Open Access Article
Open Access ArticleOleuropein and its peracetylated derivative negatively regulate osteoclastogenesis by controlling the expression of genes involved in osteoclast differentiation and function
Maria Angeles
Rosillo
 *ab,
Sergio
Montserrat-de-la-Paz
*ab,
Sergio
Montserrat-de-la-Paz
 c,
Rocio
Abia
a,
Maria Luisa
Castejon
c,
Rocio
Abia
a,
Maria Luisa
Castejon
 b,
Maria Carmen
Millan-Linares
b,
Maria Carmen
Millan-Linares
 d,
Catalina
Alarcon-de-la-Lastra
b,
Jose G.
Fernandez-Bolaños
d,
Catalina
Alarcon-de-la-Lastra
b,
Jose G.
Fernandez-Bolaños
 e and
Francisco J. G.
Muriana
*a
e and
Francisco J. G.
Muriana
*a
aLaboratory of Cellular and Molecular Nutrition, Instituto de la Grasa, CSIC, 41013 Seville, Spain. E-mail: rosillo@ig.csic.es
bDepartment of Pharmacology, School of Pharmacy, University of Seville, 41012 Seville, Spain
cDepartment of Medical Biochemistry, Molecular Biology and Immunology, School of Medicine, University of Seville, 41009 Seville, Spain
dCell Biology Unit, Instituto de la Grasa, CSIC, 41013 Seville, Spain
eDepartment of Organic Chemistry, School of Chemistry, University of Seville, 41012 Seville, Spain
First published on 17th April 2020
Abstract
During chronic inflammation, macrophage colony-stimulating factor (M-CSF) and receptor activator of nuclear factor-κB ligand (RANKL) have well established effects on gene networks that stimulate osteoclastogenesis, which is the culprit of several bone diseases. In this study, we investigated the anti-osteoclastogenic effects in vitro of oleuropein (OL) and its peracetylated derivative (Per-OL) by exploring the expression level of key hub genes involved in fate decision and lineage commitment, differentiation, and function of human blood monocyte-derived osteoclasts. Monocytes were purified from peripheral blood mononuclear cells of healthy individuals using commercial antibodies coated with magnetic beads and treated with M-CSF/RANKL in the presence or absence of OL or Per-OL (25 and 50 μM) for 6 days. We demonstrated that OL and especially Per-OL impair transcriptional gene circuits able to support osteoclastogenesis from human blood monocytes. Our results indicate that OL and notably Per-OL are promising candidates to control osteoclastogenesis.
Introduction
Bone homeostasis is a dynamic complex process involving equilibrium of opposite driving forces for bone resorption by osteoclasts and bone formation by osteoblasts. The disruption of bone homeostasis in favour of osteoclasts leads to manifestation of several pathological conditions, such as osteoporosis, rheumatoid arthritis, lytic bone metastases, and Paget's bone disease.1–3 Osteoclastogenesis is primarily mediated by cells of the monocyte/macrophage lineage, which act as upstream precursors of osteoclasts upon stimulation with the macrophage colony-stimulating factor (M-CSF) and the ligand for the receptor activator of nuclear factor-κB (RANKL). M-CSF/RANKL signalling dictates the transcriptional activation of genes involved in the commitment of pre-osteoclasts, including the TNFRSF11A gene that encodes the receptor for RANKL (RANK) providing an autocrine loop to amplify the input signal of RANKL for further differentiation into mononucleated pre-osteoclasts and then into mature and functional multinucleated osteoclasts.4,5 Therefore, osteoclasts are target cells to arrest their differentiation and/or function for the treatment of osteoclastogenic disorders.Antiresorptive medication is often associated with many safety concerns.6 Owing to these limitations, natural products, mainly phytochemicals are becoming an important research field to explore new therapeutic choices for bone diseases. Oleuropein (OL) is a secoiridoid glycoside that typically represents the most prominent hydrophilic phenolic compound in olives and olive leaves. By enzymatic or chemical hydrolysis, OL can degrade to yield its aglyconic form and hydroxytyrosol, which may be found naturally in olive oils.7 On the other hand, full acetylated or peracetylated OL (Per-OL) has been recently shown to be a safe and biologically active semi-synthetic stable derivative with better bioavailability and sometimes even with better biological activity than OL.8 After consumption, this approach designed to preserve OL integrity allows the OL to be released in the surrounding extracellular milieu or inside cells upon extensive deacetylation by secreted or cytosolic esterases.9 Pharmacological effects of OL such as antioxidant, antihypertensive, hypoglycaemic, cardioprotective, neuroprotective, antimicrobial, and anti-inflammatory, and as coadjuvant in multitarget anti-cancer treatments have already been outlined.10–13 OL also elicits osteoprotective effects by promoting differentiation of human bone marrow mesenchymal stem cells into osteoblasts14,15 and by reducing tartrate-resistant acid phosphatase (TRAP) activity in osteoclasts formed from mouse spleen cells.16 In addition, Per-OL has been confirmed to lessen carrageenan-induced paw oedema in rats17 and metabolic abnormalities in a mouse model of high-fat induced obesity,18 and also to exhibit antioxidant and antiproliferative effects in human breast19 and thyroid20 cancer cell lines. However, the effects of OL and Per-OL on any stage of osteoclast development of the human monocyte/macrophage lineage have not yet been reported. In the present study, Per-OL was prepared by environmentally friendly protocols, and the effects of OL and Per-OL on osteoclast differentiation and function were investigated. We put the focus on the transcriptional activity of genes related to the differentiation process from committed pre-osteoclasts to mature and functional osteoclasts.
Materials and methods
Chemicals
OL was extracted from olive leaves according to the reported literature21 and then purified using silica gel column chromatography (CH2Cl2/MeOH, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 5
1 to 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) to give a yellow solid (Scheme 1A). For obtaining Per-OL, OL (1 g, 1.85 mmol) was solved in pyridine/acetic anhydride (1
1 v/v) to give a yellow solid (Scheme 1A). For obtaining Per-OL, OL (1 g, 1.85 mmol) was solved in pyridine/acetic anhydride (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) at 0 °C for 10 min and then the reaction was kept at room temperature overnight. After hydrolysing the acetic anhydride, the solution was concentrated to dryness and the crude residue was purified using silica gel column chromatography (ethyl acetate–hexane, 1
1 v/v) at 0 °C for 10 min and then the reaction was kept at room temperature overnight. After hydrolysing the acetic anhydride, the solution was concentrated to dryness and the crude residue was purified using silica gel column chromatography (ethyl acetate–hexane, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Spectroscopy data of the resultant brown solid (Scheme 1B) was identical to that reported in literature.22
1). Spectroscopy data of the resultant brown solid (Scheme 1B) was identical to that reported in literature.22
Human monocytes
This study was conducted according to Good Clinical Practice Guidelines and in line with the principles outlined in the Helsinki Declaration of the World Medical Association. Study subjects were recruited at Virgen del Rocio University Hospital (VRUH, Seville, Spain). Informed consent for the study was obtained. All protocols were approved by the local institutional review board (Ethics Committee of the VRUH, #OLNAMS 08/11). Venous blood samples were obtained from healthy adult volunteers (<35 years old) non-smokers and not taking any medication. Donors were recognized as healthy, according to medical history and routine laboratory tests. Peripheral blood mononuclear cells (MNCs) were isolated by centrifugation over a Ficoll-Histopaque (Sigma-Aldrich, St Louis, MO, USA) gradient.23 Monocytes were isolated from peripheral blood MNCs using CD14 microbeads and LS columns on a midiMACS system (Miltenyi Biotec, Madrid, Spain). Monocyte (CD14+) purity was routinely >90% by flow cytometry analysis (FACSCanto II flow cytometer and FACSDiva software; Becton Dickinson, San Jose, CA, USA) and cell viability >95% by trypan blue exclusion (Sigma-Aldrich). The monocytes were seeded in 24-well culture plates at a density of 1 × 106 cells per mL and cultured in ultra-low attachment flasks in RPMI 1640 medium supplemented with L-glutamine, penicillin, streptomycin, and 10% heat-inactivated foetal bovine serum (complete culture medium).Osteoclast differentiation and viability
Monocytes were induced to differentiate into osteoclasts for 6 days with human recombinant M-CSF (50 ng mL−1) and RANKL (50 ng mL−1) in the absence or presence OL or Per-OL (25 and 50 μM). These concentrations were within the range of concentrations consistent with the in vitro beneficial effects of oleuropein or Per-OL on different cell types.16,24–27 Complete culture medium was replaced every 2 days with fresh medium, the cytokines, and OL or Per-OL. For cell viability, monocytes were seeded in 96-well plates (1 × 105 cells per well) and differentiated into osteoclasts as indicated above, in the absence or presence of OL or Per-OL (in the range of 12.5–50 μM). At day 6, methylthiazolyldiphenyl-tetrazolium bromide (MTT) solution (Sigma-Aldrich) was added to cells for 2 h until a purple precipitate was visible. MTT-formazan crystals were then solubilized with DMSO (Sigma-Aldrich) and measured with a microplate reader at 570 nm corrected to 650 nm. Cell survival was expressed as the percentage of absorbance compared with that of control, nontreated cells.TRAP staining and activity assay
TRAP staining was performed using an acid phosphatase leukocyte kit (Sigma-Aldrich) in osteoclasts fixed with 4% paraformaldehyde. TRAP-positive multinuclear cells containing more than three nuclei were scored as osteoclasts and examined using an Olympus microscope (40×) (Model IX71, Olympus, Center Valley, PA, USA). TRAP activity was determined using a TRAP assay kit (Takara Bio, Shiga, Japan). Briefly, 50 μL of cell extract and 50 μL of p-nitro-phenyl phosphate (pNPP) were mixed with a sodium tartrate solution and incubated at 37 °C for 25 min. The reaction was stopped with 0.5 N NaOH and the absorbance was measured at 405 nm. A solution containing acid phosphatase and pNPP, but not the sodium tartrate solution, was used as a control.RNA isolation and quantitative PCR
Total RNA was extracted by using Trisure Reagent (Bioline, London, UK). RNA quality was assessed by A260/A280 ratio in a NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Greenville, SC, USA). Briefly, RNA (250 ng) was subjected to reverse transcription (iScript, Bio-Rad, Madrid, Spain). An amount of 20 ng of the resulting cDNA was used as a template for real-time PCR amplifications. The mRNA levels for specific genes were determined in a CFX96 system (Bio-Rad). For each PCR reaction, cDNA template was added to Brilliant SYBR green QPCR Super-mix (Bio-Rad) containing the primer pairs for either gene or for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a housekeeping gene (Table 1). All amplification reactions were performed in triplicate and average threshold cycle (Ct) numbers of the triplicates were used to calculate the relative mRNA expression of candidate genes. The magnitude of change of mRNA expression for candidate genes was calculated by using the standard 2−(ΔΔCt) method. All data were normalized to endogenous reference (GAPDH) gene content and expressed as relative fold-change of control.| Target | Accession no. | Direction | Sequence (5′ → 3′) |
|---|---|---|---|
| ACP5 | NM_001111034 | Forward | CTTTCTACCGCCTGCACTTC |
| Reverse | GTTTCTTGAGCCAGGACAGC | ||
| ATP5PO | NM_001697.3 | Forward | TCCTGAAGGAACCCAAAGTG |
| Reverse | ATCGACCATTTTCAGCAAGC | ||
| ATP6V0D2 | NM_152565.1 | Forward | TGGCACTGAATTGAGCAAAG |
| Reverse | CACTGCCACCTACAGCTTCA | ||
| CTSK | NM_000396 | Forward | TTCTGCTGCTACCTGTGGTG |
| Reverse | CCAGGTGGTTCATAGCCAGT | ||
| CYCS | NM_018947.5 | Forward | TTGGCAATCCGTCATCAGTA |
| Reverse | CCCGACAGTGCCTAGAAGAG | ||
| DCSTAMP | NM_030788.4 | Forward | CACTTGAAACTGCACGGAGA |
| Reverse | AGGACAACAGTCCCAGCATC | ||
| FOS | NM_005252.4 | Forward | CTTCCTGTTCCCAGCATCAT |
| Reverse | GTACAGGTGACCACCGGAGT | ||
| GAPDH | NM_001289746 | Forward | CACATGGCCTCCAAGGAGTAAG |
| Reverse | CCAGCAGTGAGGGGTCTCTCT | ||
| GNA13 | NM_006572.6 | Forward | TCCGTGAAATCATGCCTGTA |
| Reverse | CGCCTTAAAATGATGGGAGA | ||
| IL10 | NM_000572 | Forward | CTGACATCAAGGAGCACGTG |
| Reverse | GGCTTTGTAGACACCCCTCT | ||
| IL1B | NM_000576 | Forward | AAGGAGAGCTCTTTCCCACC |
| Reverse | GCCTCTGGTCTCCTTGGATT | ||
| IL6 | NM_000600_ | Forward | TAACCATCCCTGTCCCAACC |
| Reverse | AGTTACATGCCCAGTGGACA | ||
| ITGB3 | NM_000212.2 | Forward | GCAATGGGACCTTTGAGTGT |
| Reverse | GTGGCAGACACATTGACCAC | ||
| MAFB | NM_005461.5 | Forward | GCCTGCGCTAATTGTAGGAG |
| Reverse | CGCACTTGAAAGTTGCAAAA | ||
| NFATC1 | NM_172390.3 | Forward | AGAAAGCGAAGCCAGTACCA |
| Reverse | CGGTCTCACTAACGGGACAT | ||
| NFE2L2 | NM_001145412.3 | Forward | GCGACGGAAAGAGTATGAGC |
| Reverse | GTTGGCAGATCCACTGGTTT | ||
| NRF1 | NM_001293163.2 | Forward | GTCCAGATCCCTGTGAGCAT |
| Reverse | CAATGTCACCACCTCCACAG | ||
| OSCAR | NM_001282349 | Forward | CCCAGCTTCATACCACCCTA |
| Reverse | GAAGAGAAGGGGAGCGATCT | ||
| PPARGC1B | NM_133263.4 | Forward | GGAGACTGAACCCTGAGCTG |
| Reverse | GTAGGAGCTGCTGGTCTTGG | ||
| TFAM | NM_001270782.1 | Forward | GTGGGAGCTTCTCACTCTGG |
| Reverse | TAGGGCTTTTTCTCCTGCAA | ||
| TNF | NM_000594 | Forward | TCCTTCAGACACCCTCCACC |
| Reverse | AGGCCCCAGTTTGAATTCTT | ||
| TNFRS11A | NM_001270949 | Forward | GGTGCAGCCTCTAACTCCTG |
| Reverse | TTGAGACCAGGCTGGGTAAC | ||
| TNFRS11B | NM_002546 | Forward | GGCAACACAGCTCACAAGAA |
| Reverse | CTGGGTTTGCATGCCTTTAT |
Cytokine quantification
Tumour necrosis factor (TNF)-α, interleukin (IL)-1β, IL-6, and IL-10 were determined by ELISA, following the indications of the manufacturer (Diaclone, Besançon, France). Cytokine concentration was expressed in pg mL−1, as calculated from the calibration curves from serial dilution of human recombinant standards in each assay.Statistical analysis
All values are expressed as mean ± SD. Data were evaluated with GraphPad Prism 5.01 (GraphPad Software, San Diego, USA). The statistical significance of any difference in each parameter among the groups was evaluated by one-way analysis of variance (ANOVA); following Newman-Keuls’ test for multiple comparisons. Differences between groups were considered to be significant at a p value of <0.05.Results and discussion
It has been well established that osteoclastogenesis plays fundamental roles in the development and progression of bone diseases. Hence, identification of new molecules effective on targets involved in the control of osteoclast differentiation and function would be crucial for maintenance of bone health and better-tailored treatments of disorders affecting the bone. Here, we designed a hypothesis-driven approach to probe if OL and Per-OL have any role in regulating transcriptional activity of selected genes that are required for commitment, differentiation, and function of osteoclasts from M-CSF/RANKL-treated human blood monocytes. This rationale was, in part, guided by previous evidence suggesting bone-protecting effects of OL in ovariectomized rats28 and antiresorptive effects of OL in TRAP positive cells derived from mouse spleen.16 A strength of our study stems from the inclusion of Per-OL, which is an OL derivative with potential enhanced ability to penetrate cell membrane.24,29OL and Per-OL disturb the instruction of M-CSF for the commitment of human blood monocytes to the osteoclast fate
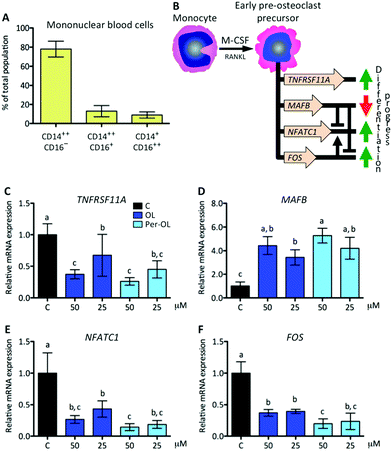
In this study, monocytes were isolated from the blood of healthy volunteers and the surface expression of CD14 and CD16 antigens was determined by FACS. As expected, classical (CD14++CD16−) and intermediate (CD14++CD16+) monocytes were the most abundant subsets (∼78% and ∼13%, respectively), with non-classical monocytes (CD14+CD16++) remaining at ∼9% (Fig. 1A). In previous studies, all of these three subsets were reported to give rise to osteoclasts.30 More recently, a systematic literature review concluded that specific features of the clinical course of bone disease can have a significant impact on commitment of human circulating monocyte subsets, with prominence of those rich in CD14, towards a bone-resorbing phenotype.31At the beginning of the differentiation process of monocytes to pre-osteoclasts, M-CSF is a cytokine that primes monocytes to gain osteoclastogenic activity by the reprogram of different target genes, which are required for a rapid response to RANKL and for successive differentiation stages. The TNFRSF11A gene is critical to sustain the RANK/RANKL loop and the transcripts of V-maf musculoaponeurotic fibrosarcoma oncogene homolog B gene (MAFB) are associated with the escape of monocytes to M-CSF-induced proliferative and osteoclast differentiation signals via repression of the master nuclear factor of activated T-cells cytoplasmic 1 (NFATC1) and FOS genes.32FOS is also involved in monocyte commitment to pre-osteoclasts via its cooperation for transcriptional activity of NFATC1 gene.33 This view is depicted in Fig. 1B. In the presence of OL and Per-OL, the transcriptional activity of TNFRSF11A gene was found to be inhibited in M-CSF/RANKL-treated human blood monocytes (Fig. 1C). In addition, the expression of MAFB gene was dramatically upregulated (Fig. 1D) while that of NFATC1 (Fig. 1E) and FOS (Fig. 1F) genes was downregulated. Differences between OL and Per-OL at 50 μM, having Per-OL higher effect than OL, were observed for FOS gene expression. However, we did not detect any dose-dependent effect of OL or Per-OL. None of the concentrations used herein (up to 50 μM) of OL and Per-OL reduced cell viability as per MTT assay (data not shown). Deficient expression of TNFRSF11A gene has been shown to impede osteoclastogenesis and to induce osteopetrosis in mice.34 A similar phenotype has been documented for patients with TNFRSF11A gene mutations associated with autosomal recessive osteopetrosis, who exhibited loss-of-function in both the extracellular and intracellular domains of RANK, and whose monocytes were unable to completely overcome the commitment to an osteoclast fate.35 In a context of OL and Per-OL inducing the repression of TNFRSF11A gene, the contrariwise effect occurring on the transcriptional activity of MAFB gene agrees with previous research showing that expression of MAFB gene is negatively regulated by the RANKL-RANK axis in M-CSF/RANKL-treated murine bone marrow monocytes.32 Gain-of-function experiments with murine bone marrow hematopoietic stem cells have also led to the identification of MAFB gene with ability to limit M-CSF instructions for commitment.36 Therefore, our findings suggest that OL and Per-OL might induce defective osteoclastogenesis during early stages in commitment of human monocyte/macrophage lineage into pre-osteoclasts.
OL and Per-OL disturb the course of M-CSF/RANKL-mediated osteoclast differentiation of human blood monocytes
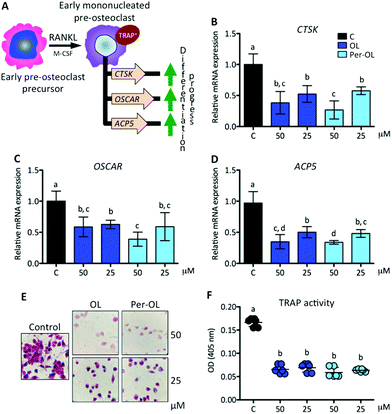
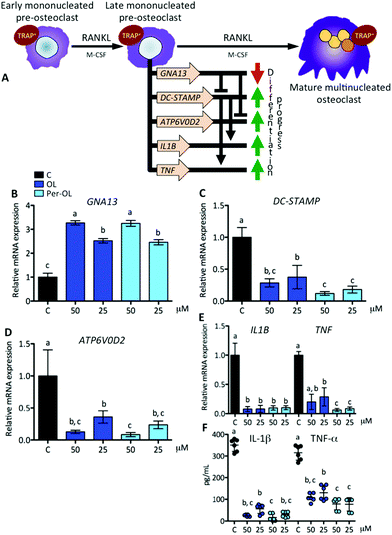
The expansion of mononucleated pre-osteoclasts from those primed with M-CSF is a consequence of RANKL making an entry into the differentiation process upon interaction with its receptor RANK. This intensifies the rise of expression of NFATC1 and FOS genes resulting in a transcriptional program of early differentiation marker genes, such as cathepsin K (CTSK),37 osteoclast-associated immunoglobulin-like receptor (OSCAR),38 and TRAP type 5 (ACP5),39 with mononucleated pre-osteoclasts ready to exhibit TRAP activity.40 This view is depicted in Fig. 2A. In the presence of OL and Per-OL, the transcriptional activity of CTSK (Fig. 2B), OSCAR (Fig. 2C), and ACP5 (Fig. 2D) genes was found to be inhibited in M-CSF/RANKL-treated human blood monocytes. In addition, TRAP-positive cells were reduced (Fig. 2E). No differences between OL and Per-OL at the same concentration were observed. However, there was a dose-dependent effect of OL on ACP5 gene and of Per-OL on CTSK and ACP5 genes. Previous studies have been reported that OL decreases the formation of TRAP in osteoclasts from mouse spleen cells16 and in periodontium of rats with experimentally induced periodontal inflammation.41 Interestingly, and coincident to what was seen above, overexpression of MAFB gene also inhibits the formation of TRAP and attenuates the expression of pro-osteoclastogenic OSCAR and ACP5 genes in M-CSF/RANKL-treated murine bone marrow monocytes.32 Furthermore, mice with deficient expression of CTSK42 and APC543 genes have been shown to suffer from osteopetrosis due to impaired osteoclastogenesis.With the progress of differentiation, RANKL drives mononucleated pre-osteoclasts to a fusion-competent state. In this late stage of osteoclast precursors, they can fuse with one another to form multinucleated osteoclasts after the transcription of master fusogenic genes, such as dendritic cell-specific transmembrane protein (DC-STAMP) and vacuolar H+ ATPase V0 subunit d2 (ATP6V0D2).44 The fusion process can also be enhanced by the expression and release of IL-1β and TNF-α functioning in an autocrine mode,45 which may depend on expression of DC-STAMP gene.46 Interestingly, recent evidence has shown that guanine nucleotide-binding protein subunit α13 gene (GNA13), a member of the G12 subfamily of the heterotrimeric G proteins, is critically integrated in the late stage of pre-osteoclast differentiation via repression of DC-STAMP and ATP6V0D2 genes for the regulation of cell–cell fusion and the maintenance of a balanced differentiation program.47 This view is depicted in Fig. 3A. In the presence of OL and Per-OL, the transcriptional activity of GNA13 gene (Fig. 3B) was found to be upregulated in M-CSF/RANKL-treated human blood monocytes. In addition, the expression of DC-STAMP (Fig. 3C) and ATP6V0D2 (Fig. 3D) genes was dramatically downregulated. Similar effects were observed on expression of IL1B and TNF genes (Fig. 3E) and on release of IL-1β and TNF-α (Fig. 3F). We detected differences between OL and Per-OL at 25 μM, having Per-OL higher effects than OL on DC-STAMP and TNF gene expression and on release of TNF-α. There was also a dose-dependent effect of OL and Per-OL on GNA13 and ATP6V0D2 genes. As a negative regulator of osteoclastogenesis, overexpression of a constitutively active form of GNA13 using lentivirus has been shown to drastically block fusion, formation of TRAP, and resorption activity of late mononucleated pre-osteoclasts derived from several M-CSF/RANKL-treated myeloid cells including murine bone marrow cells and those of the macrophage-like cell line RAW264.7.48 The authors also found that gain-of-function of GNA13 using adenoviral transfection was protective for bone loss in different animal models of bone disease. The relevance of DC-STAMP gene in the fusion step in osteoclastogenesis has been well established in vitro using an osteoclast precursor clone from RAW264.7 cells when exposed to RANKL49 and in vivo using DC-STAMP−/− mice in which multinucleation of osteoclast was entirely abolished.50 High-resolution microcomputed tomography studies in ATP6V0D2−/− mice have also revealed a direct relationship between the extremely low mRNA levels of ATP6V0D2 and the mineral density of bones, mainly explained by reduced cell–cell fusion and thereby lowered formation of mature multinucleated osteoclasts in late stages of osteoclast differentiation.51 The inhibitory effects of OL on release of IL-1β and TNF-α have been previously described in human neutrophils52 and synovial fibroblasts.26 Per-OL was also shown to inhibit the concentrations of IL-1β and TNF-α in the supernatants of LPS-treated murine peritoneal macrophages.24 Therefore, our findings underscore the notion that OL and Per-OL might induce defective osteoclastogenesis during late stages of differentiation of human blood monocyte-derived pre-osteoclasts.
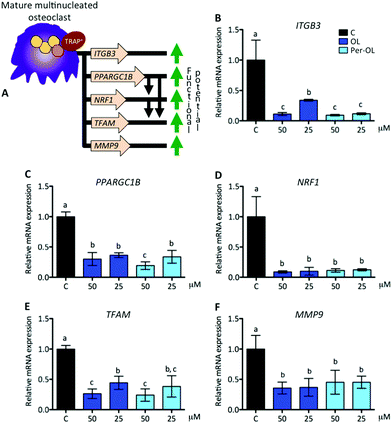
OL and Per-OL disturb the function of human blood monocyte-derived osteoclasts
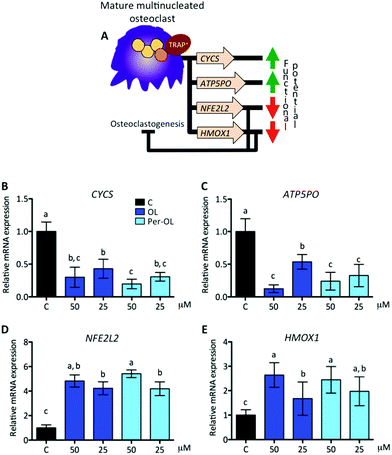
Functional mature multinucleated osteoclasts can attach to mineralized surfaces for which they are given novel cytoskeletal organization that requires mitochondrial biogenesis probably due to high energetic needs. Expression of adhesion receptor αvβ3 integrin-encoding gene (ITGB3)53 and of peroxisome proliferator-activated receptor gamma coactivator 1-beta (PGC1β) gene (PPARGC1B)54 is considered essential in the organization of osteoclast cytoskeleton for mineralized matrix recognition. This is of interest because PPARGC1B gene is also a master regulator of mitochondrial biogenesis via stimulation gene expression of nuclear respiratory factor 1 (NRF1)55 and then of mitochondrial transcription factor A (TFAM).56 In this scenario of cytoskeletal rearrangements, the matrix metalloproteinase 9 gene (MMP9) has been shown to be entailed in osteoclast motility and bone resorption activity,57 and in efficient transcription of key osteoclastogenic genes.58 This view is depicted in Fig. 4A. In the presence of OL and Per-OL, the transcriptional activity of ITGB3 (Fig. 4B), PPARGC1B (Fig. 4C), NRF1 (Fig. 4D), TFAM (Fig. 4E), and MMP9 (Fig. 4F) genes was found to be dramatically downregulated in M-CSF/RANKL-treated human blood monocytes. We detected differences between OL and Per-OL at 50 μM, having Per-OL higher effects than OL on PPARGC1B gene expression. There was also a dose-dependent effect of OL on ITGB3 and TFAM genes, and of Per-OL on PPARGC1B gene. M-CSF/RANKL-treated bone marrow cells from mice with deficient expression of ITGB3 gene have been reported to exhibit apparent normal differentiation, but severe morphological and functional abnormalities that led to diminished bone resorptive capacity.59 Similar results of a role for PGC1β in osteoclast function but not in osteoclast differentiation irrespective of the presence of M-CSF and RANKL have been recently shown in bone marrow cells from PPARGC1B-deficient mice.54 In keeping the notion that ITGB3 and PPARGC1B genes mediate osteoclast cytoskeleton organization in a canonical and non-canonical manner, respectively, OL and Per-OL could therefore be acting to interfere both pathways during cytoskeleton remodelling of osteoclast from human blood monocytes. The inhibitory effects of OL and Per-OL on expression of NRF1 and TFAM genes also suggest a dampening of mitochondrial function. This ability of OL to induce mitochondrial dysfunction has been observed in human bone marrow mesenchymal stem cells during adipogenic differentiation.14 Also importantly, engineered mice lacking the MMP9 gene were shown to have abnormal ossification60 and IL-1β-treated chondrocytes of patients with osteoarthritis pre-incubated with OL were shown to have lower mRNA levels of MMP1 and MMP3 genes than control cells.25The pattern of excessive mitochondrial biogenesis in functional osteoclasts is associated with high expression of oxidative phosphorylation genes, such as cytochrome c (CYCS) and ATP synthase subunit O (ATP5PO).47 Likewise, a disproportionate oxidative phosphorylation produces intramitochondrial reactive oxygen species and free radicals, which are essential for osteoclastogenesis and osteoclast function.61 To protect against oxidative stress and deregulation of cellular redox homeostasis, the nuclear factor-erythroid 2-related factor 2 gene (NFE2L2) is involved in the transcription of powerful anti-oxidant genes including heme oxygenase-1 (HMOX1). NFE2L2 and HMOX1 genes also take part in osteoclastogenesis, the first as negative regulator of early pre-osteoclasts by its role in the upregulation of MAFB gene,62 the second as negative regulator of late pre-osteoclasts by its role in the downregulation of RANKL-dependent osteoclastogenic genes.63 This view is depicted in Fig. 5A. In the presence of OL and Per-OL, the transcriptional activity of CYCS (Fig. 5B) and ATP5PO (Fig. 5C) genes was found to be downregulated in M-CSF/RANKL-treated human blood monocytes. In addition, the expression of NFE2L2 gene (Fig. 5D) and that of its downstream target HMOX1 gene (Fig. 5E) were upregulated. Differences between OL and Per-OL at 25 μM, having Per-OL higher effect than OL, were observed for ATP5PO gene expression. There was also a dose-dependent effect of OL on ATP5PO and HMOX1 genes, and of Per-OL on NFE2L2 gene. Consistent with these findings, recent studies have documented that OL suppresses oxidative stress by targeting the transcriptional activity of renal NFE2L2 and HMOX1 genes in mice with experimentally-induced acute kidney injury.64
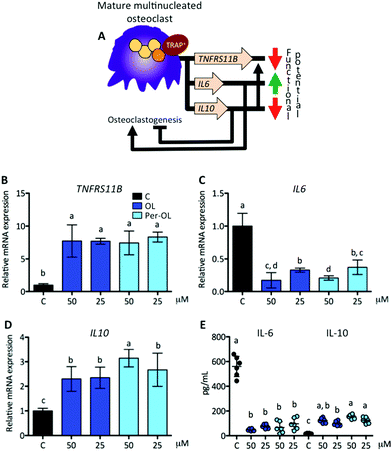
Interestingly, osteoclasts themselves can limit osteoclastogenesis and osteoclast function through the activation of TNFRS11B gene, which encodes osteoprotegerin (OPG).65 OPG is a member of the superfamily of TNF receptors that interferes with the RANK/RANKL system, and inhibits bone-resorbing activity and promotes apoptosis of mature osteoclasts. Fully differentiated osteoclasts have also been shown to exhibit a cytokine phenotype that resembles that of non-classically activated M2 macrophages, with an inverse modulation in the secretion of IL-6 and IL-10.66 IL-6 is pro-osteoclastogenic presumably by the upregulation of TNFRS11A gene,67 whereas IL-10 has the opposite effect by different mechanisms, among which the upregulation of TNFRS11B gene.68 This view is depicted in Fig. 6A. In the presence of OL and Per-OL, the transcriptional activity of TNFRS11B gene (Fig. 6B) was found to be dramatically upregulated in M-CSF/RANKL-treated human blood monocytes. In addition, the expression of IL6 gene (Fig. 6C) was downregulated, whereas IL10 gene (Fig. 6D) was upregulated. Similar effects were observed on release of IL-6 (decreasing) and IL-10 (increasing) (Fig. 6E). We detected differences between OL and Per-OL, having Per-OL higher effects than OL on IL10 gene expression (at 50 μM) and on release of IL-10 (at 25 μM). There was also a dose-dependent effect of OL and Per-OL on IL6 gene, and of Per-OL on IL10 gene. Previous studies have addressed the ability of OL and Per-OL to decrease the production and release of IL-6 in LPS-treated murine peritoneal macrophages,26 and of OL to increase the concentration of IL-10 in the colon tissue with experimentally-induced colitis.69 These findings and those highlighted above are indicative that OL and Per-OL might induce dysfunction in human blood monocyte-derived osteoclasts.
Taken together, this study demonstrates for the first time that OL and Per-OL (Per-OL > OL) strongly interfered with M-CSF/RANKL-induced osteoclast commitment, differentiation, and function from human blood monocytes, suggesting a novel inhibitory role of OL and Per-OL in multiple stages of osteoclastogenesis. Given that OL and Per-OL may be subjected to phase I and phase II hepatic metabolism, a major limitation of our study is that included only in vitro experiments of direct interactions between of OL or Per-OL and monocytes/osteoclasts. Previous studies have shown that intact OL may circulate in blood after administration of a single oral dose in humans.70,71 It is possible that treatment with OL or Per-OL maintained for long periods could have cumulative effects in specific anatomical sites. Further studies on this topic and on novel administration routes,72 and on protection strategies to avoid intense exposure to metabolic detoxification pathways73,74 of OL and Per-OL are needed to substantiate our findings. This will pave the way for novel strategies to protect bone health, opening up new directions in research against the development and progression of osteoclast-related diseases.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by grants from the MCYT and MEC (AGL2011-29008, AGL2016-80852-R, and AGL2017-89342-P). The authors thank Cell Biology Unit at the Instituto de la Grasa (CSIC) for its assistance during the fulfilment of this study. MAR acknowledges financial support from the Spanish Research Council (CSIC)/Juan de la Cierva (FJCI-2017-33132).References
- L. Gennari, D. Rendina, A. Falchetti and D. Merlotti, Paget's disease of bone, Calcif. Tissue Int., 2019, 104, 483–500 CrossRef CAS PubMed.
- M. B. Madel, L. Ibáñez, A. Wakkach, T. J. De Vries, A. Teti, F. Apparailly and C. Blin-Wakkach, Immune function and diversity of osteoclasts in normal and pathological conditions, Front. Immunol., 2019, 10, 1408 CrossRef CAS PubMed.
- A. Maurizi and N. Rucci, The osteoclast in bone metastasis: Player and target, Cancer, 2018, 10, E218 CrossRef PubMed.
- T. Ono and T. Nakashima, Recent advances in osteoclast biology, Histochem. Cell Biol., 2018, 149, 325–341 CrossRef CAS PubMed.
- K. H. Park-Min, Mechanisms involved in normal and pathological osteoclastogenesis, Cell. Mol. Life Sci., 2018, 75, 2519–2528 CrossRef CAS PubMed.
- M. K. Skjødt, M. Frost and B. Abrahamsen, Side effects of drugs for osteoporosis and metastatic bone disease, Br. J. Clin. Pharmacol., 2019, 85, 1063–1071 CrossRef PubMed.
- A. Romani, F. Ieri, S. Urciuoli, A. Noce, G. Marrone, C. Nediani and R. Bernini, Health effects of phenolic compounds found in extra-virgin olive oil, by-products, and leaf of Olea europaea L., Nutrients, 2019, 11, E1776 CrossRef PubMed.
- S. Bonacci, R. Paonessa, P. Costanzo, R. Salerno, J. Maiuolo, M. Nardi, A. Procopio and O. Manuela, Peracetylation as a strategy to improve oleuropein stability and its affinity to fatty foods, Food Funct., 2018, 9, 5759–5767 RSC.
- T. Fukami and T. Yokoi, The emerging role of human esterases, Drug Metab. Pharmacokinet., 2012, 27, 466–477 CrossRef CAS PubMed.
- M. Celano, V. Maggisano, S. M. Lepore, D. Russo and S. Bulotta, Secoiridoids of olive and derivatives as potential coadjuvant drugs in cancer: A critical analysis of experimental studies, Pharmacol. Res., 2019, 142, 77–86 CrossRef CAS PubMed.
- A. K. Marković, J. Torić, M. Barbarić and C. J. Brala, Hydroxytyrosol, tyrosol and derivatives and their potential effects on human health, Molecules, 2019, 24, E2001 CrossRef PubMed.
- G. Marcelino, P. A. Hiane, K. de C. Freitas, L. F. Santana, A. Pott, J. R. Donadon and R. de C. A. Guimarães, Effects of olive oil and its minor components on cardiovascular diseases, inflammation, and gut microbiota, Nutrients, 2019, 11, E1826 CrossRef PubMed.
- P. Przychodzen, R. Wyszkowska, M. Gorzynik-Debicka, T. Kostrzewa, A. Kuban-Jankowska and M. Gorska-Ponikowska, Anticancer potential of oleuropein, the polyphenol of olive oil, with 2-methoxyestradiol, separately or in combination, in human osteosarcoma cells, Anticancer Res., 2019, 39, 1243–1251 CrossRef PubMed.
- A. Casado-Diaz, J. Anter, S. Müller, P. Winter, J. M. Quesada-Gomez and G. Dorado, Transcriptomic analyses of the anti-adipogenic effects of oleuropein in human mesenchymal stem cells, Food Funct., 2017, 8, 1254–1270 RSC.
- R. Santiago-Mora, A. Casado-Diaz, M. D. De Castro and J. M. Quesada-Gomez, Oleuropein enhances osteoblastogenesis and inhibits adipogenesis: the effect on differentiation in stem cells derived from bone marrow, Osteoporosis Int., 2011, 22, 675–684 CrossRef CAS PubMed.
- K. Hagiwara, T. Goto, M. Araki, H. Miyazaki and H. Hagiwara, Olive polyphenol hydroxytyrosol prevents bone loss, Eur. J. Pharmacol., 2011, 662, 78–84 CrossRef CAS PubMed.
- A. Procopio, S. Alcaro, M. Nardi, M. Oliverio, F. Ortuso, P. Sacchetta, D. Pieragostino and G. Sindona, Synthesis, biological evaluation, and molecular modeling of oleuropein and its semisynthetic derivatives as cyclooxygenase inhibitors, J. Agric. Food Chem., 2009, 57, 11161–11167 CrossRef CAS PubMed.
- S. M. Lepore, V. M. Morittu, M. Celano, F. Trimboli, M. Oliverio, A. Procopio, C. Di Loreto, G. Damante, D. Britti, S. Bulotta and D. Russo, Oral administration of oleuropein and its semisynthetic peracetylated derivative prevents hepatic steatosis, hyperinsulinemia, and weight gain in mice fed with high fat cafeteria diet, Int. J. Endocrinol., 2015, 2015, 431453 Search PubMed.
- S. Bulotta, R. Corradino, M. Celano, M. D'Agostino, J. Maiuolo, M. Oliverio, A. Procopio, M. Iannone, D. Rotiroti and D. Russo, Antiproliferative and antioxidant effects on breast cancer cells of oleuropein and its semisynthetic peracetylated derivatives, Food Chem., 2011, 127, 1609–1614 CrossRef CAS.
- S. Bulotta, R. Corradino, M. Celano, J. Maiuolo, M. D'Agostino, M. Oliverio, A. Procopio, S. Filetti and D. Russo, Antioxidant and antigrowth action of peracetylated oleuropein in thyroid cancer cells, J. Mol. Endocrinol., 2013, 51, 181–189 CAS.
- K. Stamatopoulos, A. Chatzilazarou and E. Katsoyannos, Optimization of multistage extraction of olive leaves for recovery of phenolic compounds at moderated temperatures and short extraction times, Foods, 2013, 3, 66–81 CrossRef PubMed.
- A. Procopio, G. Sindona, N. Costa, M. Gaspari and M. Nardi, Chemical-catalytic method for the peracylation of oleuropein and its products of hydrolysis, WO Pat, PCT/IT2008/000303, 2008 Search PubMed.
- M. C. Naranjo, I. Garcia, B. Bermudez, S. Lopez, M. P. Cardelo, R. Abia, F. J. G. Muriana and S. Montserrat-de la Paz, Acute effects of dietary fatty acids on osteclastogenesis via RANKL/RANK/OPG system, Mol. Nutr. Food Res., 2016, 60, 2505–2513 CrossRef CAS PubMed.
- M. L. Castejon, M. Sanchez-Hidalgo, M. Aparicio-Soto, A. Gonzalez-Benjumea, J. G. Fernandez-Bolaños and C. Alarcon-de-la-Lastra, Olive secoiridoid oleuropein and its semisynthetic acetyl-derivatives reduce LPS-induced inflammatory response in murine peritoneal macrophages via JAK-STAT and MAPKs signaling pathways, J. Funct. Foods, 2019, 58, 95–104 CrossRef CAS.
- Z. Feng, X. Li, J. Lin, W. Zheng, Z. Hu, J. Xuan, W. Ni and X. Pan, Oleuropein inhibits the IL-1β-induced expression of inflammatory mediators by suppressing the activation of NF-κB and MAPKs in human osteoarthritis chondrocytes, Food Funct., 2017, 8, 3737–3744 RSC.
- M. L. Castejon, M. A. Rosillo, T. Montoya, A. Gonzalez-Benjumea, J. G. Fernandez-Bolaños and C. Alarcon-de-la-Lastra, Oleuropein down-regulated IL-1β-induced inflammation and oxidative stress in human synovial fibroblast cell line SW982, Food Funct., 2017, 8, 1890–1898 RSC.
- S. J. Ryu, H. S. Choi, K. Y. Yoon, O. H. Lee, K. J. Kim and B. Y. Lee, Oleuropein suppresses LPS-induced inflammatory responses in RAW 264.7 Cell and zebrafish, J. Agric. Food Chem., 2015, 63, 2098–2105 CrossRef CAS PubMed.
- K. Y. Chin and S. Ima-Nirwana, Olives and bone: A green osteoporosis prevention option, Int. J. Environ. Res. Public Health, 2016, 13, E755 CrossRef PubMed.
- M. Rizzo, D. Ventrice, F. Giannetto, S. Cirinnà, N. A. Santagati, A. Procopio, V. Mollace and C. Muscoli, Antioxidant activity of oleuropein and semisynthetic acetyl-derivatives determined by measuring malondialdehyde in rat brain, J. Pharm. Pharmacol., 2017, 69, 1502–1512 CrossRef CAS PubMed.
- S. Sprangers, T. Schoenmaker, Y. Cao, V. Everts and T. J. de Vries, Different blood-borne human osteoclast precursors respond in distinct ways to IL-17A, J. Cell. Physiol., 2016, 231, 1249–1260 CrossRef CAS PubMed.
- T. J. de Vries, I. El Bakkali, T. Kamradt, G. Schett, I. D. C. Jansen and P. D'Amelio, What are the peripheral blood determinants for increased osteoclast formation in the various inflammatory diseases associated with bone loss?, Front. Immunol., 2019, 10, 505 CrossRef CAS PubMed.
- K. Kim, J. H. Kim, J. Lee, H. M. Jin, H. Kook, K. K. Kim, S. Y. Lee and N. Kim, MafB negatively regulates RANKL-mediated osteoclast differentiation, Blood, 2007, 109, 3253–3259 CrossRef CAS PubMed.
- M. P. Yavropoulou and J. G. Yovos, Osteoclastogenesis - Current knowledge and future perspectives, J. Musculoskeletal Neuronal Interact., 2008, 8, 204–216 CAS.
- K. Okamoto, T. Nakashima, M. Shinohara, T. Negishi-Koga, N. Komatsu, A. Terashima, S. Sawa, T. Nitta and H. Takayanagi, Osteoimmunology: The conceptual framework unifying the immune and skeletal systems, Physiol. Rev., 2017, 97, 1295–1349 CrossRef CAS PubMed.
- M. M. Guerrini, C. Sobacchi, B. Cassani, M. Abinun, S. S. Kilic, A. Pangrazio, D. Moratto, E. Mazzolari, J. Clayton-Smith, P. Orchard, F. P. Coxon, M. H. Helfrich, J. C. Crockett, D. Mellis, A. Vellodi, I. Tezcan, L. D. Notarangelo, M. J. Rogers, P. Vezzoni, A. Villa and A. Frattini, Human osteoclast-poor osteopetrosis with hypogammaglobulinemia due to TNFRSF11A (RANK) mutations, Am. J. Hum. Genet., 2008, 83, 64–76 CrossRef CAS PubMed.
- S. Sarrazin, N. Mossadegh-Keller, T. Fukao, A. Aziz, F. Mourcin, L. Vanhille, L. Kelly Modis, P. Kastner, S. Chan, E. Duprez, C. Otto and M. H. Sieweke, MafB restricts M-CSF-dependent myeloid commitment divisions of hematopoietic stem cells, Cell, 2009, 138, 300–313 CrossRef CAS PubMed.
- M. Pang, M. Rodriguez-Gonzalez, M. Hernandez, C. C. Recinos, K. L. Seldeen and B. R. Troen, AP-1 and Mitf interact with NFATc1 to stimulate cathepsin K promoter activity in osteoclast precursors, J. Cell. Biochem., 2019, 120, 12382–12392 CrossRef CAS PubMed.
- M. Asagiri and H. Takayanagi, The molecular understanding of osteoclast differentiation, Bone, 2007, 40, 251–264 CrossRef CAS PubMed.
- B. Faulkner, K. Astleford and K. C. Mansky, Regulation of osteoclast differentiation and skeletal maintenance by histone deacetylases, Molecules, 2007, 12, 2519–2529 Search PubMed.
- Q. Zhao, J. Shao, W. Chen and Y. P. Li, Osteoclast differentiation and gene regulation, Front. Biosci., 2007, 12, 2519–2529 CrossRef CAS PubMed.
- M. M. Taskan, H. Balci Yuce, O. Karatas, F. Gevrek and H. Toker, Evaluation of the effect of oleuropein on alveolar bone loss, inflammation, and apoptosis in experimental periodontitis, J. Periodontal Res., 2019, 54, 624–632 CrossRef CAS PubMed.
- P. Saftig, E. Hunziker, O. Wehmeyer, S. Jones, A. Boyde, W. Rommerskirch, J. D. Moritz, P. Schu and K. Von Figura, Impaired osteoclastic bone resorption leads to osteopetrosis in cathepsin-K-deficient mice, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 13453–13458 CrossRef CAS PubMed.
- K. Hollberg, K. Hultenby, A. Hayman, T. Cox and G. Andersson, Osteoclasts from mice deficient in tartrate-resistant acid phosphatase have altered ruffled borders and disturbed intracellular vesicular transport, Exp. Cell Res., 2002, 279, 227–238 CrossRef CAS PubMed.
- K. Kim, S.-H. Lee, J. Ha Kim, Y. Choi and N. Kim, NFATc1 induces osteoclast fusion via up-regulation of Atp6v0d2 and the dendritic cell-specific transmembrane protein (DC-STAMP), Mol. Endocrinol., 2008, 22, 176–185 CrossRef CAS PubMed.
- K. Søe, T. L. Andersen, M. Hinge, L. Rolighed, N. Marcussen and J.-M. Delaisse, Coordination of fusion and trafficking of pre-osteoclasts at the marrow-bone interface, Calcif. Tissue Int., 2019, 105, 430–445 CrossRef PubMed.
- A. Sanecka, M. Ansems, A. C. Prosser, K. Danielski, K. Warner, M. H. den Brok, B. J. H. Jansen, D. Eleveld-Trancikova and G. J. Adema, DC-STAMP knock-down deregulates cytokine production and T-cell stimulatory capacity of LPS-matured dendritic cells, BMC Immunol., 2011, 12, 57 CrossRef CAS PubMed.
- S. Nakano, K. Inoue, C. Xu, Z. Deng, V. Syrovatkina, G. Vitone, L. Zhao, X.-Y. Huang and B. Zhao, G-protein Gα13 functions as a cytoskeletal and mitochondrial regulator to restrain osteoclast function, Sci. Rep., 2019, 9, 4236 CrossRef PubMed.
- M. Wu, W. Chen, Y. Lu, G. Zhu, L. Hao and Y. P. Li, Gα13 negatively controls osteoclastogenesis through inhibition of the Akt-GSK3β-NFATc1 signalling pathway, Nat. Commun., 2017, 8, 1–13 CrossRef PubMed.
- T. Kukita, N. Wada, A. Kukita, T. Kakimoto, F. Sandra, K. Toh, K. Nagata, T. Iijima, M. Horiuchi, H. Matsusaki, K. Hieshima, O. Yoshie and H. Nomiyama, RANKL-induced DC-STAMP is essential for osteoclastogenesis, J. Exp. Med., 2004, 200, 941–946 CrossRef CAS PubMed.
- M. Yagi, T. Miyamoto, Y. Sawatani, K. Iwamoto, N. Hosogane, N. Fujita, K. Morita, K. Ninomiya, T. Suzuki, K. Miyamoto, Y. Oike, M. Takeya, Y. Toyama and T. Suda, DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells, J. Exp. Med., 2005, 202, 345–351 CrossRef CAS PubMed.
- S.-H. Lee, J. Rho, D. Jeong, J.-Y. Sul, T. Kim, N. Kim, J.-S. Kang, T. Miyamoto, T. Suda, S.-K. Lee, R. J. Pignolo, B. Koczon-Jaremko, J. Lorenzo and Y. Choi, v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation, Nat. Med., 2006, 12, 1403–1409 CrossRef CAS PubMed.
- A. K. Kiss, B. Michalak, A. Patyra and M. Majdan, UHPLC-DAD-ESI-MS/MS and HPTLC profiling of ash leaf samples from different commercial and natural sources and their in vitro effects on mediators of inflammation, Phytochem. Anal., 2020, 31, 57–67 CrossRef CAS PubMed.
- K. P. McHugh, K. Hodivala-Dilke, M. H. Zheng, N. Namba, J. Lam, D. Novack, X. Feng, F. P. Ross, R. O. Hynes and S. L. Teitelbaum, Mice lacking beta3 integrins are osteosclerotic because of dysfunctional osteoclasts, J. Clin. Invest., 2000, 105, 433–440 CrossRef CAS PubMed.
- Y. Zhang, N. Rohatgi, D. J. Veis, J. Schilling, S. L. Teitelbaum and W. Zou, PGC1β organizes the osteoclast cytoskeleton by mitochondrial biogenesis and activation, J. Bone Miner. Res., 2018, 33, 1114–1125 CrossRef CAS.
- R. C. Scarpulla, Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network, Biochim. Biophys. Acta, 2011, 1813, 1269–1278 CrossRef CAS.
- J. Lin, C. Handschin and B. M. Spiegelman, Metabolic control through the PGC-1 family of transcription coactivators, Cell Metab., 2005, 1, 361–370 CrossRef CAS PubMed.
- O. Ishibashi, S. Niwa, K. Kadoyama and T. Inui, MMP-9 antisense oligodeoxynucleotide exerts an inhibitory effect on osteoclastic bone resorption by suppressing cell migration, Life Sci., 2006, 79, 1657–1660 CrossRef CAS.
- Y. Shin, N. B. Ghate, B. Moon, K. Park, W. Lu and W. An, DNMT and HDAC inhibitors modulate MMP-9-dependent H3 N-terminal tail proteolysis and osteoclastogenesis, Epigenet. Chromatin, 2019, 12, 25 CrossRef.
- W. Zou and S. L. Teitelbaum, Absence of Dap12 and the αvβ3 integrin causes severe osteopetrosis, J. Cell Biol., 2015, 208, 125–136 CrossRef CAS.
- K. B. S. Paiva and J. M. Granjeiro, Matrix metalloproteinases in bone resorption, remodeling, and repair, Prog. Mol. Biol. Transl. Sci., 2017, 148, 203–303 Search PubMed.
- T. S. Agidigbi and C. Kim, Reactive oxygen species in osteoclast differentiation and possible pharmaceutical targets of ROS-mediated osteoclast diseases, Int. J. Mol. Sci., 2019, 20, E3576 CrossRef PubMed.
- E. Sakai, M. Morita, M. Ohuchi, M. A. Kido, Y. Fukuma, K. Nishishita, K. Okamoto, K. Itoh, M. Yamamoto and T. Tsukuba, Effects of deficiency of Kelch-like ECH-associated protein 1 on skeletal organization: a mechanism for diminished nuclear factor of activated T cells cytoplasmic 1 during osteoclastogenesis, FASEB J., 2017, 31, 4011–4022 CrossRef CAS.
- U. Florczyk-Soluch, E. Józefczuk, J. Stępniewski, K. Bukowska-Strakova, M. Mendel, M. Viscardi, W. N. Nowak, A. Józkowicz and J. Dulak, Various roles of heme oxygenase-1 in response of bone marrow macrophages to RANKL and in the early stage of osteoclastogenesis, Sci. Rep., 2018, 8, 10797 CrossRef PubMed.
- M. Yin, N. Jiang, L. Guo, Z. Ni, A. Y. Al-Brakati, M. S. Othman, A. E. Abdel Moneim and R. B. Kassab, Oleuropein suppresses oxidative, inflammatory, and apoptotic responses following glycerol-induced acute kidney injury in rats, Life Sci., 2019, 232, 116634 CrossRef CAS PubMed.
- J. H. Kang, H. M. Ko, J. S. Moon, H. I. Yoo, J. Y. Jung, M. S. Kim, J. T. Koh, W. J. Kim and S. H. Kim, Osteoprotegerin expressed by osteoclasts: an autoregulator of osteoclastogenesis, J. Dent. Res., 2014, 93, 1116–1123 CrossRef CAS PubMed.
- H.-C. Tseng, K. Kanayama, K. Kaur, S.-H. Park, S. Park, A. Kozlowska, S. Sun, C. E. McKenna, I. Nishimura and A. Jewett, Bisphosphonate-induced differential modulation of immune cell function in gingiva and bone marrow in vivo: role in osteoclast-mediated NK cell activation, Oncotarget, 2015, 6, 20002–20025 Search PubMed.
- D. Harmer, C. Falank and M. R. Reagan, Interleukin-6 interweaves the bone marrow microenvironment, bone loss, and multiple myeloma, Front. Endocrinol., 2018, 9, 788 CrossRef.
- D. S. Amarasekara, H. Yun, S. Kim, N. Lee, H. Kim and J. Rho, Regulation of osteoclast differentiation by cytokine networks, Immune Netw., 2018, 18, e8 CrossRef.
- E. Giner, M.-C. Recio, J.-L. Rios and R.-M. Giner, Oleuropein protects against dextran sodium sulfate-induced chronic colitis in mice, J. Nat. Prod., 2013, 76, 1113–1120 CrossRef CAS PubMed.
- M. de Bock, E. B. Thorstensen, J. G. B. Derraik, H. V. Henderson, P. L. Hofman and W. S. Cutfield, Human absorption and metabolism of oleuropein and hydroxytyrosol ingested as olive (Olea europaea L.) leaf extract, Mol. Nutr. Food Res., 2013, 57, 2079–2085 CrossRef PubMed.
- N. Lemonakis, V. Mougios, M. Halabalaki, A.-L. Skaltsounis and E. Gikas, A novel bioanalytical method based on UHPLC-HRMS/MS for the quantification of oleuropein in human serum. Application to a pharmacokinetic study, Biomed. Chromatogr., 2016, 30, 2016–2023 CrossRef CAS PubMed.
- S. Palagati, S. Sv and B. R. Kesavan, Application of computational tools for the designing of Oleuropein loaded nanostructured lipid carrier for brain targeting through nasal route, Daru, 2019, 27, 695–708 CrossRef CAS.
- C. Bonechi, A. Donati, G. Tamasi, A. Pardini, H. Rostom, G. Leone, S. Lamponi, M. Consumi, A. Magnani and C. Rossi, Chemical characterization of liposomes containing nutraceutical compounds: Tyrosol, hydroxytyrosol and oleuropein, Biophys. Chem., 2019, 246, 25–34 CrossRef CAS.
- A. M. Nassir, I. A. A. Ibrahim, S. Mdc, M. Waris, Tanuja, M. R. Ain, I. Ahmad and N. Shahzad, Surface functionalized folate targeted oleuropein nano-liposomes for prostate tumor targeting: In vitro and in vivo activity, Life Sci., 2019, 220, 136–146 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |