Open Access Article
Open Access ArticlePerspectives of genetically engineered microbes for groundwater bioremediation
Dick B.
Janssen
 *a and
Gerhard
Stucki
*a and
Gerhard
Stucki
 b
b
aBiotransformation and Biocatalysis, Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, Nijenborgh 4, 9747AG Groningen, The Netherlands. E-mail: d.b.janssen@rug.nl; Tel: +31-50-363-4008
bOrewa GmbH, Oberer Eggrainweg 11, CH-4410 Ormalingen, Switzerland. E-mail: gerhard.stucki@breitband.ch; Tel: +41-79-849-8857
First published on 25th February 2020
Abstract
Biodegradation is the main process for the removal of organic compounds from the environment, but proceeds slowly for many synthetic chemicals of environmental concern. Research on microbial biodegradation pathways revealed that recalcitrance is – among other factors – caused by biochemical blockages resulting in dysfunctional catabolic routes. This has raised interest in the possibility to construct microorganisms with improved catabolic activities by genetic engineering. Although this goal has been pursued for decades, no full-scale applications have emerged. This perspective explores the lagging implementation of genetically engineered microorganisms in practical bioremediation. The major technical and scientific issues are illustrated by comparing two examples, that of 1,2-dichloroethane where successful full-scale application of pump-and-treat biotreatment processes has been achieved, and 1,2,3-trichloropropane, for which protein and genetic engineering yielded effective bacterial cultures that still await application.
Environmental significanceA frequent groundwater pollutant is 1,2,3-trichloropropane (TCP). It has been used as a degreasing agent and solvent, was formed as a side product during epichlorohydrin synthesis and occurred as a contaminant in nematicides. TCP contamination is frequently detected at sites where chemical waste has been inappropriately disposed of. It is highly toxic, recalcitrant in the environment and can spread via groundwater flows. Whereas anaerobic and oxidative biotransformations are possible, no natural microorganisms are known that can mineralize TCP under oxic conditions. Since some structurally similar compounds are biodegradable, TCP was chosen as a target for constructing bacteria that use it as a growth substrate. This was indeed achieved by a combination of protein- and metabolic engineering. The manuscript evaluates the scientific approaches used to obtain TCP-degrading bacteria and the perspective of using bioaugmentation with genetically engineered microorganisms for removing synthetic pollutants from groundwater. |
Introduction
Recalcitrance to biodegradation of synthetic chemicals remains an issue of serious concern. Recent years have seen increasing attention on xenobiotic compounds such as diclofenac or cabamazepine (pharmaceuticals), sucralose (artificial sweetener), and polyfluorinated alkanes and alkanoates (PFAS). Their common use and lack of biodegradation causes widespread occurrence and persistence in soils, surface water and groundwater. In outlets of chemical treatment plants, compounds such as dimethoxymethane or ethyl dimethylcarbamate may be detected even after effluents have passed different biological treatment steps. Classical pollutants that are poorly degradable such as PCBs or chlorinated pesticides have been prohibited but they are still present at many waste dumps. Although the use and release of several chlorinated solvents has been stopped or reduced, they remain a major concern at sites that were contaminated decades ago, but have not been cleaned up. Chlorinated solvents are relatively water soluble and can be distributed over large distances by groundwater flows.The main process by which environmental chemicals should be removed is biodegradation, and many catabolic pathways and biotic reactions have been discovered in the last decades of the previous century, especially in microorganisms.1–3 Organic compounds released into the environment are subject to biological transformation and elimination as long as microorganisms with the required catabolic routes are present and their activity or proliferation is not hindered by unfavorable redox conditions, the absence of oxygen in case of petroleum hydrocarbons, extremes of pH and temperature, hostile salt concentrations, sorption of compounds to the solid matter, sub-threshold concentrations, or high toxicity of a chemical itself. In the absence of organisms possessing a complete catabolic pathway, degradation may still occur by cometabolism, which includes fortuitous biotransformation by non-dedicated enzymes that happen to have a broad substrate specificity.4
Our past work and this review focus on two closely related highly important compounds, 1,2-dichloroethane (DCA) and 1,2,3-trichloropropane (TCP). Both are synthetic chemicals, not known to occur naturally at concentrations that have biological effects, and are only detected at polluted sites. DCA and TCP also have similar physical properties such as aqueous solubility, low hydrophobicity (reflected in adsorption to activated carbon), modest volatility and air–water partitioning, like some other important pollutants (Table 1). Both DCA and TCP are used or formed in large quantities worldwide, and of both compounds polluted areas have been reported due to accidental spills or inappropriate local waste disposal. Known sites are for example in the Botlek area (the Netherlands); near Bremerhaven and Lübeck (Germany) and the Tyson's dump Superfund site not far from the Philadelphia area (Pennsylvania, USA). Since liquid DCA and TCP have a higher density than water, they sink in the groundwater body, occasionally causing a subsurface liquid phase in case of large spills (DNAPL). The relatively high water solubility causes spreading by flowing groundwater which may lead to contamination of large groundwater systems after local discharge. This necessitates isolation and remediation measures, which may be based on (accelerated) in situ treatment or pump-and-treat methodologies. In both cases the use of microorganisms is an option. Dependent on local circumstances, anaerobic treatment, such as reductive dechlorination, or aerobic processes may be used for bioremediation. Our work focused on microbial cultures that are applicable for on-site aerobic treatment of contaminated groundwater in bioreactors and was stimulated by the need to prevent spread of DCA and TCP to surface water and groundwater in urban areas.
| Parameters | Units | 1,2-Dichloro-ethane | 1,2,3-Trichloro-propane | Tetrachloro-ethene | 2-Chlorophenol |
|---|---|---|---|---|---|
| a Data from http://www.pubchem.ncbi.nlm.nih.gov | |||||
| Density | g cm−3 | 1.25 | 1.39 | 1.62 | 1.26 |
| Aqueous solubility (25 °C) | g l−1 | 8.6 | 1.8 | 0.2 | pH dependent |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kow (activated carbon sorption) Kow (activated carbon sorption) |
— | 1.48–1.76 | 2.27 | 3.4 | 2.15 |
| Henry law constant (stripping) | atm m3 mol−1 | 2.51 × 10−4 | 3.43 × 10−4 | 1.77 × 10−2 | 1.12 × 10−5 |
| Double bond (ozone/UV light) | — | Absent | Absent | Present | Present |
| Aerobic bacterial cultures available | — | Yes | Yes, genetically engineered | No | Yes |
| Anaerobic bacterial conversion known | — | Yes | Yes | Yes | Yes |
Microorganisms capable of aerobic growth on 1,2-dichloroethane were obtained some 35 years ago by classical microbial enrichment.5,6 Knowing that biodegradation is possible, a full scale groundwater treatment system was developed by one of us (GS).7 It was operated for 18 years, after which the site was found to be sufficiently clean to stop the process. Confronted with sites that were seriously polluted with TCP, and in view of the similar physico-chemical properties, we initiated research on TCP biodegradation with the idea to apply TCP-degrading microorganisms in a similar process as for DCA remediation. However, various efforts to obtain aerobic biodegradation or enrich cultures that would grow at the expense of TCP failed.
Since the later 1980s, the use of genetic engineering to construct catabolic pathways for recalcitrant compounds was pursued by different research groups.8–14 The use of protein- and metabolic engineering for this purpose has been contemplated ever since.15–17 However, this research did not lead to full scale applications. The example of TCP seemed particularly interesting since short and simple catabolic pathways that look reasonable for the aerobic conversion of TCP to normal cellular metabolites could be postulated on basis of biodegradation studies with related compounds.18 Consequently, the design of enzymes and pathways that would allow organisms to degrade or even recycle TCP was investigated by different groups, with results that convincingly showed that biodegradation can be achieved.19,20 The results of this work provide insight in the causes of success and failure as well as provide indications for future directions.
