk and P. Habibovic, Acta Biomater., 2016, 34, 1–20 CrossRef CAS PubMed.
- T. P. Kraehenbuehl, R. Langer and L. S. Ferreira, Nat. Methods, 2011, 8, 731–736 CrossRef CAS PubMed.
- D. Loessner, K. S. Stok, M. P. Lutolf, D. W. Hutmacher, J. A. Clements and S. C. Rizzi, Biomaterials, 2010, 31, 8494–8506 CrossRef CAS PubMed.
- G. Rossi, A. Manfrin and M. P. Lutolf, Nat. Rev. Genet., 2018, 19, 671–687 CrossRef CAS PubMed.
- B. Alberts, A. Johnson, J. Lewis, M. Raff, K. Roberts and P. Walter, in Molecular Biology of the Cell, 2002, pp. 1131–1204 Search PubMed.
- M. Cavo, M. Caria, I. Pulsoni, F. Beltrame, M. Fato and S. Scaglione, Sci. Rep., 2018, 8, 5333 CrossRef PubMed.
- M. Cavo, M. Fato, L. Peñuela, F. Beltrame, R. Raiteri and S. Scaglione, Sci. Rep., 2016, 6, 35367 CrossRef CAS PubMed.
- V. Kumar and S. Varghese, Adv. Healthcare Mater., 2019, 8, e1801198 Search PubMed.
- K. Chitcholtan, E. Asselin, S. Parent, P. H. Sykes and J. J. Evans, Exp. Cell Res., 2013, 319, 75–87 CrossRef CAS PubMed.
- T. E. Kim, C. G. Kim, J. S. Kim, S. Jin, S. Yoon, H. R. Bae, J.-H. Kim, Y. H. Jeong, J. Y. Kwak and K. J. Kim, Int. J. Nanomed., 2016, 11, 823–835 CAS.
- S. Cai, H. Xu, Q. Jiang and Y. Yang, Langmuir, 2013, 29, 2311–2318 CrossRef CAS PubMed.
- M. Rabionet, M. Yeste, T. Puig and J. Ciurana, Polymers, 2017, 9, E328 CrossRef PubMed.
- E. L. S. Fong, S. E. Lamhamedi-Cherradi, E. Burdett, V. Ramamoorthy, A. J. Lazar, F. K. Kasper, M. C. Farach-Carson, D. Vishwamitra, E. G. Demicco, B. A. Menegaz, H. M. Amin, A. G. Mikos and J. A. Ludwig, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 6500–6505 CrossRef CAS PubMed.
- A. A. Bulysheva, G. L. Bowlin, S. P. Petrova and W. A. Yeudall, Biomed. Mater., 2013, 8, 55009 CrossRef PubMed.
- C. Ricci, C. Mota, S. Moscato, D. D'Alessandro, S. Ugel, S. Sartoris, V. Bronte, U. Boggi, D. Campani, N. Funel, L. Moroni and S. Danti, Biomatter, 2014, 4, e955386 CrossRef PubMed.
- H. Moghadas, M. S. Saidi, N. Kashaninejad, A. Kiyoumarsioskouei and N.-T. Nguyen, Biomed. Microdevices, 2017, 19, 74 CrossRef PubMed.
- I. W. Y. Mak, N. Evaniew and M. Ghert, Am. J. Transl. Res., 2014, 6, 114–118 Search PubMed.
- G. Rijal and W. Li, Biomaterials, 2016, 81, 135–156 CrossRef CAS PubMed.
- C. Frantz, K. M. Stewart and V. M. Weaver, J. Cell Sci., 2010, 123, 4195–4200 CrossRef CAS PubMed.
- J. A. DeQuach, V. Mezzano, A. Miglani, S. Lange, G. M. Keller, F. Sheikh and K. L. Christman, PLoS One, 2010, 5, e13039 CrossRef PubMed.
- C. Rianna, P. Kumar and M. Radmacher, Semin. Cell Dev. Biol., 2018, 73, 107–114 CrossRef CAS PubMed.
- N. F. Boyd, Q. Li, O. Melnichouk, E. Huszti, L. J. Martin, A. Gunasekara, G. Mawdsley, M. J. Yaffe and S. Minkin, PLoS One, 2014, 9, e100937 CrossRef PubMed.
- K. R. Levental, H. Yu, L. Kass, J. N. Lakins, M. Egeblad, J. T. Erler, S. F. T. Fong, K. Csiszar, A. Giaccia, W. Weninger, M. Yamauchi, D. L. Gasser and V. M. Weaver, Cell, 2009, 139, 891–906 CrossRef CAS PubMed.
- C. A. Sherman-Baust, A. T. Weeraratna, L. B. A. Rangel, E. S. Pizer, K. R. Cho, D. R. Schwartz, T. Shock and P. J. Morin, Cancer Cell, 2003, 3, 377–386 CrossRef CAS PubMed.
- T. Sethi, R. C. Rintoul, S. M. Moore, A. C. MacKinnon, D. Salter, C. Choo, E. R. Chilvers, I. Dransfield, S. C. Donnelly, R. Strieter and C. Haslett, Nat. Med., 1999, 5, 662–668 CrossRef CAS PubMed.
- A. Aung, V. Kumar, J. Theprungsirikul, S. K. Davey and S. Varghese, Cancer Res., 2020, 80, 263–275 CrossRef CAS PubMed.
- H. K. Kleinman and G. R. Martin, Semin. Cancer Biol., 2005, 15, 378–386 CrossRef CAS PubMed.
- G. Benton, H. K. Kleinman, J. George and I. Arnaoutova, Int. J. Cancer, 2011, 128, 1751–1757 CrossRef CAS PubMed.
- V. Härmä, J. Virtanen, R. Mäkelä, A. Happonen, J. P. Mpindi, M. Knuuttila, P. Kohonen, J. Lötjönen, O. Kallioniemi and M. Nees, PLoS One, 2010, 5, e10431 CrossRef PubMed.
- S. Chen, B. Liu, M. A. Carlson, A. F. Gombart, D. A. Reilly and J. Xie, Nanomedicine, 2017, 12, 1335–1352 CrossRef CAS PubMed.
- J. Coburn, M. Gibson, P. A. Bandalini, C. Laird, H. Q. Mao, L. Moroni, D. Seliktar and J. Elisseeff, Smart Struct. Syst., 2011, 7, 213–222 CrossRef PubMed.
- S. Chen, S. K. Boda, S. K. Batra, X. Li and J. Xie, Adv. Healthcare Mater., 2018, 7, 1701024 CrossRef PubMed.
- R. L. Siegel, K. D. Miller and A. Jemal, Ca-Cancer J. Clin., 2019, 69, 7–34 CrossRef PubMed.
- S. Saha, X. Duan, L. Wu, P. K. Lo, H. Chen and Q. Wang, Langmuir, 2012, 28, 2028–2034 CrossRef CAS PubMed.
- Y. K. Girard, C. Wang, S. Ravi, M. C. Howell, J. Mallela, M. Alibrahim, R. Green, G. Hellermann, S. S. Mohapatra and S. S. Mohapatra, PLoS One, 2013, 8, e75345 CrossRef CAS PubMed.
- J. Du, P. L. Che, Z. Y. Wang, U. Aich and K. J. Yarema, Biomaterials, 2011, 32, 5427–5437 CrossRef CAS PubMed.
- K. Guiro, S. A. Patel, S. J. Greco, P. Rameshwar and T. L. Arinzeh, PLoS One, 2015, 10, e0118724 CrossRef PubMed.
- R. Siegel, D. Naishadham and A. Jemal, Ca-Cancer J. Clin., 2012, 62, 10–29 CrossRef PubMed.
- C. Feig, A. Gopinathan, A. Neesse, D. S. Chan, N. Cook and D. A. Tuveson, Clin. Cancer Res., 2012, 18, 4266–4276 CrossRef CAS PubMed.
- Q. He, X. Wang, X. Zhang, H. Han, B. Han, J. Xu, K. Tang, Z. Fu and H. Yin, Int. J. Nanomed., 2013, 8, 1167–1176 Search PubMed.
- X. Wang, X. Zhang, Z. Fu and H. Yin, J. Biotechnol., 2013, 166, 166–173 CrossRef CAS PubMed.
- Y. Yamaguchi, D. Deng, Y. Sato, Y. Te Hou, R. Watanabe, K. Sasaki, M. Kawabe, E. Hirano and T. Morinaga, Anticancer Res., 2013, 33, 5301–5309 CAS.
- J. A. Ludwig, S. E. Lamhamedi-Cherradi, H. Y. Lee, A. Naing and R. Benjamin, Cancers, 2011, 3, 3029–3054 CrossRef CAS PubMed.
- M. Santoro, S. E. Lamhamedi-Cherradi, B. A. Menegaz, J. A. Ludwig and A. G. Mikos, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 10304–10309 CrossRef CAS PubMed.
- A. C. Bellail, S. B. Hunter, D. J. Brat, C. Tan and E. G. Van Meir, Int. J. Biochem. Cell Biol., 2004, 36, 1046–1069 CrossRef CAS PubMed.
- A. Jain, M. Betancur, G. D. Patel, C. M. Valmikinathan, V. J. Mukhatyar, A. Vakharia, S. B. Pai, B. Brahma, T. J. MacDonald and R. V. Bellamkonda, Nat. Mater., 2014, 13, 308–316 CrossRef CAS PubMed.
- M. Alfano, M. Nebuloni, R. Allevi, P. Zerbi, E. Longhi, R. Lucianò, I. Locatelli, A. Pecoraro, M. Indrieri, C. Speziali, C. Doglioni, P. Milani, F. Montorsi and A. Salonia, Sci. Rep., 2016, 6, 36128 CrossRef CAS PubMed.
- J. Hogge, D. Krasner, H. Nguyen, L. B. Harkless and D. G. Armstrong, J. Am. Podiatr. Med. Assoc., 2000, 90, 57–65 CrossRef CAS PubMed.
- Y. Wang, J. Qian, T. Liu, W. Xu, N. Zhao and A. Suo, Mater. Sci. Eng., C, 2017, 76, 313–318 CrossRef CAS PubMed.
- D. H. Kim and D. Wirtz, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 6693–6694 CrossRef CAS PubMed.
- S. Ali, B. Nathalie, C. Judith and H. Dietmar, Front. Bioeng. Biotechnol., 2016, 4, 03011–03011 Search PubMed.
- A. Mantovani, Nature, 2009, 457, 36–37 CrossRef CAS PubMed.
- A. S. Thakor and S. S. Gambhir, Ca-Cancer J. Clin., 2013, 63, 395–418 CrossRef PubMed.
- M. S. Aslam, S. Naveed, A. Ahmed, Z. Abbas, I. Gull and M. A. Athar, J. Cancer Ther., 2014, 05, 817–822 CrossRef.
- M. Zamani, M. P. Prabhakaran and S. Ramakrishna, Int. J. Nanomed., 2013, 8, 2997–3017 Search PubMed.
- R. Langer, Science, 1990, 249, 1527–1533 CrossRef CAS PubMed.
- S. K. Vakkalanka, C. S. Brazel and N. A. Peppas, J. Biomater. Sci., Polym. Ed., 1996, 8, 119–129 CrossRef CAS PubMed.
- L. Brannon-Peppas, Med. Plast. Biomater, 1997, 4, 34–44 Search PubMed.
- N. Bölgen, I. Vargel, P. Korkusuz, Y. Z. Menceloǧlu and E. Pişkin, J. Biomed. Mater. Res., Part B, 2007, 81, 530–543 CrossRef PubMed.
- X. Xu, X. Chen, P. Ma, X. Wang and X. Jing, Eur. J. Pharm. Biopharm., 2008, 70, 165–170 CrossRef CAS PubMed.
- X. Xu, X. Chen, Z. Wang and X. Jing, Eur. J. Pharm. Biopharm., 2009, 72, 18–25 CrossRef CAS PubMed.
- J. W. Gatti, M. C. Smithgall, S. M. Paranjape, R. J. Rolfes and M. Paranjape, Biomed. Microdevices, 2013, 15, 887–893 CrossRef CAS PubMed.
- T. Kowalczyk, A. Nowicka, D. Elbaum and T. A. Kowalewski, Biomacromolecules, 2008, 9, 2087–2090 CrossRef CAS PubMed.
- K. Kanawung, K. Panitchanapan, S. O. Puangmalee, W. Utok, N. Kreua-Ongarjnukool, R. Rangkupan, C. Meechaisue and P. Supaphol, Polym. J., 2007, 39, 369–378 CrossRef CAS.
- D. Puppi, A. M. Piras, N. Detta, D. Dinucci and F. Chiellini, Acta Biomater., 2010, 6, 1258–1268 CrossRef CAS PubMed.
- S. Y. Chew, R. Mi, A. Hoke and K. W. Leong, Adv. Funct. Mater., 2007, 17, 1288–1296 CrossRef CAS PubMed.
- H. Cao, X. Jiang, C. Chai and S. Y. Chew, J. Controlled Release, 2010, 144, 203–212 CrossRef CAS PubMed.
- S. H. Ranganath and C. H. Wang, Biomaterials, 2008, 29, 2996–3003 CrossRef CAS PubMed.
- S. Liu, G. Zhou, D. Liu, Z. Xie, Y. Huang, X. Wang, W. Wu and X. Jing, J. Mater. Chem. B, 2013, 1, 101–109 RSC.
- S. Zong, X. Wang, Y. Yang, W. Wu, H. Li, Y. Ma, W. Lin, T. Sun, Y. Huang, Z. Xie, Y. Yue, S. Liu and X. Jing, Eur. J. Pharm. Biopharm., 2015, 93, 127–135 CrossRef CAS PubMed.
- R. Ramachandran, V. R. Junnuthula, G. S. Gowd, A. Ashokan, J. Thomas, R. Peethambaran, A. Thomas, A. K. K. Unni, D. Panikar, S. V. Nair and M. Koyakutty, Sci. Rep., 2017, 7, 43271 CrossRef PubMed.
- J. Zeng, X. Xu, X. Chen, Q. Liang, X. Bian, L. Yang and X. Jing, J. Controlled Release, 2003, 92, 227–231 CrossRef CAS PubMed.
- K. Kim, Y. K. Luu, C. Chang, D. Fang, B. S. Hsiao, B. Chu and M. Hadjiargyrou, J. Controlled Release, 2004, 98, 47–56 CrossRef CAS PubMed.
- S. Y. Chew, J. Wen, E. K. F. Yim and K. W. Leong, Biomacromolecules, 2005, 5, 2017–2024 CrossRef PubMed.
- J. A. Henry, M. Simonet, A. Pandit and P. Neuenschwander, J. Biomed. Mater. Res., Part A, 2007, 82, 669–679 CrossRef PubMed.
- K. Kim, M. Yu, X. Zong, J. Chiu, D. Fang, Y. S. Seo, B. S. Hsiao, B. Chu and M. Hadjiargyrou, Biomaterials, 2003, 24, 4977–4985 CrossRef CAS PubMed.
- X. Zhou, L. Chen, W. Wang, Y. Jia, A. Chang, X. Mo, H. Wang and C. He, RSC Adv., 2015, 5, 65897–65904 RSC.
- X. Zhang, X. Xie, H. Wang, J. Zhang, B. Pan and Y. Xie, J. Am. Chem. Soc., 2013, 135, 18–21 CrossRef CAS PubMed.
- B. Song, C. Wu and J. Chang, Acta Biomater., 2012, 8, 1901–1907 CrossRef CAS PubMed.
- F. Zheng, S. Wang, S. Wen, M. Shen, M. Zhu and X. Shi, Biomaterials, 2013, 34, 1402–1412 CrossRef CAS PubMed.
- P. Balakrishnan, L. Gardella, M. Forouharshad, T. Pellegrino and O. Monticelli, Colloids Surf., B, 2018, 161, 488–496 CrossRef CAS PubMed.
- M. Yoshida, R. Langer, A. Lendlein and J. Lahann, Polym. Rev., 2006, 46, 347–375 CAS.
- M. Deminsky, V. Jivotov, B. Potapkin and V. Rusanov, Pure Appl. Chem., 2002, 74, 413–418 CAS.
- Y. Il Cho, J. S. Choi, S. Y. Jeong and H. S. Yoo, Acta Biomater., 2010, 6, 4725–4733 CrossRef PubMed.
- F. Zomer Volpato, J. Almodóvar, K. Erickson, K. C. Popat, C. Migliaresi and M. J. Kipper, Acta Biomater., 2012, 8, 1551–1559 CrossRef CAS PubMed.
- J. S. Im, J. Yun, Y. M. Lim, H. Il Kim and Y. S. Lee, Acta Biomater., 2010, 6, 102–109 CrossRef CAS PubMed.
- G. Ma, Y. Liu, C. Peng, D. Fang, B. He and J. Nie, Carbohydr. Polym., 2011, 86, 505–512 CrossRef CAS.
- P. Zhao, H. Jiang, H. Pan, K. Zhu and W. Chen, J. Biomed. Mater. Res., Part A, 2007, 83, 372–382 CrossRef PubMed.
- H. Jiang, Y. Hu, Y. Li, P. Zhao, K. Zhu and W. Chen, J. Controlled Release, 2005, 108, 237–243 CrossRef CAS PubMed.
- Y. Z. Zhang, X. Wang, Y. Feng, J. Li, C. T. Lim and S. Ramakrishna, Biomacromolecules, 2006, 7, 1049–1057 CrossRef CAS PubMed.
- A. Saraf, L. S. Baggett, R. M. Raphael, F. K. Kasper and A. G. Mikos, J. Controlled Release, 2010, 143, 95–103 CrossRef CAS PubMed.
- A. Mickova, M. Buzgo, O. Benada, M. Rampichova, Z. Fisar, E. Filova, M. Tesarova, D. Lukas and E. Amler, Biomacromolecules, 2012, 13, 952–962 CrossRef CAS PubMed.
- T. Okuda, K. Tominaga and S. Kidoaki, J. Controlled Release, 2010, 143, 258–264 CrossRef CAS PubMed.
- Y. J. Kim, M. Ebara and T. Aoyagi, Adv. Funct. Mater., 2013, 23, 5753–5761 CrossRef CAS.
- G. D. Fu, L. Q. Xu, F. Yao, G. L. Li and E. T. Kang, ACS Appl. Mater. Interfaces, 2009, 1, 2424–2427 CrossRef CAS PubMed.
- S. Kato, K. Ozasa, M. Maeda, Y. Tanno, S. Tamaki, M. Higuchi-Takeuchi, K. Numata, Y. Kodama, M. Sato, K. Toyooka and T. Shinomura, Plant J., 2019, 14576 Search PubMed.
- M. Chen, Y. F. Li and F. Besenbacher, Adv. Healthcare Mater., 2014, 3, 1721–1732 CrossRef CAS PubMed.
- Y. J. Kim, M. Ebara and T. Aoyagi, Angew. Chem., Int. Ed., 2012, 51, 10537–10541 CrossRef CAS PubMed.
- X. Zhao, Z. Yuan, L. Yildirimer, J. Zhao, Z. Y. Lin, Z. Cao, G. Pan and W. Cui, Small, 2015, 11, 4284–4291 CrossRef CAS PubMed.
- Y. J. Kim, M. Ebara and T. Aoyagi, Sci. Technol. Adv. Mater., 2012, 13, 064203 CrossRef PubMed.
- X. Fang and D. H. Reneker, J. Macromol. Sci., Part B: Phys., 1997, 36, 169–173 CrossRef.
- H. S. Kim and H. S. Yoo, J. Controlled Release, 2010, 145, 264–271 CrossRef CAS PubMed.
- C. Achille, S. Sundaresh, B. Chu and M. Hadjiargyrou, PLoS One, 2012, 7, e52356 CrossRef CAS PubMed.
- C. Lei, Y. Cui, L. Zheng, P. Kah-Hoe Chow and C. H. Wang, Biomaterials, 2013, 34, 7483–7494 CrossRef CAS PubMed.
- J. Jiang, M. Ceylan, Y. Zheng, L. Yao, R. Asmatulu and S.-Y. Yang.
- H.-L. Che, H. J. Lee, K. Uto, M. Ebara, W. J. Kim, T. Aoyagi and I.-K. Park, J. Nanosci. Nanotechnol., 2015, 15, 7971–7975 CrossRef CAS PubMed.
- J. Zhang, Y. Duan, D. Wei, L. Wang, H. Wang, Z. Gu and D. Kong, J. Biomed. Mater. Res., Part A, 2011, 96, 212–220 CrossRef PubMed.
- U. Menon and I. J. Jacobs, in International Journal of Gynecological Cancer, 2001, pp. 3–6 Search PubMed.
- I. Kirman, G. F. Kalantarov, L. I. Lobel, H. Hibshoosh, A. Estabrook, R. Canfield and I. Trakht, Hybridoma Hybridomics, 2002, 21, 405–414 CrossRef CAS PubMed.
- H. Y. Son, J. H. Ryu, H. Lee and Y. S. Nam, ACS Appl. Mater. Interfaces, 2013, 5, 6381–6390 CrossRef CAS PubMed.
- M. A. Ali, K. Mondal, C. Singh, B. Dhar Malhotra and A. Sharma, Nanoscale, 2015, 7, 7234–7245 RSC.
- K. B. Paul, V. Singh, S. R. K. Vanjari and S. G. Singh, Biosens. Bioelectron., 2017, 88, 144–152 CrossRef CAS PubMed.
- J. C. Soares, L. E. O. Iwaki, A. C. Soares, V. C. Rodrigues, M. E. Melendez, J. H. T. G. Fregnani, R. M. Reis, A. L. Carvalho, D. S. Correa and O. N. Oliveira, ACS Omega, 2017, 2, 6975–6983 CrossRef CAS PubMed.
- L. L. del Mercato, M. Moffa, R. Rinaldi and D. Pisignano, Small, 2015, 22, 6417–6424 CrossRef PubMed.
- S. Hou, L. Zhao, Q. Shen, J. Yu, C. Ng, X. Kong, D. Wu, M. Song, X. Shi, X. Xu, W.-H. H. Ouyang, R. He, X.-Z. Z. Zhao, T. Lee, F. C. Brunicardi, M. A. Garcia, A. Ribas, R. S. Lo and H.-R. R. Tseng, Angew. Chem., Int. Ed., 2013, 52, 3379–3383 CrossRef CAS PubMed.
- N. Zhang, Y. Deng, Q. Tai, B. Cheng, L. Zhao, Q. Shen, R. He, L. Hong, W. Liu, S. Guo, K. Liu, H. R. Tseng, B. Xiong and X. Z. Zhao, Adv. Mater., 2012, 24, 2756–2760 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2020 |