Use and biodegradation of 1,2-dichloroethane
DCA is by production volume the largest chlorinated industrial chemical, most of it being used for synthesis of vinyl chloride and smaller amounts for ethylene diamine and other chemicals. It was also used as a solvent. Groundwater contamination is mainly due to leakages and improper waste disposal.Bacterial degradation of DCA under oxic conditions was investigated by us in the 1980s.5,6 A pure culture of Xanthobacter autotrophicus was isolated and the catabolic pathway was elucidated (Fig. 1).6 First a haloalkane dehalogenase hydrolyzes one C–Cl bond of DCA to give 2-chloroethanol, which is oxidized in 2 steps by dehydrogenase enzymes to produce chloroacetic acid, which in turn is converted by a second hydrolytic dehalogenase to glycolic acid. The initial dehalogenase was isolated and crystallized, revealing for the first time the enzymatic mechanism of a fundamental reaction in organic chemistry: the SN2 nucleophilic displacement reaction of an alkylhalide with water.21 The structures also showed that the haloalkane dehalogenases are members of a large group of related enzymes, the α/β-hydrolase fold family, which also includes lipases and epoxide hydrolases. Later, identical haloalkane dehalogenases acting on DCA were discovered in DCA-degrading Xanthobacter and Ancylobacter strains isolated from different geographic locations, incl. South Korea, Australia, South Africa, and Germany.22–26 The dehalogenase gene is plasmid localized, and a plasmid was also found in other organisms that grow on DCA.23–26 These dehalogenases catalyze cofactor-independent hydrolytic reactions. An oxidative route for aerobic bacterial DCA metabolism was described by Hage and Hartmans (Fig. 1).27 Furthermore, DCA can also be degraded anaerobically under different redox conditions.28 A pure culture converting DCA anaerobically with nitrate as electron acceptor was described by Dinglasan-Panlilio et al.29
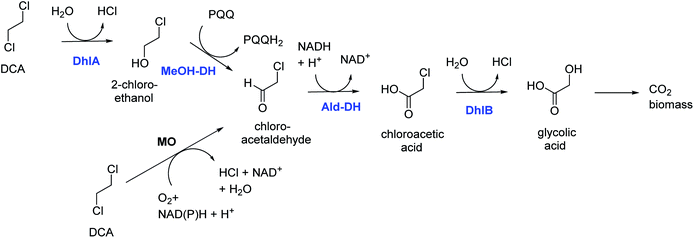 | ||
| Fig. 1 Aerobic catabolic pathways for 1,2-dichloroethane.6,27 Enzymes of the Xanthobacter autotrophicus pathway are indicted in blue: DhlA, haloalkane dehalogenase; MeOH-DH, methanol dehydrogenase; Ald-DH, chloroacetaldehyde dehydrogenase; DhlB, haloacid dehalogenase. Monooxygenase (MO) – mediated conversion leads to chloroacetaldehyde. | ||
Biochemical and genetic studies on X. autotrophicus GJ10 and related strains revealed signs of recent genetic adaptation, which is not surprising as industrial production of DCA started only in the 1920s. First, the genes for enzymes catalyzing the most critical steps (haloalkane dehalogenase for the initial dechlorination, chloroacetaldehyde dehydrogenase for converting the very toxic and bifunctionally reactive intermediate chloroacetaldehyde) are plasmid encoded.30 A chloroacetate dehalogenase gene has also been found on a plasmid.26 Genes encoding catabolic enzymes for synthetic compounds are often located on plasmids, frequently associated with transposons or insertion elements which enable gene transfer or gene activation. Insertion elements also flank regions encoding haloalkane dehalogenase.24,26,30 Second, the haloalkane dehalogenase is constitutively expressed, which is unusual for enzymes active in a growth-supporting catabolic pathway and indicates there has been insufficient time to evolve a regulatory system. Control of haloalkane dehalogenase gene expression by its substrate would require a second protein that recognizes DCA, in addition to the haloalkane dehalogenase itself. Third, there are indications of recent mutations in the haloalkane dehalogenase protein itself. The enzyme's cap domain harbors tandem repeats of short stretches of sequence that would disappear in the course of evolutionary time due to neutral genetic drift.22 The repeats are lacking in close homologs of the haloalkane dehalogenase discovered by genome mining in bacteria with no reputation of 1,2-dichloroethane degradation. Laboratory evolution experiments in which the DCA dehalogenase gene was expressed in a Pseudomonas strain and placed under selection pressure to accept another substrate (1-chlorohexane) could mimic the appearance of such mutations in the cap domain.31,32 The isolated DCA-degrading bacteria grew well aerobically with DCA as sole carbon source in the presence of small amounts of vitamins or yeast extract. Growth in the absence of vitamins was better in consortia where cross-feeding is possible. These observations suggested that a growth-associated DCA treatment process was feasible. Indeed, Stucki et al.33 demonstrated that DCA degrading bacterial cultures (Pseudomonas DE1,5X. autotrophicus GJ10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) inoculated in lab-scale fixed-bed bioreactors could remove DCA under groundwater conditions. Furthermore, Freitas dos Santos and Livingston34,35 at Imperial College, London, demonstrated laboratory-scale bioprocesses for waste gas and waste water treatment using X. autotrophicus GJ10.
6) inoculated in lab-scale fixed-bed bioreactors could remove DCA under groundwater conditions. Furthermore, Freitas dos Santos and Livingston34,35 at Imperial College, London, demonstrated laboratory-scale bioprocesses for waste gas and waste water treatment using X. autotrophicus GJ10.
Full-scale processes for 1,2-dichloroethane removal from groundwater
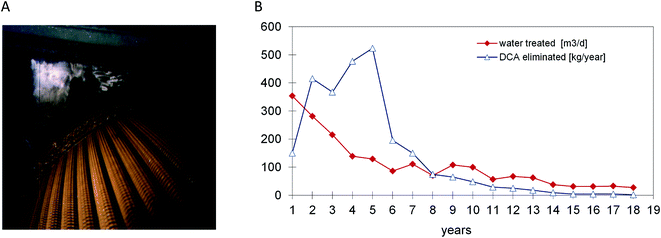
Based on the experience with DCA-degrading bacteria and their availability for inoculation, a full scale process for DCA remediation was realized by Ciba Specialty Chemicals Inc. and its successor Novartis AG. At a site near Lübeck (Germany) the source of groundwater pollution was a former pharmaceutical production plant. DCA had replaced petroleum ether as a non-flammable solvent and was used from the 1950s until 1987 for the extraction of pancreatin from dried and grained calf's stomach tissue. When pollution by DCA was detected during deconstruction of the site, rapid action was deemed necessary. The sources were removed and a gallery of extraction wells was installed for pumping and treatment of groundwater, which contained DCA concentrations of 1–200 mg l−1. The plant was initially designed to treat about 20 m3 h−1 of groundwater by a conventional sand filtration unit followed by carbon adsorption. After start up in 1993 the process turned out to be extremely costly as carbon adsorption of DCA is poor, necessitating frequent replacement. Other remediation technologies such as air-stripping of DCA or oxidation by UV-ozone (DCA lacks a double bond) were also considered inefficient and costly. The lack of efficient classical alternatives and the low financial risk of modifying the plant such that it would work biologically led to the decision to inoculate the sand and carbon filters with DCA-degrading bacteria and to equip the plant with dosing stations for H2O2 and ammonium phosphate. Later, with increasing DCA feed concentrations, the capacity of the biological step was extended by installment of a rotating disk biological contactor (Fig. 2), which was also inoculated with DCA-degrading bacteria (laboratory grown X. autotrophicus, Fig. 1).6 At this stage, several uncertainties existed: would bacteria selected in the laboratory remove the chemical under groundwater conditions (8–12 °C) with low buffer capacity, down to below the limit needed to allow surface discharge (10 μg l−1)? H2O2 was added as oxygen source instead of aeration to avoid stripping of DCA, but would it not inhibit microbial activity when used at large scale? Would the amount of energy associated with microbial DCA oxidation be sufficient to sustain a viable biofilm? After startup, the analytical data showed that the system performed remarkably well.7,36 The service life of the activated carbon columns initially increased from 3 to 6 months, and later the formation of a stable microbial population enabled the use of the same activated carbon columns for 14 years without any carbon replacement. About 4 years after inoculation, the X. autotrophicus strain was re-isolated from the yellow biofilm formed on the solid support in the bioreactor (Fig. 2). The organism was apparently unchanged, including the sequence of the haloalkane dehalogenase gene.The addition of H2O2 was stopped when the DCA feed had dropped to a level of 10–20 g daily, which was still sufficient for the system to maintain an active microbial community. During the final years, the advantage of the biological system became even more apparent as the plant's operational costs were very low since neither oxygen nor fertilizer had to be added. The main running costs consisted of electricity to pump the groundwater, heating of the plant during the winter season, supply of H2O2 and fertilizer and removal of the mineral sludge and were ca. 0.26 € per m3. About 85% of the costs were for electricity to operate the pumps. Additional expenditures were costs for investigating the polluted site, installation of about 60 groundwater observation and extraction wells, for construction (ca. 200 k€) and maintenance of the treatment plant, and for expenses of management and consultation with authorities. Overall, the total costs to remediate the Lübeck site were in the range of about 5 M€. Whereas costs of anaerobic in situ treatment may be lower, the urgency to clean up the site and prevent spreading of the mobile pollutants to nearby surface water and groundwater wells triggered the decision to quickly install a groundwater extraction and treatment plant, and the choice for an aerobic process was based on previous lab-scale results which showed that a very short adaptation time was sufficient and that rapid degradation was possible.
With average feed concentrations dropping to 246 μg l−1, outlet concentrations of less than 0.5 μg l−1 were achieved. After 18 years of pump and treat activity, concentrations in all pumping wells dropped to below the required limit of 10 μg l−1. In agreement with the local government, active groundwater treatment was stopped and the remaining observation wells in the area still containing trace concentrations of DCA were monitored for another 5 years.36 DCA concentrations in the sandy aquifer material dropped further due to natural attenuation.
In 2002, a second bioprocess for DCA removal was established at Akzo-Nobel and is currently operated by Remondis Aqua B.V. in the Botlek area (the Netherlands) (Fig. 3). On the site, ShinEtsu PVC B.V. is using DCA as a precursor for vinyl chloride production. Earlier, over a long period, serious groundwater contamination with DCA had happened due to leakage. Liquid DCA had entered the groundwater body due to its higher density than water. Groundwater treatment was initially carried out by extraction and stripping, using excess steam from a nearby plant. The liquid condensate collected from the stripper contained DCA, which was incinerated. When the steam supply was no longer freely available, economically more attractive alternatives were examined. In view of the experience with the Lübeck site, one of us (GS) working at BMG Engineering AG (now Arcadis Switzerland AG) was contacted to design a groundwater treatment process for this site (Fig. 3A). The plant was designed to treat 15 m3 h−1 groundwater contaminated with 100 mg DCA per l, a capacity that was 25-fold higher than that of the plant operated at Lübeck. The biological part consisted of two packed-bed bioreactors. To prevent inhibition of bacterial activity, the first of the two reactors was equipped with a caustic soda addition loop to neutralize HCl generated as a product of DCA mineralization. Investment and operational costs were estimated much lower than for alternative processes, and the plant was built in a few months. For inoculation, about 40 kg of back washing sludge from the Lübeck plant was used. The two slightly aerated fixed-film bioreactors (total volume 100 m3) were inoculated with this biomass together with 2 l of laboratory-grown DCA mineralizing microorganisms and about 100 l of diluted sludge from the on-site wastewater treatment plant, which had also received DCA. Two months later, the full DCA elimination capacity was gained with DCA outlet concentrations of <10 μg l−1. The concomitant quantitative increase of inorganic chloride indicated that DCA was completely mineralized. In 2020, the process is still in operation (Fig. 3B).
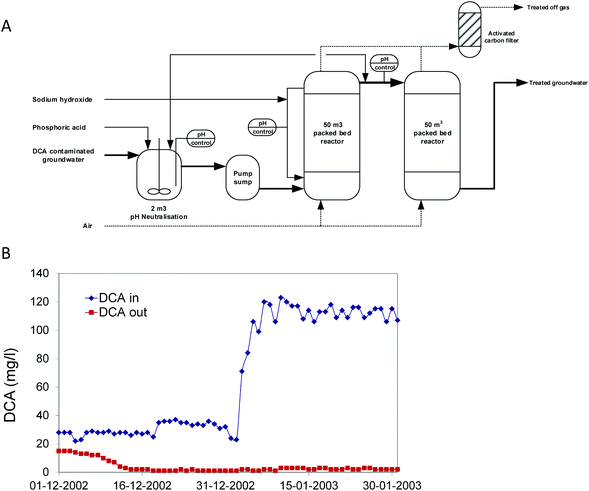 | ||
| Fig. 3 Full scale treatment of 1,2-dichloroethane-contaminated groundwater in a packed bed bioreactor. (A) Process scheme. (B) Influent and effluent concentrations after startup. | ||
Why were these two cases of groundwater treatment with specialized bacteria degrading a synthetic chlorinated hydrocarbon successful? First, the availability of aerobic microorganisms, either as pure cultures or enriched sludge, that rapidly grow on DCA and use it as a carbon source for growth. These are naturally evolved bacteria, with an effective combination of plasmid-encoded and host genes and gene expression for detoxification. The organism was isolated from samples of polluted soil and sediment and apparently could establish itself in a biofilm and survive groundwater-treatment conditions. We consider the ability to grow at the expense of DCA and synthesize new biomass when DCA is the only organic compound in the feed essential for the establishment of a stable active biofilm. Second, the properties of DCA made the usual classical chemical–physical treatment technologies such as extraction, carbon absorption, air stripping or UV peroxide treatment costly and less effective. At the same time, the properties of DCA facilitate biological removal, e.g. by avoiding loss and spread of contaminants by stripping and preventing strong absorption to organic carbon. Third, DCA has a modest tendency to adsorb to soil and is reasonably water soluble, allowing removal of subsurface contaminants by extraction of groundwater. Fourth, at the sites discussed above, DCA is the major contaminant, giving organisms that can biodegrade it and thrive on it a competitive advantage in the artificial ecosystem of a bioreactor. Fifth, in view of the location of the sites, near urban areas and with a risk of surface- or drinking water contamination, there was a real quest for swift cleanup. Often, treatment is postponed or left to natural attenuation because there is no risk of spreading or (eco)toxicological hazards. Sixth, the initial concentrations of pollutants were quite high, facilitating establishment of a biofilm by organisms that could grow on the contaminant. Extremely low levels (ng l−1) likely cannot be easily removed biologically since degradation would yield insufficient metabolic energy for cell growth and establishment of a stable population in the biofilm.
Occurrence of 1,2,3-trichloropropane
TCP emerges as a side product and environmental pollutant from past industrial processes for the synthesis of epichlorohydrin.37 These are nowadays replaced by the less problematic glycerol to epichlorohydrin (GTE) process, e.g. at the Dow Chemical Company.38 In the 1990s, interest in a biological process for TCP conversion emerged at Dow from the desire to feed the side product back into the classical epichlorohydrin synthesis process. A single hydrolytic dechlorination of TCP would yield one of the dichloropropanol isomers, both of which are easily converted to epichlorohydrin at alkaline pH. The option to use a haloalkane dehalogenase for this initial dehalogenation was pursued in a research project involving Dow, Diversa Corp. (later Verenium, now incorporated in BASF), and the Terwilliger protein crystallography group at Los Alamos National Laboratory.At about the same time, working with Ciba Specialty Chemicals Inc., now BASF too, and the University of Groningen, we investigated TCP degradation because of the need to remediate sites where TCP from epichlorohydrin manufacturing processes had been improperly disposed. A prominent example is the so-called Tyson's dump site near Philadelphia, PA, where a stone quarry was filled with such waste by Franklin P. Tyson and Fast Pollution Treatment Inc.39 TCP had leaked into sediments and rock fissures, and a pump-and-treat installation was used by Ciba to stop contamination of nearby river water and to reduce spreading of pollutants beyond the site. The activated carbon needed frequent replacement because of the low binding capacity, due to the low hydrophobicity (Table 1). In view of the positive experience with the biological treatment process for DCA-contaminated groundwater, we considered that demonstrating the aerobic biodegradability of TCP and isolating TCP-degrading bacteria would be key steps in the development of a bioremediation process for this extracted groundwater.
Engineering TCP degrading enzymes and organisms
Initial biodegradation studies on TCP performed by us included enrichment with soil and sediment samples from various polluted sites, as well as shake-flask experiments with activated sludge. The results indicated that TCP was highly recalcitrant under aerobic conditions. Enrichment cultures failed, and continuous flow experiments in laboratory-scale bioreactors performed by Ciba Specialty Chemicals Inc. in Switzerland never indicated any adaptation. To date, still no natural organisms growing on TCP as sole carbon source under oxic conditions have been described. This may well change as natural adaptation processes by mutations in structural and regulatory genes as well as exchange of genes between organisms can lead to evolution of new catabolic activities, similar to what happened within 100 years for DCA.Concerning TCP biodegradation, both anaerobic conversions (partial reductive dehalogenation)40–44 and cometabolic oxygenation reactions45,46 have been reported for isolated microbial cultures.40 Thermodynamic and quantum mechanical calculations indeed predict that diverse reactions are possible.47 Anaerobic transformation of TCP by strains of Chloroflexi gave allyl chloride as a main product, which hydrolyzed to allylalcohol.43 Cometabolic biooxidation yields various products, including toxic metabolites.45,46 Also abiotic conversions have been explored, especially the reductive conversion of TCP by metals.49,50 Both during abiotic and biotic transformation dehydrodehalogenation is important. Recent quantum chemical calculations show that β-elimination of TCP to allyl chloride is the energetically most favorable abiotic transformation.48
We considered that a metabolic route that involves an initial energy-requiring monooxygenation reaction would not be the best route. An initial hydrolytic step seemed more attractive, since studies by others and by us had identified bacteria that could aerobically grow on dichloropropanols.51,52 Furthermore, a hydrolytic haloalkane dehalogenase catalyzed the first step in DCA degradation, and the development or discovery of a dehalogenase with a suitably expanded (or shifted) substrate range could yield an enzyme catalyzing hydrolysis of TCP to a dichloropropanol.
The haloalkane dehalogenase called DhaA from Rhodococcus (UniProtKB/Swiss-Prot: P0A3G2.1)53,54 was used as the starting point for developing improved enzymes, both by us19 and by investigators from Diversa in collaboration with the scientists at Dow Chemical and Los Alamos. It has very low but detectable activity with TCP, much lower than its activity with 1-chlorobutane or 1-chlorohexane.53 Newman et al.55 solved the structure of this haloalkane dehalogenase (PDB ID: 1BN6), which is different from the X. autotrophicus enzyme called DhlA described above. Is has a broader substrate range but unlike DhlA with its small occluded active site it does not hydrolyze DCA. Diversa developed variants that have increased activity towards TCP and were much more thermostable, as reported by Gray et al.56 They selected and characterized a 5-fold mutant (D89G + F91S + T159L + G182Q + I220L) and an 8-fold mutant (+N238T + W251Y + P302A). Independently, Bosma et al.19 improved the same haloalkane dehalogenase by two rounds of directed evolution, which gave a 2-position mutant called DhaAM2 (C176Y + Y273F) with improved TCP hydrolysis activity. Pavlova et al.57 further improved the activity of DhaAM2 by inserting mutations in the substrate access/product exit tunnel that connects solvent and active site, yielding among other variants a mutant called DhaA31, with 3 more mutations (I135F + V245F + L246I). Finally, van Leeuwen et al.58 performed a further directed evolution study producing enantiocomplementary dehalogenase mutants carrying multiple mutations around the active site region, including variant DhaA90R (13 mutations in comparison to DhaA) producing (R)-2,3-dichloro-1-propanol and variant DhaA97S (17 mutations) producing mainly (S)-2,3-dichloro-1-propanol from TCP. The latter enantioselective variants had lower activity than DhaA31 but offered the opportunity to direct TCP hydrolysis towards a dichloropropanol enantiomer that can be better converted by subsequent enzymes, i.e. by the (R)-selective 2,3-dichloro-1-propanol dehalogenase termed HheC and the (S)-epichlorohydrin active epoxide hydrolase called EchA, both from a strain of the Gram-negative bacterium Agrobacterium radiobacter.59,60 This latter organism was isolated in Groningen in the 1980s from sediment collected in a ditch near an epichlorohydrin manufacturing plant called Chemische Fabriek Zaltbommel (now Sachem B.V.), the Netherlands.51,59
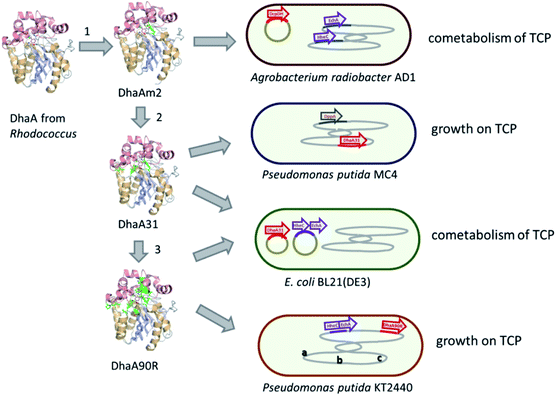
Introduction of the improved mutant dehalogenases in host organisms to construct bacteria that degrade TCP was pursued various times (Fig. 5). Initially, Bosma et al.18,19 used the 2,3-dichloro-1-propanol utilizing strain of A. radiobacter mentioned above as a host, introducing the improved dehalogenase DhaA-M2 on a transmissible plasmid derived from the broad-host range cloning vector pLAFR3, which also carries a tetracycline antibiotic resistance marker (Fig. 4, lower pathway; Fig. 5, top). TCP degradation by the resulting engineered strain was indeed observed in shake flasks with addition of pulses of TCP, but the organism still failed to grow with TCP as a sole carbon source.
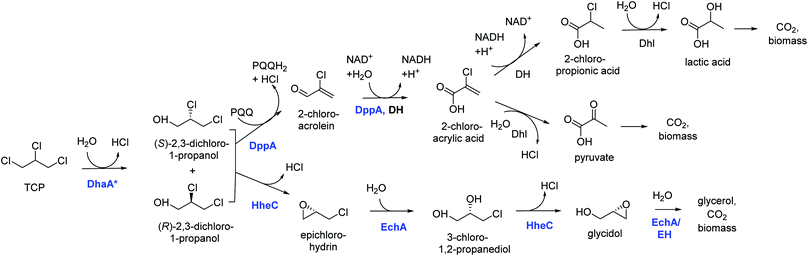 | ||
| Fig. 4 Growth-supporting catabolic pathways for aerobic TCP mineralization. The upper route is based on the pathway for dichloropropanol mineralization by Pseudomonas MC4,60 the lower route on the pathway in Agrobacterium radiobacter AD1.51,59 The pathways for 2-chloroacrylic acid metabolism in MC4 are hypothetical. Experimentally confirmed enzymes (in blue font): DhaA*, engineered variants of DhaA-type haloalkane dehalogenase;19,57,58 DppA, non-stereoselective dehalogenating quinohemoprotein alcohol dehydrogenase;61 HheC, halohydrin dehalogenase, a protein related to dehydrogenases that converts vicinal halohydrins to epoxides;59 EchA, epoxide hydrolases for conversion of epoxides to diols.60 Hypothetical enzymes (black font): DH, dehydrogenase; Dhl, dehalogenase; EH, alternative epoxide hydrolase. | ||
The DhaA31 variant57 was introduced in the 2,3-dichloro-1-propanol degrader Pseudomonas MC4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 61 using chromosomal integration instead of a plasmid and without co-introduction of an antibiotic resistance marker or plasmid mobilization genes.20 Insertion into the chromosome looks more attractive for bioremediation applications, and should relieve concerns related to distribution in the environment of genes encoding engineered enzymes or antibiotic resistance proteins. The Pseudomonas MC4 host was obtained earlier by classical enrichment with 2,3-dichloro-1-propanol from a site in the Botlek area mentioned above.61 That site was polluted with side products from epichlorohydrin synthesis. Strain MC4 possessed a different dichloropropanol catabolic pathway as A. radiobacter, using in the first step a periplasmic quinohemoprotein alcohol dehydrogenase (DppA) rather than a cytoplasmic halohydrin dehalogenase (Fig. 4, top). The enzyme catalyzes both an oxidation and a dehalogenation reaction on 2,3-dichloro-1-propanol, is not stereoselective, and can also convert 2-chloroacrolein to 2-chloroacrylic acid, which is a growth substrate. How this latter compound is dehalogenated to either pyruvate or lactate is unknown. The periplasmic localization of DppA keeps formation of the reactive product 2-chloroacrolein out of the cytoplasm. The recombinant strain with the engineered haloalkane dehalogenase on the chromosome was termed MC4-5222 and utilized TCP as sole carbon source for growth, although the growth rate was low.20 The genome was partially sequenced, revealing the location of the engineered haloalkane dehalogenase in the host DNA (GenBank: JOJW00000000.1). Strain MC4-5222 was introduced in laboratory-scale oxic packed bed reactor (ceramic Raschig rings or sintered glass beads as packing material) that was used for continuous degradation of TCP present in influent water at 0.3 mM with a water residence time of 116 h. TCP removal increased from 87% to 97% during a 2 month test period.20 Additional experiments showed that the organism continued to grow well in this system, and occasionally it would grow back into the TCP supply vessel – a gratifying observation of what normally should be avoided in continuous-flow bioreactor experiments. Different reactor setups and microbial cultures were tested, including co-inoculation of the packed bed bioreactor with a mixture of similarly engineered strains capable of degrading TCP (MC4-5221, MC4-5221, MC4-1331) and A. radiobacter AD1 as a natural dichloropropanol degrader. The results clearly demonstrated the possibility of continuous TCP biodegradation under aerobic conditions by genetically modified microorganisms (GEMs) (Fig. 6).62
61 using chromosomal integration instead of a plasmid and without co-introduction of an antibiotic resistance marker or plasmid mobilization genes.20 Insertion into the chromosome looks more attractive for bioremediation applications, and should relieve concerns related to distribution in the environment of genes encoding engineered enzymes or antibiotic resistance proteins. The Pseudomonas MC4 host was obtained earlier by classical enrichment with 2,3-dichloro-1-propanol from a site in the Botlek area mentioned above.61 That site was polluted with side products from epichlorohydrin synthesis. Strain MC4 possessed a different dichloropropanol catabolic pathway as A. radiobacter, using in the first step a periplasmic quinohemoprotein alcohol dehydrogenase (DppA) rather than a cytoplasmic halohydrin dehalogenase (Fig. 4, top). The enzyme catalyzes both an oxidation and a dehalogenation reaction on 2,3-dichloro-1-propanol, is not stereoselective, and can also convert 2-chloroacrolein to 2-chloroacrylic acid, which is a growth substrate. How this latter compound is dehalogenated to either pyruvate or lactate is unknown. The periplasmic localization of DppA keeps formation of the reactive product 2-chloroacrolein out of the cytoplasm. The recombinant strain with the engineered haloalkane dehalogenase on the chromosome was termed MC4-5222 and utilized TCP as sole carbon source for growth, although the growth rate was low.20 The genome was partially sequenced, revealing the location of the engineered haloalkane dehalogenase in the host DNA (GenBank: JOJW00000000.1). Strain MC4-5222 was introduced in laboratory-scale oxic packed bed reactor (ceramic Raschig rings or sintered glass beads as packing material) that was used for continuous degradation of TCP present in influent water at 0.3 mM with a water residence time of 116 h. TCP removal increased from 87% to 97% during a 2 month test period.20 Additional experiments showed that the organism continued to grow well in this system, and occasionally it would grow back into the TCP supply vessel – a gratifying observation of what normally should be avoided in continuous-flow bioreactor experiments. Different reactor setups and microbial cultures were tested, including co-inoculation of the packed bed bioreactor with a mixture of similarly engineered strains capable of degrading TCP (MC4-5221, MC4-5221, MC4-1331) and A. radiobacter AD1 as a natural dichloropropanol degrader. The results clearly demonstrated the possibility of continuous TCP biodegradation under aerobic conditions by genetically modified microorganisms (GEMs) (Fig. 6).62
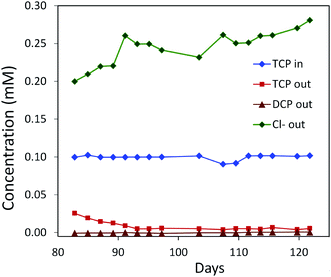 | ||
| Fig. 6 Degradation of TCP by a mixed culture of P. putida strains MC4-5221, MC4-5221, MC4-1331 and A. radiobacter AD1 in a laboratory-scale aerated packed bed bioreactor operated in a continuous mode. Packing material was sintered glass and data were collected after an initial 80 day acclimation period. TCP loading rate 0.40–0.44 mg h−1 l−1; residence time 23 h; removal increasing to 95%.40,62 | ||
An artificial TCP catabolic pathway was also constructed in E. coli strain BL21(DE3). Kurumbang et al.63 tested DhaA31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 57 and the enantioselective derivative DhaA90R58 in combination with the halohydrin dehalogenase (HheC) and epoxide hydrolase (EchA) genes from A. radiobacter,59,60 the latter two combined on a separate dual-gene expression plasmid (Fig. 4 and 5). For degradation experiments, cells were cultivated in rich medium (Luria Broth) with antibiotics and synthesis of the catabolic enzymes was induced by adding IPTG (isopropyl β-D-1-thiogalactopyranoside). Unfortunately, the engineered E. coli strain did not grow on TCP as sole carbon source, and TCP did not stimulate growth on another carbon source, even though enzyme expression levels were carefully balanced to optimize the flux through the pathway to glycerol.63 The lack of growth and requirement of rich medium components make successful application unlikely in case of a treatment system into which competing organisms can enter from air or via the influent. Probably E. coli is quite sensitive to TCP and some of its transformation products,64,65 which can be formed by various unspecific biotic reactions.40 It was proposed that the haloalkane dehalogenase activity was still too low to produce sufficient glycerol to generate energy for maintenance and growth. Furthermore, there is a mismatch in stereoselectivity between the most active haloalkane dehalogenase (DhaA31, makes mainly (S)-2,3-dichloro-1-propanol) and the halohydrin dehalogenase (HheC is selective for (R)-2,3-dichloro-1-propanol), explaining the accumulation of dichloropropanol in these cultures. E. coli probably is a less suitable host for development of bioremediation organisms, since the same initial haloalkane dehalogenase allowed growth of Pseudomonas strain mc-5222 and Pseudomonas putida.
57 and the enantioselective derivative DhaA90R58 in combination with the halohydrin dehalogenase (HheC) and epoxide hydrolase (EchA) genes from A. radiobacter,59,60 the latter two combined on a separate dual-gene expression plasmid (Fig. 4 and 5). For degradation experiments, cells were cultivated in rich medium (Luria Broth) with antibiotics and synthesis of the catabolic enzymes was induced by adding IPTG (isopropyl β-D-1-thiogalactopyranoside). Unfortunately, the engineered E. coli strain did not grow on TCP as sole carbon source, and TCP did not stimulate growth on another carbon source, even though enzyme expression levels were carefully balanced to optimize the flux through the pathway to glycerol.63 The lack of growth and requirement of rich medium components make successful application unlikely in case of a treatment system into which competing organisms can enter from air or via the influent. Probably E. coli is quite sensitive to TCP and some of its transformation products,64,65 which can be formed by various unspecific biotic reactions.40 It was proposed that the haloalkane dehalogenase activity was still too low to produce sufficient glycerol to generate energy for maintenance and growth. Furthermore, there is a mismatch in stereoselectivity between the most active haloalkane dehalogenase (DhaA31, makes mainly (S)-2,3-dichloro-1-propanol) and the halohydrin dehalogenase (HheC is selective for (R)-2,3-dichloro-1-propanol), explaining the accumulation of dichloropropanol in these cultures. E. coli probably is a less suitable host for development of bioremediation organisms, since the same initial haloalkane dehalogenase allowed growth of Pseudomonas strain mc-5222 and Pseudomonas putida.
The well-studied bacterium Pseudomonas putida strain KT2440![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 66 was more recently investigated as a host for TCP degradation.67 The constructed pathway for 2,3-dichloropropanol degradation was the same as the natural pathway of A. radiobacter,23,51,59 again employing the halohydrin dehalogenase (HheC)51 and epoxide hydrolase (EchA)60 obtained from that organism, just like the plasmid-based route studied in E. coli by Kurumbang et al.63 However, Gong et al. inserted the catabolic genes into the chromosome, avoiding the presence of transmissible plasmids and resistance markers (Fig. 5). They also carried out further genetic engineering to improve the strain, which may be a better host for heterologous catabolic pathways than E. coli since it is an established biodegradation organism that can be equipped with additional beneficial features.66 Two haloalkane dehalogenases were tested: DhaA31
66 was more recently investigated as a host for TCP degradation.67 The constructed pathway for 2,3-dichloropropanol degradation was the same as the natural pathway of A. radiobacter,23,51,59 again employing the halohydrin dehalogenase (HheC)51 and epoxide hydrolase (EchA)60 obtained from that organism, just like the plasmid-based route studied in E. coli by Kurumbang et al.63 However, Gong et al. inserted the catabolic genes into the chromosome, avoiding the presence of transmissible plasmids and resistance markers (Fig. 5). They also carried out further genetic engineering to improve the strain, which may be a better host for heterologous catabolic pathways than E. coli since it is an established biodegradation organism that can be equipped with additional beneficial features.66 Two haloalkane dehalogenases were tested: DhaA31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 57 and DhaA90R.58 The latter variant was found to be more useful since the chirality of the main product of TCP degradation ((R)-1,2-dichloro-3-propanol) better matched that of the subsequent HheC and EchA enzymes than the (S)-enantiomer mainly produced by DhaA31. Besides introduction of the pathway into the chromosome, the glpR gene, which encodes a negative regulator of glycerol utilization in Pseudomonas KT2440, was deleted. Furthermore, a gene encoding a hemoglobin-like protein was introduced to improve oxygen utilization under low-oxygen conditions. Finally, flagella synthesis genes were omitted. The resulting strain (P. putida KTU-TGVF) could use TCP as a growth substrate, and the growth rate was better than that of previously reported constructs. About 20 h were required for doubling of the cell density in a batch culture. As with Pseudomonas MC4-5222, P. putida KTU-TGVF was tested in an aerated packed bed bioreactor. Continuous degradation of 0.2 mM TCP was achieved at a hydraulic retention time of 133 h with 95–97% TCP removal, accompanied by quantitative release of chloride.67
57 and DhaA90R.58 The latter variant was found to be more useful since the chirality of the main product of TCP degradation ((R)-1,2-dichloro-3-propanol) better matched that of the subsequent HheC and EchA enzymes than the (S)-enantiomer mainly produced by DhaA31. Besides introduction of the pathway into the chromosome, the glpR gene, which encodes a negative regulator of glycerol utilization in Pseudomonas KT2440, was deleted. Furthermore, a gene encoding a hemoglobin-like protein was introduced to improve oxygen utilization under low-oxygen conditions. Finally, flagella synthesis genes were omitted. The resulting strain (P. putida KTU-TGVF) could use TCP as a growth substrate, and the growth rate was better than that of previously reported constructs. About 20 h were required for doubling of the cell density in a batch culture. As with Pseudomonas MC4-5222, P. putida KTU-TGVF was tested in an aerated packed bed bioreactor. Continuous degradation of 0.2 mM TCP was achieved at a hydraulic retention time of 133 h with 95–97% TCP removal, accompanied by quantitative release of chloride.67
The use of isolated enzymes and cell lysates for TCP dechlorination has also been considered, and this in fact originally triggered the dehalogenase research at Dow.68 However, the dehalogenation rates that have been achieved so far are too low for practical implementation in a TCP to epichlorohydrin recycling process. Other enzyme-based systems have been reported69–71 but these are not discussed in detail here, since the use of isolated enzymes will not allow the development of cost-effective robust processes required for prolonged groundwater bioremediation like the processes described above for DCA bioremediation. Such systems are operated under non-sterile conditions, and long-term survival of enzymes would require separate enzyme production and a process allowing for physical separation of the biocatalyst from groundwater. There may be attractive application opportunities if biotransformation of synthetic chemicals in waste streams leads to products that can be recycled, e.g. when a waste product can be converted to a chiral building block for chemical synthesis.58 Protein engineering of dehalogenases has already led to fascinating applications. Codexis and Pfizer engineered the A. radiobacter HheC by directed evolution to obtain a biocatalyst for statin side chain synthesis,72 and researchers at Promega Corp. engineered the Rhodococcus DhaA to develop the HaloTag system that is used to investigate cellular localization of proteins.73
An issue that received little attention during the development of strains growing on TCP (or other xenobiotic compounds) is the role of compartmentalization. The DCA catabolic pathway of X. autotrophicus6 generates the most toxic intermediate 2-chloroacetaldehyde in the periplasm, since the reaction is carried out by methanol dehydrogenase, a common periplasmic quinoprotein enzyme of methylotrophs like Xanthobacter. The 2,3-dichloropropanol-dehalogenating dehydrogenase that allows growth of Pseudomonas MC4 with that substrate (DppA in Fig. 4) produces the very toxic compound 2-chloroacrolein. The enzyme also resides in the periplasm. Introduction of the DCA dehalogenase gene from X. autotrophicus into a 2-chloroethanol mineralizing Pseudomonas strain unexpectedly did not allow the resulting recombinant strain to use DCA for growth, even though the introduced gene is well expressed and the pathway seems complete.20 A periplasmic localization of enzymes that produce the most reactive and toxic intermediates (cf. chloroacetaldehyde, chloroacrolein) would prevent unbalanced release of the toxic products in the cytoplasm, where reactions with nucleophilic sites on DNA, RNA and proteins may happen.
Opportunities for genetically engineered microbes
The application of GEMs requires strains that really solve problems with the implementation of bioremediation processes for removal of recalcitrant compounds that are of environmental concern. The TCP-degrading Pseudomonas strains described above are probably the best examples of genetically engineered (or metabolically engineered) bacteria that grow on a recalcitrant chlorinated chemical.20–67 These strains were obtained by a combination of classical microbiology, protein engineering including directed evolution, and metabolic engineering. Furthermore, laboratory-scale studies with these GEMs provided proof of principle for continuous treatment of TCP-contaminated water in packed bed bioreactors.20,67 Other groups have also investigated the development of bioremediation organisms by genetic engineering.8–13 However, to our knowledge, full-scale applications of genetically engineered strains for the bioremediation of xenobiotic compounds have not been reported so far. What hinders full scale implementation of these and other GEMs for practical cleanup operations?First, in comparison to pure or mixed natural cultures with a reputation of exceptional catabolic activity, engineered strains with better biodegradation potential towards important environmental chemicals are rare. A major problem is the technical difficulty of constructing microorganisms that degrade compounds which are really recalcitrant and for which no catabolic activity can be found in pure or mixed cultures that are conveniently obtained by classical adaptation or enrichment methods. TCP is a relatively simple compound, with only one hydrolytic reaction separating it from 2,3-dichloro-1-propanol for which microorganisms can be easily isolated. The structure differs only by one chloromethyl substituent from DCA, for which microorganisms also are found all over the globe. Yet, a decent body of protein engineering and metabolic engineering was required to obtain GEMs that degrade TCP and use it for growth.20,67 In case of important compounds like the trichloroethanes, trichloroethene or various chlorinated pesticides, the task becomes increasingly complicated, and most past achievements with less recalcitrant compounds (like chlorinated aromatics) are scientifically impressive but of low practical value. A serious issue is that GEMs for bioremediation do not always utilize the target substrate as well as expected, even if the pathway seems well designed biochemically. Unwanted side reactions of chlorinated compounds can easily yield reactive products, and toxic effects of such reactive intermediates formed during metabolism of halogenated aliphatics are reported, also for the compounds discussed in this perspective.40,65,74 The development of rapid tools for synthetic biology and genome engineering (e.g. DNA synthesis, combinatorial and multiplex methods for gene recombination and integration) will certainly accelerate the pace at which known bottlenecks can be solved, but the identification of these bottlenecks is still a tedious and time consuming task. Like with directed evolution, the use of high-throughput approaches in metabolic engineering may circumvent some of these design problems since the rate at which functional genetic diversity can be created will continue to increase.
Sometimes, biosafety is mentioned as a cause of modest progress with the application of genetically engineered organisms (GMOs) for environmental cleanup.16,43,67 There are different aspects to this: ecological, health-related and regulatory. Regarding ecological effects, one should consider the functional properties associated with the genetic changes that are introduced in engineered bioremediation organisms, including the way such changes could impact ecological behavior. It seems impossible that introduction of a few catabolic genes encoding enzymes that act specifically on xenobiotic compounds that should not occur in nature anyway (like TCP) would have ecological effects beyond persistence of such organisms in polluted environments and improved degradation of the target compounds. The main concern may be the formation of reactive side- or end products, like it occurs during stepwise anaerobic reductive dechlorination of tetra- and trichloroethene. Introduction of the catabolic genes of interest in the chromosome of a GEM and avoiding transposons, plasmids, and antibiotic resistance markers, is recommended and possible. This should prevent additional mobilization of genetic material between replicons and organisms, a process that widely happens in nature. Avoiding pathogenic hosts and antibiotic resistance markers will also prevent health-related side effects. The regulatory context is very much dependent on regional, national and international provisions. In most European countries, release of GMOs is prohibited unless permission is obtained. However, we are not aware of any granted or non-granted request for permission to use genetically engineered microorganisms in bioremediations schemes. The only well documented case seems to be the release of a bioluminescent GEMs for monitoring purposes.75 Acceptance of the use of GMOs in the environment – at least in Europe – is low. Consequently, procedures require detailed risk assessment studies and are time-consuming, probably not considered worth the effort if there is no large gain. However, a real benefit of using recombinant organisms can only be expected if these GEMs outperform natural organisms – of which there are probably still no proven examples, apart from the laboratory scale TCP removal by engineered strains described above.
Regulatory and responsibility issues can delay innovative clean-up actions for up to tens of years. US-EPA legislation demands positive results of pilot tests for a given technology to be implemented. To the best of our knowledge and not surprisingly, bioremediation of TCP-contaminated groundwater by engineered microbes has not been considered so far. At Tyson's dump site, groundwater that is apparently free of suspended solids is currently pumped and treated using two 20 m3 granular activated carbon columns. Over the 5 year treatment period an average of 700 g of volatile organic carbon was removed daily, with TCP being the primary chlorinated organic chemical. Dense non-aqueous phase liquids (DNAPLs) are present at very deep levels, including in rock fissures, so spreading of contaminants may continue for a long time. Apparently, the costs for running a groundwater treatment plant at the relatively low extraction rate of 5 m3 h−1 are still economically bearable.
Bioaugmentation in the sense of introducing cultures with favorable degradation abilities is an established technology for in situ groundwater treatment by reductive dehalogenation, e.g. in case of pollution with chlorinated solvents such as trichloroethene and tetrachloroethene.76–78 It was also used for on-site groundwater treatment as explained above for the removal of DCA from extracted groundwater. In general, introducing organisms will be helpful in case of xenobiotic compounds which are not rapidly degraded by individual strains or consortia of naturally evolved organisms that are already present. In situ and bioreactor bioaugmentation can be effective in case cultures are available that use the target compound as a growth-stimulating substrate, in case of aerobic processes either as carbon source or electron donor. Introduction of host organisms carrying transmissible genetic material that can spread in the natural microbial communities may also stimulate biodegradation but may be less acceptable from a regulatory point of view. Stable and long-term establishment of a population of GEMs will require an ecological niche for these organisms. To avoid the need of repeated inoculation with large amounts of cultures, introduced organisms should not only persist but also proliferate as long as pollutants are present. Such a niche that allows growth will exist if the target compounds are present at concentrations that support growth and maintenance of the GEMs. If a specific recalcitrant compound is a minor component in a stream of polluted groundwater, a dedicated GEM will have difficulty establishing itself in a bioreactor, especially if the host is not an organism adapted to survival in a complex ecosystem. Compounds often occurring as predominant pollutants are solvents and intermediates in chemical synthesis at places where leakages of storage tanks have occurred. Examples are DCA, TCP, chloroform, and trichloroethanes. Improper waste disposal of side products from chemical synthesis tends to lead to more complex mixtures of pollutants, where only a battery of organisms would help. Obviously, the genetic construction of such strain collections would be an enormous task.
The presence of recalcitrant chemicals, including solvents and pharmaceuticals in groundwater and surface water used for drinking water preparation is a problem of increasing concern. Biological removal of such compounds, which often occur at trace concentrations, will likely remain problematic even in case suitable microorganisms would be found or constructed. Such decorated organisms will have little competitive advantage to establish themselves in complex microbial communities. Low concentrations such as defined in Alaska (groundwater cleanup standard for TCP is 0.0075 μg l−1) are unlikely to be met by biological methods. Furthermore, feed concentrations of below 1 mg l−1 will delay the development of good aerobic biofilms unless the system is fed with huge quantities of water with considerable mass flow of the chemicals under consideration. This fact is even more important for chlorinated chemicals, as a large fraction of the compound's mass (TCP is 71% chlorine by mass) does not contribute to growth. It is conceivable that high initial concentrations of a contaminant and large amounts of inoculum would allow the development of an active biofilm in which desired activity stays long enough for prolonged treatment of low concentrations, but there is little experimental evidence for such a scenario. The prospects of application of specialized organisms, such as the recently evolved DCA degraders and the TCP degrading GEMs described above, are best in case of concentrated waste streams and spills, with pollutants that have a high water solubility and can be extracted via a pump-and-treat approach, and in the absence of more cost and energy efficient technologies. The combination of activated carbon filters inoculated with special microorganisms that can degrade micropollutants is also an interesting prospect,79 especially if concentrating effects of activated carbon and positive effects of immobilization on population dynamics would act synergistically.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Prof. Thomas Leisinger for initiating and supporting key steps of these studies.References
- M. H. van Agteren, S. Keuning and D. B. Janssen, Handbook on Biodegradation and Biological Treatment of Hazardous Organic Compounds, Kluwer/Springer, Dordrecht, the Netherlands, 1998 Search PubMed.
- L. P. Wackett and C. D. Hershberger, Biocatalysis and Biodegradation: Microbial Transformation of Organic Compounds, ASM Press, Washington, DC, USA, 2001 Search PubMed.
- S. Fetzner, Biodegradation of xenobiotics, in Biotechnology, ed. H. W. Doelle, S. Rokem and M. Berovic, Unesco, EoLSS Publishers, Oxford, UK, 2009, Vol X Search PubMed.
- T. C. Hazen, Cometabolic Bioremediation, in Handbook of Hydrocarbon and Lipid Microbiology, ed. K. N. Timmis, Springer, Berlin, Germany, 2010 Search PubMed.
- G. Stucki, U. Krebser and T. Leisinger, Bacterial growth on 1,2-dichloroethane, Experientia, 1983, 39, 1271–1273 CrossRef CAS PubMed.
- D. B. Janssen, A. Scheper, L. Dijkhuizen and B. Witholt, Degradation of halogenated aliphatic compounds by Xanthobacter autotrophicus GJ10, Appl. Environ. Microbiol., 1985, 49, 673–677 CrossRef CAS PubMed.
- G. Stucki and M. Thüer, Experiences of a large-scale application of 1,2-dichloroethane degrading microorganisms for groundwater treatment, Environ. Sci. Technol., 1995, 29, 2339–2345 CrossRef CAS PubMed.
- I. Cases and V. de Lorenzo, Genetically modified organisms for the environment: stories of success and failure and what we have learned from them, Int. Microbiol., 2005, 8, 213–222 CAS.
- W. Chen, F. Brühlmann, R. D. Richins and A. Mulchandani, Engineering of improved microbes and enzymes for bioremediation, Curr. Opin. Biotechnol., 1999, 10, 137–141 CrossRef CAS PubMed.
- P. C. K. Lau and V. de Lorenzo, Genetic engineering: the frontier of bioremediation, Environ. Sci. Technol., 1999, 33, 124A–128A CrossRef CAS PubMed.
- K. N. Timmis and D. H. Pieper, Bacteria designed for bioremediation, Trends Biotechnol., 1999, 5, 201–204 CrossRef.
- D. H. Pieper and W. Reineke, Engineering bacteria for bioremediation, Curr. Opin. Biotechnol., 2000, 11, 262–270 CrossRef CAS PubMed.
- K. Furukawa, ‘Super bugs’ for bioremediation, Trends Biotechnol., 2003, 21, 187–190 CrossRef CAS.
- J. S. Singh, P. C. Abhilash, H. B. Singh, R. P. Singh and D. P. Singh, Genetically engineered bacteria: an emerging tool for environmental remediation and future research perspectives, Gene, 2011, 480, 1–9 CrossRef CAS PubMed.
- V. de Lorenzo, Cleaning up behind us. The potential of genetically modified bacteria to break down toxic pollutants in the environment, EMBO Rep., 2001, 5, 357–359 CrossRef PubMed.
- G. S. Sayler and S. Ripp, Field applications of genetically engineered microorganisms for bioremediation processes, Curr. Opin. Biotechnol., 2000, 11, 286–289 CrossRef CAS PubMed.
- P. Dvorak, P. I. Nikel, J. Damborský and V. de Lorenzo, Bioremediation 3.0: Engineering pollutant-removing bacteria in the times of systemic biology, Biotechnol. Adv., 2017, 35, 845–866 CrossRef CAS PubMed.
- T. Bosma, E. Kruizinga, E. J. D. Bruin, G. J. Poelarends and D. B. Janssen, Utilization of trihalogenated propanes by Agrobacterium radiobacter AD1 through heterologous expression of the haloalkane dehalogenase from Rhodococcus sp. strain m15–3, Appl. Environ. Microbiol., 1999, 65, 4575–4581 CrossRef CAS PubMed.
- T. Bosma, J. Damborsky, G. Stucki and D. B. Janssen, Biodegradation of 1,2,3-trichloropropane through directed evolution and heterologous expression of a haloalkane dehalogenase gene, Appl. Environ. Microbiol., 2002, 68, 3582–3587 CrossRef CAS PubMed.
- G. Samin, M. Pavlova, M. I. Arif, C. P. Postema, J. Damborsky and D. B. Janssen, A Pseudomonas putida strain genetically engineered for 1,2,3-trichloropropane bioremediation, Appl. Environ. Microbiol., 2014, 80, 5467–5476 CrossRef PubMed.
- K. H. Verschueren, F. Seljée, H. J. Rozeboom, K. H. Kalk and B. W. Dijkstra, Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase, Nature, 1993, 693–698 CrossRef CAS PubMed.
- D. B. Janssen, Evolving haloalkane dehalogenases, Curr. Opin. Chem. Biol., 2004, 8, 150–159 CrossRef CAS PubMed.
- A. J. van den Wijngaard, K. W. H. J. van der Kamp, J. van der Ploeg, F. Pries, B. Kazemier and D. B. Janssen, Degradation of 1,2-dichloroethane by Ancylobacter aquaticus and other facultative methylotrophs, Appl. Environ. Microbiol., 1992, 58, 976–983 CrossRef CAS PubMed.
- J. S. Song, D. H. Lee, K. Lee and C. K. Kim, Genetic organization of the dhlA gene encoding 1,2-dichloroethane dechlorinase from Xanthobacter flavus UE15, Int. J. Microbiol., 2004, 42, 188–193 CAS.
- G. Algasan and B. Pillay, Biochemical activities of 1, 2-dichloroethane (DCA) degrading bacteria, Afr. J. Biotechnol., 2011, 10, 11567–11573 Search PubMed.
- J. Munro, E. Liew, M. Ly and N. Coleman, A new catabolic plasmid in Xanthobacter and Starkeya spp. from a 1,2-dichloroethane-contaminated site, Appl. Environ. Microbiol., 2016, 82, 5298–5308 CrossRef CAS PubMed.
- H. C. Hage and S. Hartmans, Monooxygenase-mediated 1,2-dichloroethane degradation by Pseudomonas sp. strain DCA1, Appl. Environ. Microbiol., 1999, 65, 2466–2470 CrossRef PubMed.
- B. van der Zaan, J. de Weert, H. Rijnaarts, W. de Vos, H. Smidt and J. Gerritse, Degradation of 1,2-dichloroethane by microbial communities from river sediment at various redox conditions, Water Res., 2009, 43, 3207–3216 CrossRef CAS PubMed.
- M. J. Dinglasan-Panlilio, S. Dworatzek, S. Mabury and E. Edwards, Microbial oxidation of 1,2-dichloroethane under anoxic conditions with nitrate as electron acceptor in mixed and pure cultures, FEMS Microbiol. Ecol., 2006, 56, 355–364 CrossRef CAS PubMed.
- G. Tardif, C. W. Greer, D. Labbé and P. C. Lau, Involvement of a large plasmid in the degradation of 1,2-dichloroethane by Xanthobacter autotrophicus, Appl. Environ. Microbiol., 1991, 57, 1853–1857 CrossRef CAS PubMed.
- F. Pries, A. J. van den Wijngaard, R. Bos, M. Pentenga and D. B. Janssen, The role of spontaneous cap domain mutations in haloalkane dehalogenase specificity and evolution, J. Biol. Chem., 1994, 269, 17490–17494 CAS.
- M. G. Pikkemaat and D. B. Janssen, Generating segmental mutations in haloalkane dehalogenase: a novel part in the directed evolution toolbox, Nucleic Acids Res., 2002, 30, e35 CrossRef PubMed.
- G. Stucki, M. Thüer and R. Bentz, Biological degradation of 1,2-dichloroethane under groundwater conditions, Water Res., 1992, 26, 273–278 CrossRef CAS.
- L. M. Freitas dos Santos and A. G. Livingston, Extraction and biodegradation of a toxic volatile organic compound (1,2-dichloroethane) from waste-waters in a membrane bioreactor, Appl. Microbiol. Biotechnol., 1994, 42, 421–431 CrossRef CAS PubMed.
- L. M. Freitas dos Santos and A. G. Livingston, Novel membrane bioreactor for detoxification of VOC wastewaters: biodegradation of 1,2-dichloroethane, Water Res., 1995, 29, 179–194 CrossRef CAS.
- G. Stucki, Ablösung einer Pump-and-treat-Maßnahme durch Monitored Natural Attenuation, Terratech, 2011, pp. 5–7 Search PubMed.
- J. W. Bijsterbosch, A. Das and F. P. J. M. Kerkhof, Clean technology in the production of epichlorohydrin, J. Cleaner Prod., 1994, 2, 181–184 CrossRef.
- B. M. Bell, J. R. Briggs, R. M. Campbell, S. M. Chambers, P. D. Gaarenstroom, J. G. Hippler, B. D. Hook, K. Kearns, J. M. Kenney, W. J. Kruper, D. J. Schreck, C. N. Theriault and C. P. Wolfe, Glycerin as a renewable feedstock for epichlorohydrin production, Clean, 2008, 36, 657–661 CAS.
- United States Environmental Protection Agency, https://cumulis.epa.gov/supercpad/cursites/csitinfo.cfm?id=0301436.
- G. Samin and D. B. Janssen, Transformation and biodegradation of 1,2,3-trichloropropane (TCP), Environ. Sci. Pollut. Res. Int., 2012, 19, 3067–3078 CrossRef CAS PubMed.
- F. E. Loffler, J. E. Champine, K. M. Ritalahti, S. J. Sprague and J. M. Tiedje, Complete reductive dechlorination of 1,2-dichloropropane by anaerobic bacteria, Appl. Environ. Microbiol., 1997, 63, 2870–2875 CrossRef CAS PubMed.
- K. S. Bowman, M. F. Nobre, M. S. da Costa, F. A. Rainey and W. M. Moe, Dehalogenimonas alkenigignens sp. nov., a chlorinated-alkane-dehalogenating bacterium isolated from groundwater, Int. J. Syst. Evol. Microbiol., 2013, 63, 1492–1498 CrossRef CAS PubMed.
- J. Yan, B. A. Rash, F. A. Rainey and W. M. Moe, Isolation of novel bacteria within the Chloroflexi capable of reductive dechlorination of 1,2,3-trichloropropane, Environ. Microbiol., 2009, 11, 833–843 CrossRef CAS PubMed.
- K. Mukherjee, K. S. Bowman, F. A. Rainey, S. Siddaramappa, J. F. Challacombe and W. M. Moe, Dehalogenimonas lykanthroporepellens BL-DC-9T simultaneously transcribes many rdhA genes during organohalide respiration with 1,2-DCA, 1,2-DCP, and 1,2,3-TCP as electron acceptors, FEMS Microbiol. Lett., 2014, 354, 111–118 CrossRef CAS PubMed.
- T. Bosma and D. B. Janssen, Conversion of chlorinated propanes by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase, Appl. Microbiol. Biotechnol., 1998, 50, 105–112 CrossRef CAS.
- B. Wang and K. H. Chu, Cometabolic biodegradation of 1,2,3-trichloropropane by propane-oxidizing bacteria, Chemosphere, 2017, 168, 1494–1497 CrossRef CAS PubMed.
- E. J. Bylaska, K. R. Glaesemann, A. R. Felmy, M. Vasiliu, D. A. Dixon and P. G. Tratnyek, Free energies for degradation reactions of 1,2,3-trichloropropane from ab initio electronic structure theory, J. Phys. Chem., 2010, 114, 12269–12282 CrossRef CAS PubMed.
- T. L. Torralba-Sanchez, E. J. Bylaska, A. J. Salter-Blanc, D. E. Meisenheimer, M. A. Lyon and P. G. Tratnyek, Reduction of 1,2,3-trichloropropane (TCP) reduction: Pathways and mechanisms from computational chemistry calculations, Environ. Sci.: Processes Impacts, 2020 Search PubMed , in press..
- V. Sarathy, A. J. Salter, J. T. Nurmi, J. G. G. O'Brien Johnson, R. L. Johnson and P. G. Tratnyek, Degradation of 1,2,3-trichloropropane (TCP): hydrolysis, elimination, and reduction by iron and zinc, Environ. Sci. Technol., 2010, 44, 787–793 CrossRef CAS PubMed.
- A. J. Salter-Blanc and P. G. Tratnyek, Effects of solution chemistry on the dechlorination of 1,2,3-trichloropropane by zero-valent zinc, Environ. Sci. Technol., 2011, 45, 4073–4079 CrossRef CAS PubMed.
- A. J. van den Wijngaard, D. B. Janssen and B. Witholt, Degradation of epichlorohydrin and halohydrins by bacterial cultures isolated from freshwater sediment, J. Gen. Microbiol., 1989, 135, 2199–2208 CAS.
- A. M. Fauzi, D. J. Hardman and A. T. Bull, Biodehalogenation of low concentrations of 1,3-dichloropropanol by mono- and mixed cultures of bacteria, Appl. Microbiol. Biotechnol., 1996, 46, 660–666 CrossRef CAS PubMed.
- T. Yokota, T. Omori and T. Kodama, Purification and properties of haloalkane dehalogenase from Corynebacterium sp. strain m15–3, J. Bacteriol., 1987, 169, 4049–4054 CrossRef CAS PubMed.
- A. N. Kulakova, M. J. Larkin and L. A. Kulakov, The plasmid-located haloalkane dehalogenase gene from Rhodococcus rhodochrous NCIMB 13064, Microbiology, 1997, 143, 109–115 CrossRef CAS PubMed.
- J. Newman, T. S. Peat, R. Richard, L. Kan, P. E. Swanson, J. A. Affholter, I. H. Holmes, J. F. Schindler, T. C. Unkefer and T. C. Terwilliger, Haloalkane dehalogenases: structure of a Rhodococcus enzyme, Biochemistry, 1999, 38, 16105–16114 CrossRef CAS PubMed.
- K. A. Gray, T. H. Richardson, K. Kretz, J. M. Short, F. Bartnek, R. Knowles, L. Kan, P. E. Swanson and D. E. Robertson, Rapid evolution of reversible denaturation and elevated melting temperature in a microbial haloalkane dehalogenase, Adv. Synth. Catal., 2001, 343, 607–617 CrossRef CAS.
- M. Pavlova, M. Klvana, Z. Prokop, R. Chaloupkova, P. Banas, M. Otyepka, R. C. Wade, M. Tsuda, Y. Nagata and J. Damborsky, Redesigning dehalogenase access tunnels as a strategy for degrading an anthropogenic substrate, Nat. Chem. Biol., 2009, 10, 727–733 CrossRef PubMed.
- J. G. van Leeuwen, H. J. Wijma, R. J. Floor, J. M. van der Laan and D. B. Janssen, Directed evolution strategies for enantiocomplementary haloalkane dehalogenases: from chemical waste to enantiopure building blocks, ChemBioChem, 2012, 13, 137–148 CrossRef CAS PubMed.
- J. E. T. van Hylckama Vlieg, L. Tang, J. H. Lutje Spelberg, T. Smilda, G. J. Poelarends, T. Bosma, A. E. J. van Merode, M. W. Fraaije and D. B. Janssen, Halohydrin dehalogenases are structurally and mechanistically related to short-chain dehydrogenases/reductases, J. Bacteriol., 2001, 183, 5058–5066 CrossRef CAS PubMed.
- R. Rink, M. Fennema, M. Smids, U. Dehmel and D. B. Janssen, Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1, J. Biol. Chem., 1997, 272, 14650–14657 CrossRef CAS PubMed.
- M. I. Arif, G. Samin, J. G. van Leeuwen, J. Oppentocht and D. B. Janssen, Novel dehalogenase mechanism for 2,3-dichloro-1-propanol utilization in Pseudomonas putida strain MC4, Appl. Environ. Microbiol., 2012, 78, 6128–6136 CrossRef CAS PubMed.
- G. Samin, Genetic Engineering for Trichloropropane Degradation, PhD thesis, Univ. of Groningen, 2012.
- N. P. Kurumbang, P. Dvorak, J. Bendl, J. Brezovsky, Z. Prokop and J. Damborsky, Computer-assisted engineering of the synthetic pathway for biodegradation of a toxic persistent pollutant, ACS Synth. Biol., 2014, 3, 172–181 CrossRef CAS PubMed.
- P. Dvorak, L. Chrast, P. I. Nikel, R. Fedr, K. Soucek, M. Sedlackova, R. Chaloupkova, V. de Lorenzo V, Z. Prokop and J. Damborsky, Exacerbation of substrate toxicity by IPTG in Escherichia coli BL21(DE3) carrying a synthetic metabolic pathway, Microb. Cell Fact., 2015, 14, 201 CrossRef PubMed.
- M. Demko, L. Chrast, P. Dvorak, J. Damborsky and D. Safranek, Computational modelling of metabolic burden and substrate toxicity in Escherichia coli carrying a synthetic metabolic pathway, Microorganisms, 2019, 11, E553 CrossRef PubMed.
- E. Martínez-García, P. I. Nikel, T. Aparicio and V. de Lorenzo, Pseudomonas 2.0: genetic upgrading of P. putida KT2440 as an enhanced host for heterologous gene expression, Microb. Cell Fact., 2014, 13, 159 CrossRef PubMed.
- T. Gong, X. Xu, Y. Che, R. Liu, W. Gao, F. Zhao, H. Yu, J. Liang, P. Xu, C. Song and C. Yang, Combinatorial metabolic engineering of Pseudomonas putida KT2440 for efficient mineralization of 1,2,3-trichloropropane, Sci. Rep., 2017, 7, 7064 CrossRef PubMed.
- P. E. Swanson, Dehalogenases applied to industrial-scale biocatalysis, Curr. Opin. Biotechnol., 1999, 10, 365–369 CrossRef CAS PubMed.
- P. Dvorak, S. Bidmanova, J. Damborsky and Z. Prokop, Immobilized synthetic pathway for biodegradation of toxic recalcitrant pollutant 1,2,3-trichloropropane, Environ. Sci. Technol., 2014, 48, 6859–6866 CrossRef CAS PubMed.
- L. Yang, E. M. Dolan, S. K. Tan, T. Lin, E. D. Sontag and S. D. Khare, Computation-guided design of a stimulus-responsive multienzyme supramolecular assembly, ChemBioChem, 2017, 18, 2000–2006 CrossRef CAS PubMed.
- T. F. Bogale, I. Gul, L. Wang, J. Deng, Y. Chen, M. Majeric-Elenkov and L. Tang, Biodegradation of 1,2,3-trichloropropane to valuable (S)-2,3-DCP using a one-pot reaction system, Catalysts, 2020, 10, 3 CrossRef CAS.
- S. K. Ma, J. Gruber, C. Davis, L. Newman, D. Gray, A. Wang, J. Grate, G. W. Huisman and R. A. Sheldon, A green-by-design biocatalytic process for atorvastatin intermediate, Green Chem., 2010, 12, 81–86 RSC.
- G. V. Los, L. P. Encell, M. G. McDougall, D. D. Hartzell, N. Karassina and C. Zimprich, HaloTag: a novel protein labeling technology for cell imaging and protein analysis, ACS Chem. Biol., 2008, 3, 373–382 CrossRef CAS PubMed.
- J. van der Ploeg, M. P. Smidt, A. S. Landa and D. B. Janssen, Identification of chloroacetaldehyde dehydrogenase involved in 1,2-dichloroethane degradation, Appl. Environ. Microbiol., 1994, 60, 1599–1605 CrossRef CAS PubMed.
- S. Ripp, D. E. Nivens, Y. Ahn, C. Werner, J. Jarrell, J. P. Easter, C. D. Cox, S. Burlage and G. S. Sayler, Controlled field release of a bioluminescent genetically engineered microorganism for bioremediation process monitoring and control, Environ. Sci. Technol., 2000, 34(5), 846–853 CrossRef CAS.
- L. Semprini, In situ bioremediation of chlorinated solvents, Environ. Health Perspect., 1995, 103, 101–105 CAS.
- Bioaugmentation for groundwater remediation, ed. H. F. Stroo, A. Leeson and C. H. Ward, Springer, New York, USA, 2013 Search PubMed.
- GeoSyntec Consultants, Bioaugmentation for Remediation of Chlorinated Solvents: (ii) Technology Development, Status, and Research Needs, DoD ESTCP Program, Washington, DC, USA, 2005, retrieved from, https://www.serdp-estcp.org Search PubMed.
- J. Benner, D. E. Helbling, H. P. Kohler, J. Wittebol, E. Kaiser, C. Prasse, T. A. Ternes, C. N. Albers, J. Aamand, B. Horemans, D. Springael, E. Walravens and N. Boon, Is biological treatment a viable alternative for micropollutant removal in drinking water treatment processes?, Water Res., 2013, 47, 5955–5976 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |