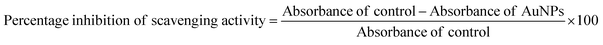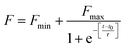Open Access Article
Open Access ArticlePolyphenol capping on a gold nanosurface modulates human serum albumin fibrillation†
Aalok
Basu
 ab,
Sonia
Kundu
c,
Aatrayee
Das
a,
Chitra
Basu
d,
Sagar
Bhayye
ab,
Sonia
Kundu
c,
Aatrayee
Das
a,
Chitra
Basu
d,
Sagar
Bhayye
 a,
Suvadra
Das
e and
Arup
Mukherjee
a,
Suvadra
Das
e and
Arup
Mukherjee
 *af
*af
aDepartment of Chemical Technology, University of Calcutta, 92 A.P.C. Road, Kolkata 700009, West Bengal, India. E-mail: arupm1234@gmail.com; Fax: +91 33 23519755; Tel: +91 33 23508387
bDr B.C. Roy College of Pharmacy and Allied Health Sciences, Bidhannagar, Durgapur 713206, West Bengal, India
cDepartment of Pharmaceutical Science and Technology, Maulana Abul Kalam Azad University of Technology, BF 142, Sector 1, Salt Lake City, Kolkata 700064, West Bengal, India
dDepartment of Biophysics, University of Calcutta, 92 A.P.C. Road, Kolkata 700009, West Bengal, India
eBasic Science and Humanities Department, University of Engineering and Management, University Area, Plot, Street Number 03, Action Area III, B/5, Newtown, Kolkata 700156, West Bengal, India
fDepartment of Biotechnology, Maulana Abul Kalam Azad University of Technology, BF 142, Sector 1, Salt Lake City, Kolkata 700064, West Bengal, India
First published on 17th June 2020
Abstract
Different small molecules and nanomaterials have been known as inhibitors of protein misfolding and subsequent fibrillation, which marks the initiation of various degenerative conditions. This work explores the effect of polyphenol-capped gold nanoparticles on the extent of human serum albumin fibrillation. Silymarin-capped (SAuNPs), quercetin-capped (QAuNPs) and gallic acid-capped gold nanoparticles (GAuNPs) were synthesized with a uniform size range and their relative antioxidant capacity was determined through DPPH assay. The fibrillation of HSA at 65 °C was inhibited by ∼15% in the presence of SAuNPs and the process was monitored through a combination of Thioflavin T fluorescence spectroscopy, circular dichroism spectroscopy and microscopic analysis. The inhibitory effect appeared much pronounced in the case of QAuNPs (∼67%) and GAuNPs (∼60%). Using SDS PAGE analysis, we demonstrated that the different inhibitory activity of SAuNPs, QAuNPs, and GAuNPs could be attributed to the antioxidant potential of the individual nanoparticles. Our work revealed that apart from protein–nanoparticle surface interactions, the antioxidant capacity has a role in determining the effectiveness of a protein fibrillation inhibitor. Cytotoxic analysis of protein–gold nanoparticle aggregates on HaCaT cell lines further confirmed that the nanoparticles were biosafe and can be considered as active therapeutics for translational use.
1. Introduction
Reactive oxygen species (ROS) are by-products of cellular metabolism and act as redox signalling molecules essential for the maintenance of physiological functions.1,2 Excessive ROS produced are promptly scavenged by molecular chaperones as part of the proteostasis mechanism,3 the failure of which can cause oxidative damage to the exquisitely arranged protein macromolecules. Under such conditions of oxidative stress, protein molecules, especially the newly synthesized ones, are susceptible to aberrant structural misfolding and subsequently experience molecular self-association to form amyloid fibrils. These fibrils are cytotoxic, and their deposition in tissues is often the pathogenic hallmark of certain degenerative conditions such as Parkinsonism, Alzheimer's disease, atherosclerosis, diabetes and several others.4A considerable number of small molecules,5 peptides,6 surfactants,7 quantum dots8 and nanoparticles are capable of binding to amyloidogenic proteins and resisting the misfolding of their tertiary structures. Nanoparticles of N-isopropylacrylamide-N-butylacrylamide copolymer,9 amino-acid based polymers,10 gold,11 graphene oxide,12 and metallic oxides13,14 have been explored as inhibitors of protein fibrillation, due to their tunable surface functionalities and high surface to volume ratio. Among them, gold nanoparticles (AuNPs) are of considerable interest in drug delivery, genetics, biosensing, and therapeutics due to their ease of synthesis with controlled dimensions, detection, facile bioconjugation, and high thermal stability.15 AuNPs have also exhibited tremendous potency in the management of oxidatively stressed conditions through quenching of free radical species.16,17 AuNPs have been strategically explored as one platform for studying the influence of nanosurface charge,18 surface chirality,19 shape and size20 on protein fibrillation.
Since bioconjugated gold nanoparticles synthesized through “green” routes are increasingly being used in a myriad of biological applications,21 polyphenols derived from indigenous plant sources have caught significant attention as reducing and capping molecules for these nanoparticles. However polyphenol-capped AuNPs, though widely established in the literature,22 have not yet been explored in the protein fibrillation domain. Therefore, elucidating the effect of different polyphenol-capped AuNPs on protein fibrillation calls for detailed investigation. Different polyphenolic compounds such as silymarin, quercetin and gallic acid have been considered in our work for the synthesis of biosafe AuNPs. These compounds with different antioxidant capacities have been selected as they have received substantial interest as alternative chemotherapeutics in degenerative conditions.23
Though 37 different proteins have been reported to cause amyloid diseases,24 human serum albumin (HSA) is extensively used as one typical α-helical (>60%) protein to understand the mechanisms of the misfolding and fibrillation process.25,26 HSA is a physiologically prolific, single chain protein containing 585 amino acids arranged in three homologous domains.27 The literature has shown that the systematic aggregation of albumin occurs through partial misfolding of its native structure and the process has been conveniently replicated in vitro through specific conditions such as high temperature, metal ions, treatment with denaturing chemicals, and lowering of pH.28 Free radical-induced fibrillation of HSA occurs when HSA in phosphate buffer is incubated at elevated temperatures.29
Our present work intends to illustrate the role of polyphenol capping molecules on the fibrillation of HSA at elevated temperatures. Silymarin, quercetin and gallic acid were used for the facile fabrication of gold nanoparticles in a controlled size range. Fibrillation inhibition capacities of the nanoparticles have been recorded through a combination of various spectroscopic and microscopic techniques. Both the nanoparticles and protein–nanoparticle complexes were separately checked for any indication of cytotoxicity at their fibrillation inhibitory concentrations. Additionally, in silico studies were performed to identify all potential interactions between the nanosurface functionalities and albumin native structure. Finally, nanoparticle-treated HSA, under oxidatively stressed conditions, was analysed through SDS PAGE to detect the presence of protein oligomers.
2. Experimental
2.1 Materials
Gold(III) chloride, quercetin, silymarin, gallic acid, HSA and trypan blue were obtained from Sigma-Aldrich, USA. Thioflavin T (ThT) dye used for fibrillation experiments was purchased from TCI chemicals, Japan. HPLC grade water and other solvents were procured from Spectrochem Pvt. Ltd, India. Potassium bromide for IR analysis was purchased from Merck, Germany. Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), and, penicillin–streptomycin and trypsin–EDTA solutions were purchased from HiMedia, India.2.2 Preparation and characterization of AuNPs
Polyphenol-capped gold nanoparticles were synthesized through methods previously developed in our laboratory. Quercetin stabilized gold nanoparticles were prepared under low temperature sonication for 1 min, where 3 mL of 1.3 mM chloroauric acid was allowed to react with 1 mL of 4 mM quercetin. Gallic acid-reduced gold nanoparticles were synthesized under similar conditions. 1 mL of a methanolic solution of 4 mM gallic acid was charged into 3 mL of chloroauric acid at 4 ± 2 °C and sonicated for 1 min to facilitate GAuNP formation. Similarly, 1 mL of a methanolic solution of 4 mM silymarin was added into 3 mL of chloroauric acid and the synthesis of silymarin-reduced gold nanoparticles was carried out at 45 ± 2 °C under sonication for 30 min. The nanoparticle suspensions were first subjected to a mild centrifugation at 6000 rpm to sediment any large particles. The supernatants were collected and centrifuged again at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 rpm to harvest the nanoparticles. A two-step centrifugation process ensured collection of the nanoparticles in a narrow size range.30 The pellets were washed, re-centrifuged and finally redispersed in water before storing them at 4 °C for further applications.
500 rpm to harvest the nanoparticles. A two-step centrifugation process ensured collection of the nanoparticles in a narrow size range.30 The pellets were washed, re-centrifuged and finally redispersed in water before storing them at 4 °C for further applications.
The surface plasmon responses of different AuNPs were recorded using a double-beam UV-vis spectrophotometer (Shimadzu-2550, Japan) using 1 cm quartz cuvettes. The samples were appropriately diluted with water and absorption spectra were obtained at medium scanning speed in the wavelength range of 200–800 nm. The hydrodynamic diameters of the samples were recorded using a Nano ZS zetasizer (Malvern Instruments, UK) equipped with a 4 mW helium–neon laser beam, 633 nm and back scattering angle of 173°. Electron microscopy was performed by placing a drop of diluted sample on a carbon-coated copper grid (Ted Pella Inc., US). Micrographs were recorded at an accelerating voltage of 200 kV using a JEOL JEM 2100 HR transmission electron microscope (JEOL, Japan). FT IR spectra over a range of 4000–400 cm−1 of pelletized samples were obtained at a resolution of 4 cm−1 using a Jasco-670 Plus FT IR spectrometer (Jasco, Japan). The particle size result, obtained from an average of 50 particles in electron micrographs, was applied to fix the working concentration of gold nanoparticles in solution (100 nM) using a spectroscopic method described by Khlebstov et al.31
2.3 DPPH radical scavenging activity
Free radical scavenging capacities of AuNPs (dispersed in water) were determined and compared by DPPH assay following a method described by Sanna et al.32 0.5 mL of samples was added to 1 mL of 0.2 mM methanolic DPPH solution such that final concentrations of AuNPs were in the range of 10–100 nM. The ratio between water and methanol in the final mixture was fixed at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 for all samples. Methanol served as the negative control. The absorbance values of the reaction mixtures were recorded at 517 nm after 30 min using a UV-vis Spectrophotometer. All experiments were performed in triplicate and the percentage inhibition of the free radical scavenging activity of the AuNPs was calculated using the following formula:
2 for all samples. Methanol served as the negative control. The absorbance values of the reaction mixtures were recorded at 517 nm after 30 min using a UV-vis Spectrophotometer. All experiments were performed in triplicate and the percentage inhibition of the free radical scavenging activity of the AuNPs was calculated using the following formula:2.4 Protein fibril preparation
A stock solution of HSA was initially prepared by dissolving HSA in 50 mM PBS (pH 7.4), and its concentration was fixed spectrophotometrically at 280 nm using a molar extinction coefficient of 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 219 M−1 cm−1. HSA (2 μM) was incubated in the presence and absence of AuNPs at 65 ± 2 °C for 24 h for the generation of matured protein fibrils.33,34 The effects of differently capped AuNPs on HSA fibrillation were studied by adding 100 μL aliquots of each type of AuNPs such that the stoichiometric ratio of AuNPs
219 M−1 cm−1. HSA (2 μM) was incubated in the presence and absence of AuNPs at 65 ± 2 °C for 24 h for the generation of matured protein fibrils.33,34 The effects of differently capped AuNPs on HSA fibrillation were studied by adding 100 μL aliquots of each type of AuNPs such that the stoichiometric ratio of AuNPs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HSA was 1
HSA was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200. Matured protein fibrils were subsequently characterized by various biophysical techniques.
200. Matured protein fibrils were subsequently characterized by various biophysical techniques.
2.5 Thioflavin T assay
Samples withdrawn after completion of the incubation period were incubated with ThT solution (20 μM)35 and fluorescence intensities were recorded using a spectrofluorimeter (PerkinElmer LS-5, PerkinElmer Inc., US) equipped with a waterbath circulator controlled by a Neslab RTE 100 thermostat. The excitation and emission wavelengths were set at 450 and 480 nm, respectively. The slit width was 5 nm. All blanks and nanoparticle samples were checked to confirm non-responsiveness to ThT fluorescence.2.6 Circular dichroism (CD)
Far-UV CD measurements of the samples (from Section 2.4) were performed on a JASCO J-815 CD spectro-polarimeter with a thermostatically controlled cell holder at 25 °C, in a cuvette of 0.1 cm path length. The scan speed was set as 100 nm min−1 and ellipticities were recorded between 190 and 250 nm from an average of three scans. The data obtained were further processed to quantify the secondary structural content using the online server BeStSel (http://bestsel.elte.hu).362.7 Microscopy
For fluorescence microscopy, HSA fibrillar samples (each 15 μL) incubated with or without AuNPs for a period of 24 h were stained with 1 mM ThT solution (15 μL) and placed on cleaned glass slides, covered with cover slips. Fluorescence micrographs of protein aggregates were captured using an Axiovert 40 CFL (Carl Zeiss, Germany) microscope.Diluted samples were applied onto carbon-coated copper grids and negatively stained with freshly prepared 2% (w/v) uranyl acetate solution. The grids were air-dried and the micrographs were obtained using transmission electron microscopy (TEM) at an operating voltage of 120 kV.
2.8 Cell culture
The HaCaT cell line was obtained from the National Centre for Cell Science (NCCS), Pune, India. Cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin–streptomycin at 37 °C and 5% CO2.2.9 Cytotoxicity assay
In vitro cytotoxicity evaluations of HSA–AuNP complexes were performed by using the trypan blue exclusion method according to a previous protocol.37 Briefly, HaCaT cells were cultured at a density of 1 × 105 per well in a 24-well plate for 24 h. Then, the cells were either left untreated or treated with different doses of HSA (2 μM)–AuNP (5–20 nM) complexes for 24 h. Then, the cells were harvested for microscopic investigations (Olympus IX70, Olympus Corp., Japan) and trypan blue solution (0.4%) was added to the cell suspensions in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Finally, the number of total and dead cells was counted using a hemocytometer. The data were averaged from a set of three replicates. The percentage of viable cells was calculated and plotted.
1. Finally, the number of total and dead cells was counted using a hemocytometer. The data were averaged from a set of three replicates. The percentage of viable cells was calculated and plotted.
2.10 Molecular modelling
Structures of silymarin, quercetin and gallic acid were prepared in LigPrep.38 Induced fit docking was performed using the IFD docking module of the Schrödinger software package. The best dock pose of the ligands based on interaction scores was selected and schematically represented as 3D figures. The amino acid sequence in HSA was obtained in the FASTA format and aggregation-prone regions were identified using the Aggrescan online server.392.11 HSA in vitro oxidation and SDS PAGE
Samples (from Section 2.4) were allowed to incubate in the presence of 50 μM H2O2 for complete oxidation of HSA. Formation of high molecular weight protein aggregates was determined by 1 D sodium dodecyl sulfate polyacrylamide gel electrophoresis. The treated and untreated HSA samples (50 μM) suspended in 20 μL of 1× loading buffer were incubated at 90 °C for 2 minutes and cooled to room temperature. Electrophoresis was then carried out using 10% polyacrylamide SDS PAGE in an electric field run under a constant voltage of 100V for 45 minutes. The gels were then fixed by Coomassie blue stain and captured on a gel documentation system (BioRad, Hercules). Freshly prepared HSA solution was also run alongside for comparative purposes.2.12 Statistical analysis
Results are presented as mean ± standard deviation (S.D.). Origin 6.0 Professional was used for the analysis of the experimental data. Student's t-test was conducted for comparative studies and the difference was considered significant when P < 0.05.3. Results and discussion
3.1 Characterization of AuNPs
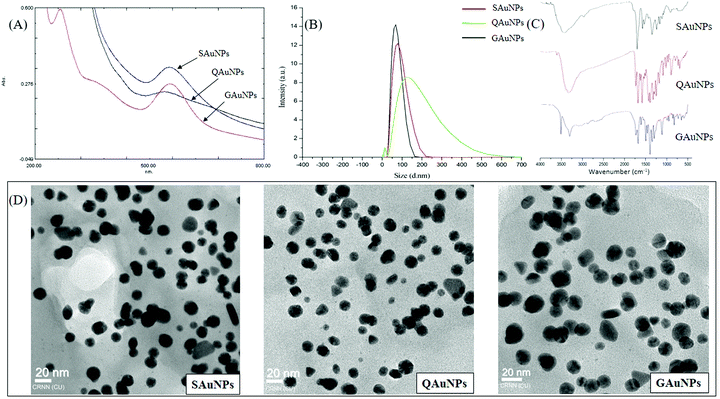
A two-step process of AuNP formation involves an initial nucleation, followed by coalescence of gold atoms40 and the phenomenon was recorded by UV-vis absorption spectral analysis. SAuNP synthesis was confirmed by a characteristic surface plasmon resonance (SPR) at 547 nm, and similar peaks were recorded at 541 nm and 539 nm for QAuNPs and GAuNPs, respectively (Fig. 1). The SPR pattern depends on the shape and size of metal nanoparticles, and also on the dielectric constant of the surrounding medium,41 and the SPR intensity and position varied with the type of capping molecules. The hydrodynamic diameters of AuNPs as recorded by DLS were within the close range of 50 to 80 nm. High resolution TEM micrographs showed that the nanoparticles capped with different polyphenols were spherical in shape. Electron microscopy also confirmed that the average particle diameters of SAuNPs, QAuNPs and GAuNPs were 15.35 ± 0.26 nm, 15.12 ± 0.21 nm and 15.05 ± 0.20 nm, respectively. Particle size is a major parameter that guides the association of protein molecules on nano-scale materials.15 Near monodispersity of gold nanoparticles as confirmed by both light scattering and microscopic techniques minimized the chances of differential interaction with HSA arising from size variation. FTIR measurements were performed to detect the surface presence of different polyphenols responsible for reduction of the gold salt (Au3+) into the elemental form (Au0). The FTIR spectra of all three samples exhibited common bands at around 3420 cm−1, 1638 cm−1 and 1508 cm−1, arising from the phenolic –OH vibrations, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching and aromatic ring stretching, respectively. These bands are characteristics of polyphenolic structures and are ubiquitous on the gold nanosurface.23,42 An additional stretching band near 1088 cm−1 was observed in the case of SAuNPs and QAuNPs, which was attributed to the benzopyran ring of the capping molecules. It is likely that the phenolic –OH groups contributed to the electrostatic stabilization of the nanoparticles in solution.41
O stretching and aromatic ring stretching, respectively. These bands are characteristics of polyphenolic structures and are ubiquitous on the gold nanosurface.23,42 An additional stretching band near 1088 cm−1 was observed in the case of SAuNPs and QAuNPs, which was attributed to the benzopyran ring of the capping molecules. It is likely that the phenolic –OH groups contributed to the electrostatic stabilization of the nanoparticles in solution.41
3.2 Free radical scavenging property of AuNPs
Hydroxyl groups present in the polyphenolic structure are not only responsible for the effective reduction of Au3+ to elemental gold, but also for imparting a free radical scavenging property to the nanoparticles.43 Discoloration of DPPH is one widely used technique to quantify the radical scavenging capacity of polyphenols.44 The DPPH radical scavenging activities of all AuNPs were observed to be a function of their concentrations and exhibited good linearity throughout (SAuNPs: R2 = 0.9989; QAuNPs: R2 = 0.9678; GAuNPs: R2 = 0.9667). Percent free radical scavenging activity of GAuNPs and QAuNPs ranged from 26% and 27% at 10 nM, to 91% and 94% at 100 nM, respectively (Fig. S1, ESI†). A significant decrease (P < 0.05) of DPPH inhibitory capacity was observed in the case of SAuNPs, with only 22% activity recorded at 100 nM. Quercetin and gallic acid are two of the best known antioxidant molecules of plant origin.45 Silymarin, however, has a comparatively lower anti-oxidation potential than the others. Previous studies have established that the antioxidant capacity of such polyphenol capping agents remains well preserved in the synthesized nanoparticles. DPPH scavenging activity of the polyphenolics is associated with the chemical structure, which influences the accessibility to the radical centre of DPPH.32,46 It is therefore understandable that the DPPH scavenging activity of SAuNPs is significantly lower than that of QAuNPs and GAuNPs at different concentrations.3.3 Effect of AuNPs on HSA fibril formation
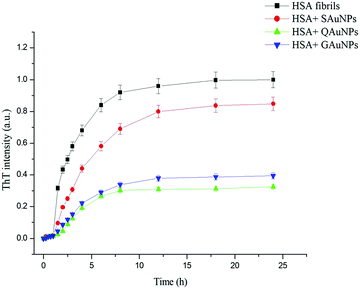
The fibrillation kinetics of HSA in the presence and absence of differently capped AuNPs have been studied using ThT fluorescence spectroscopy (Fig. 2). Thioflavin T (ThT) is a fluorescent dye capable of binding specifically to amyloid fibrils, and not to precursor proteins and aggregates.47 HSA fibrillation was instituted at ∼65 °C. Hydrogen peroxide is a major product generated when phosphate buffer is heated at elevated temperatures. This induced oxidative stress and initiated protein fibrillation. The ThT responses from HSA samples withdrawn at different time intervals increased in a sigmoidal manner after an initial lag phase of 0.6 h and achieved saturation at around 6 h. The fibril growth curve was observed to be in agreement with the nucleation-dependent polymerization model for amyloidogenic proteins.48 While studying the inhibitory effects of different AuNPs on HSA fibrillation, the maximum ThT intensity obtained from HSA fibrils was used to normalize the ThT readings. The HSA fibril growth profile showed significant deviation when HSA was incubated in the presence of SAuNPs. All the ThT data were fitted into the following sigmoidal equation (Microcal Origin 6.0) to obtain the lag time informationwhere Y is ThT fluorescence intensity, Fmin is the minimal fluorescence intensity, Fmax is the maximum intensity, t is incubation time, and t0 is the time required to attain 50% maximal fluorescence. The apparent first order rate constant for protein fibril growth is expressed as 1/τ and its lag time is calculated as t0 − 2τ.
Upon co-incubation of SAuNPs, the ThT temporal response value at steady state was decreased by 15.2%. Conversely, the ThT response diminished greatly throughout the incubation period when HSA was incubated with QAuNPs. Though the ThT value at equilibrium was reduced by 67.6%, no change in lag phase was observed. Similar effects on the lag phase of HSA were recorded when GAuNPs were co-incubated under identical conditions. A decrease in the ThT response by 60.2% detected after 24 h of incubation indicated that GAuNPs delayed the formation of HSA fibrils. Multivalent polyphenol-capped nanoparticles likely achieved concentration-dependent access to HSA fibrillation zones. It was inferred that though the presence of different polyphenol-capped AuNPs in HSA solution did not interfere with the nucleation of protein monomers, a diminution in the fibril content was, however, observed in the elongation and saturation stages in the case of HSA solutions co-incubated with different AuNPs.
3.4 Effect of AuNPs on protein secondary conformation
Conformational changes in serum albumin secondary structure during fibrillation were studied in detail using far UV-circular dichroism. The CD spectra of native HSA and HSA fibrils in the presence and absence of polyphenol-capped AuNPs are presented in Fig. 3a. A freshly prepared HSA solution at pH 7.4 exhibited the two obvious negative peaks at 208 and 222 nm, owing to its α-helical structure.14 The negative peaks at 208 and 222 nm arise from the π → π* and n → π* transitions of carbonyl groups present in the polypeptide chains. Samples incubated under fibrillating conditions for 24 hours showed a disappearance of the negativity at 208 and 222 nm due to loss of α-helicity. The appearance of the single negative peak at 215 nm indicated an increase of β-sheet content, which leads to protein fibrillation.49,50 Co-incubation with SAuNPs resulted in a significant change in the CD spectra as compared to that of the HSA fibrils. However, more negative mdeg values as observed in the case of GAuNPs and QAuNPs revealed that GAuNPs and QAuNPs were more effective in retaining the α-helical features of the HSA structures. Nanoparticles apparently hindered HSA hydrophobic interactions and catalysed the α-helical transformations.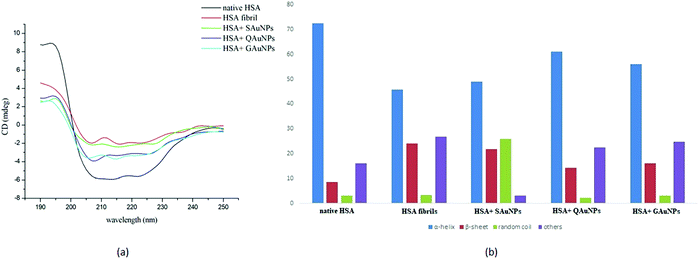 | ||
| Fig. 3 (A) CD spectra and (B) histograms of secondary structural contents of HSA incubated in the absence and presence of AuNPs for 24 h. | ||
The histograms in Fig. 3b show that while native HSA in the absence of nanoparticles contained ∼72.4% α-helical structure and ∼8.5% β-sheets, the samples collected after 24 h of incubation exhibited ∼45.8% α-helical structure and 24.1% β-sheet contents. It was observed that the α-helical content of HSA fibril in the presence of SAuNPs improved to ∼49%, although changes in the β-sheet content were minimal. In contrast, quantitative estimation of secondary structures of GAuNP-co-incubated HSA samples revealed a notable amount of α-helix (∼56%) and a low β-sheet content (∼16%). Similarly, it was found that HSA samples incubated with QAuNPs contained ∼61.2% α-helix and only ∼14% β-sheets. The α-helix structural retention capacity was most pronounced in the case of QAuNPs, closely followed by GAuNPs, whereas β-sheets constituted the majority of the secondary structural content of SAuNP-co-incubated HSA samples.
3.5 Morphological analysis of HSA aggregates
The formation and distribution of ThT stained protein fibril structures were preliminarily monitored by using fluorescence microscopy. HSA fibrils formed in the absence of AuNPs appeared as irregular patches with high fluorescence intensities scattered throughout the micrograph (Fig. 4a). A substantial number of fluorescent patches were observed in the case of HSA samples co-incubated with SAuNPs for 24 hours (Fig. 4b). However, the numbers were found to be significantly smaller as compared to the control HSA fibril solution. The fluorescent patches disappeared even further in incubated HSA samples containing QAuNPs and GAuNPs (Fig. 4c and d), which indicated that even lower amounts of fibrils were formed as compared to the control and SAuNP co-incubated HSA solution.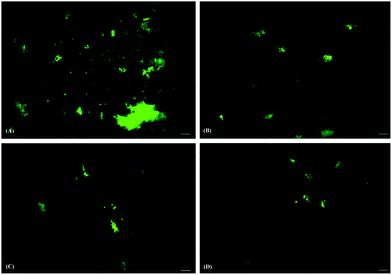 | ||
| Fig. 4 Fluorescence micrographs of HSA incubated (A) alone and in the presence of (B) SAuNPs, (C) QAuNPs, and (D) GAuNPs. Scale bars represent 100 μm. | ||
Electron microscopy is a powerful technique to study the fibrillar structures of amyloid proteins.51Fig. 5 shows the electron micrographs of HSA fibrils incubated in the absence or presence of different polyphenol-capped gold nanoparticles. Images of HSA solutions when incubated alone at 65 °C displayed entangled networks of long fibrils, and these observations are found to be similar to those of earlier works.26,52,53 Co-incubation of SAuNPs resulted in protein absorption on the nanoparticle surface and the generation of a disorganised network of protein aggregates. A further reduction of fibrillar networks and the presence of amorphous protein aggregates on gold nanosurfaces were observed in the case of QAuNP and GAuNP co-incubated HSA samples. It was also noted that the sample containing QAuNPs contained significantly fewer aggregates compared to the HSA solutions incubated with GAuNPs. These observations were found to be in accordance with the spectroscopic studies.
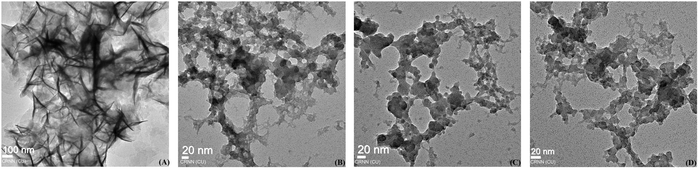 | ||
| Fig. 5 Electron micrographs of HSA incubated (A) alone and in the presence of (B) SAuNPs, (C) QAuNPs, and (D) GAuNPs. | ||
3.6 Effect of HSA–AuNP complexes on cell viability
Apart from considering fibrillation inhibitory efficacy, cell viability conditions should also be taken into account when selecting an appropriate inhibitor.18 Although serum albumin has been reported to be non-cytotoxic,54 understanding the effects of AuNP-co-incubated HSA solutions on cell viability is essential prior to claiming polyphenol-capped AuNPs as protein fibrillation inhibitors. Cell viability analysis (Fig. 6) on HaCaT cells revealed that no significant cell death was recorded for HSA fibril solutions incubated with varying concentrations of AuNPs (5–20 nM). The results were further confirmed through microscopy, which showed no apparent changes in cellular morphology. The cytotoxicity of all the synthesized nanoparticles was found to be minimal even at 2 fold the working concentration (Fig. S2, ESI†). Polyphenolic coating on the nanosurface contributed to the biocompatibility of the gold nanoparticles,21 which otherwise are known for their cytotoxic properties.55,56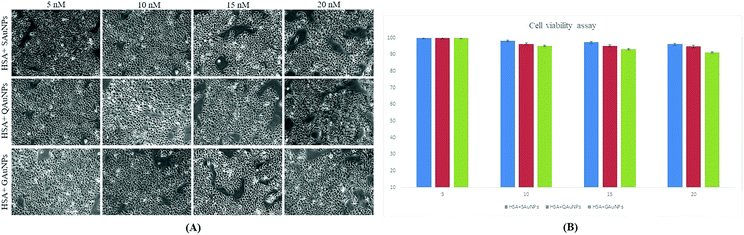 | ||
| Fig. 6 (A) Microscopic views of HaCaT cell viability at 20× magnification and (B) percentage cell viability of HSA–AuNP complexes at varying concentrations of AuNPs (5–20 nM). | ||
3.7 Influence of anti-oxidative capacity of AuNPs on HSA fibrillation
The inhibition capacity of gold nanoparticles as examined by a combination of spectroscopic and microscopic techniques was found to differ for the differently capped nanoparticles. As the physical characteristics of the synthesized nanoparticles were similar, it can be hypothesized that polyphenol molecules on the gold nanosurface dictated the fibril inhibitory capacity of the nanoparticles. The inhibition mechanism caused by the polyphenolic compounds constitutes complex pathways involving both specific molecular interactions and free radical scavenging activities.57 IR experiments on the gold nanoparticles previously revealed that the surface hydroxyl groups were available for interaction with albumin macromolecules (Section 3.1). Molecular docking studies on quercetin, gallic acid and silymarin demonstrated that the interactions of these capping molecules with the amyloidogenic regions of HSA were different in each case (ESI†). While gallic acid failed to form interactions with any of the amyloidogenic spots, silymarin formed one hydrogen bond with the GLU 153 residue of HSA. Quercetin, on the other hand, formed two hydrogen bonds with the GLU 230 and ASP 237 residues individually. These interactions alone could not explain the strong inhibitory activity of GAuNPs and milder inhibitory capacity of SAuNPs. Gold nanoparticles are known to enhance polyphenol solubilisation and diffusive adsorption at protein interfaces.58,59 Polyphenols like quercetin ionize extensively at pH 7.4, but that was minimal in the case of silymarin. The different inhibitory effect of AuNPs on HSA fibrillation was likely due to similar effects. A prominent correlation between the DPPH scavenging activity and fibrillation inhibitory capacity of capped AuNPs could also be outlined. The QAuNPs bearing the highest DPPH scavenging capacity restricted fibril formation to the maximum extent. This observation further demanded an understanding of the role of the polyphenol-tethered nanosurface in the free radical-induced protein fibrillation inhibition. Samples were subjected to hydrogen peroxide treatment to ensure complete oxidation of HSA. Similar studies reported that oxidised protein aggregates could be conveniently detected through SDS PAGE.60 While gel electropherograms (Fig. 7) of hydrogen peroxide-treated HSA showed the presence of different oxidation products, AuNPs effectively preserved the protein from oxidant-induced aggregation. It was recorded that QAuNPs effectively conserved a majority of the native HSA monomers in the presence of oxidants. Similarly, it was observed that GAuNPs prevented the formation of oxidation species through conservation of native HSA structure. SAuNPs, possessing the least scavenging activity, allowed much of the HSA molecules to lose their original configuration. This specific experimental design supported the hypothesis that the polyphenol-capped nanoparticles quenched the free radicals in the medium, thereby maintaining the integrity of the native protein structures.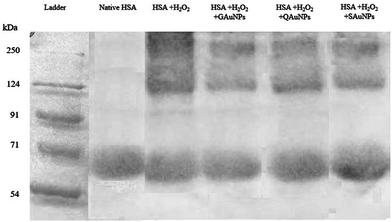 | ||
| Fig. 7 Resolved SDS PAGE of (i) untreated HSA solution, (ii) HSA + H2O2, (iii) HSA + H2O2 + SAuNPs, (iv) HSA + H2O2 + QAuNPs and (v) HSA + H2O2 + GAuNPs. | ||
4. Conclusion
Silymarin, quercetin and gallic acid were used as reducing and stabilizing agents for the synthesis of biosafe gold nanoparticles of uniform size range. The influence of different polyphenol-capped gold nanoparticles on human serum albumin fibril formation was studied by a combination of various biophysical techniques. Fluorescence spectroscopy revealed that although the ThT intensity of the HSA fibril sample incubated with SAuNPs reduced by ∼15%, a considerable decrease of the signals was observed in the case of QAuNP(∼67%)- and GAuNP (∼60%)-co-incubated HSA samples. Comparable observations of the secondary structural content of AuNP-co-incubated HSA samples were recorded using CD spectroscopy. QAuNPs and GAuNPs effectively conserved the native α-helical features of albumin incubated at high temperatures. Microscopic investigations showed that HSA incubated alone formed well-defined regular fibrils, but formed amorphous aggregates in the presence of AuNPs. The difference of polyphenol molecule tethering thus caused a dissimilarity in fibrillation modulatory effects of the structurally identical AuNPs. Non-covalent interactions between nanosurface capping moeities and HSA as perceived through in silico experiments essentially remained unrelated to the difference in nanoparticle fibrillation inhibitory potential. However, an association between the fibrillation inhibitory capacities of the AuNPs and their DPPH scavenging activities was well perceived. SDS PAGE experiments corroborated that free radical generation in aqueous medium diminished in the presence of polyphenol-capped AuNPs, thereby restricting protein misfolding. This work provided the first evidence that that antioxidant capacity of nanoparticles contributes to their fibrillation inhibitory activity. It is therefore expected that this report will contribute to the better design of nanomaterials for therapeutic interventions under amyloidogenic conditions.Conflicts of interest
The authors would like to declare that there are no conflicts of interest in the publication of this article.Acknowledgements
The authors would like to acknowledge the TEQIP project at the University of Calcutta for instrumental facilities.References
- D. B. Zorov, M. Juhaszova and S. J. Sollott, Physiol. Rev., 2014, 94, 909–950 CrossRef CAS.
- M. Schieber and N. S. Chandel, Curr. Biol., 2014, 24, R453–R462 CrossRef CAS PubMed.
- F. U. Hartl, A. Bracher and M. Hayer-Hartl, Nature, 2011, 475, 324–332 CrossRef CAS PubMed.
- S. K. Chaturvedi, M. K. Siddiqi, P. Alam and R. H. Khan, Process Biochem., 2016, 51, 1183–1192 CrossRef CAS.
- C. Wang, A. Yang, X. Li, D. Li, M. Zhang, H. Du, C. Li, Y. Guo, X. Mao, M. Dong, F. Besenbacher, Y. Yang and C. Wang, Nanoscale, 2012, 4, 1895 RSC.
- A. J. Doig and P. Derreumaux, Curr. Opin. Struct. Biol., 2015, 30, 50–56 CrossRef CAS PubMed.
- E. K. Kumar and N. P. Prabhu, Phys. Chem. Chem. Phys., 2014, 16, 24076–24088 RSC.
- L. Xiao, D. Zhao, W.-H. Chan, M. M. F. Choi and H.-W. Li, Biomaterials, 2010, 31, 91–98 CrossRef CAS PubMed.
- C. Cabaleiro-Lago, F. Quinlan-Pluck, I. Lynch, S. Lindman, A. M. Minogue, E. Thulin, D. M. Walsh, K. A. Dawson and S. Linse, J. Am. Chem. Soc., 2008, 130, 15437–15443 CrossRef CAS PubMed.
- H. Skaat, R. Chen, I. Grinberg and S. Margel, Biomacromolecules, 2012, 13, 2662–2670 CrossRef CAS PubMed.
- S. Sardar, S. Pal, S. Maity, J. Chakraborty and U. C. Halder, Int. J. Biol. Macromol., 2014, 69, 137–145 CrossRef CAS PubMed.
- M. Mahmoudi, O. Akhavan, M. Ghavami, F. Rezaee and S. M. A. Ghiasi, Nanoscale, 2012, 4, 7322 RSC.
- S. Mirsadeghi, S. Shanehsazzadeh, F. Atyabi and R. Dinarvand, Mater. Sci. Eng., C, 2016, 59, 390–397 CrossRef CAS PubMed.
- S. Konar, S. Sen and A. Pathak, J. Phys. Chem. B, 2017, 121, 11437–11448 CrossRef CAS PubMed.
- S. H. De Paoli Lacerda, J. J. Park, C. Meuse, D. Pristinski, M. L. Becker, A. Karim and J. F. Douglas, ACS Nano, 2010, 4, 365–379 CrossRef CAS PubMed.
- S. BarathManiKanth, K. Kalishwaralal, M. Sriram, S. Pandian, H. Youn, S. Eom and S. Gurunathan, J. Nanobiotechnol., 2010, 8, 16 CrossRef PubMed.
- A. P. Muller, G. K. Ferreira, A. J. Pires, G. de Bem Silveira, D. L. de Souza, J. de, A. Brandolfi, C. T. de Souza, M. M. S. Paula and P. C. L. Silveira, Mater. Sci. Eng., C, 2017, 77, 476–483 CrossRef CAS PubMed.
- S. Sudhakar, P. Kalipillai, P. B. Santhosh and E. Mani, J. Phys. Chem. C, 2017, 121, 6339–6348 CrossRef CAS.
- S. Sen, S. Dasgupta and S. DasGupta, J. Phys. Chem. C, 2017, 121, 18935–18946 CrossRef CAS.
- Y. Kim, J. H. Park, H. Lee and J. M. Nam, Sci. Rep., 2016, 6, 1–14 CrossRef PubMed.
- S. Fazal, A. Jayasree, S. Sasidharan, M. Koyakutty, S. V. Nair and D. Menon, ACS Appl. Mater. Interfaces, 2014, 6, 8080–8089 CrossRef CAS PubMed.
- Y. Lee and T. G. Park, Langmuir, 2011, 27, 2965–2971 CrossRef CAS PubMed.
- S. Das, P. Roy, R. G. Auddy and A. Mukherjee, Int. J. Nanomed., 2011, 6, 1291–1301 CrossRef CAS PubMed.
- F. Chiti and C. M. Dobson, Annu. Rev. Biochem., 2017, 86, 1–42 CrossRef PubMed.
- J. Juárez, S. G. López, A. Cambón, P. Taboada and V. Mosquera, J. Phys. Chem. B, 2009, 113, 10521–10529 CrossRef PubMed.
- S. Bag, R. Mitra, S. DasGupta and S. Dasgupta, J. Phys. Chem. B, 2017, 121, 5474–5482 CrossRef CAS PubMed.
- L. Xu, Y. X. Hu, Y. C. Li, L. Zhang, H. X. Ai, H. S. Liu, Y. F. Liu and Y. L. Sang, J. Mol. Struct., 2017, 1149, 645–654 CrossRef CAS.
- P. Taboada, S. Barbosa, E. Castro and V. Mosquera, J. Phys. Chem. B, 2006, 110, 20733–20736 CrossRef CAS PubMed.
- V. I. Bruskov, L. V. Malakhova, Z. K. Masalimov and A. V. Chernikov, Nucleic Acids Res., 2002, 30, 1354–1363 CrossRef CAS PubMed.
- F. Bonaccorso, M. Zerbetto, A. C. Ferrari and V. Amendola, J. Phys. Chem. C, 2013, 117, 13217–13229 CrossRef CAS.
- N. G. Khlebtsov, Anal. Chem., 2008, 80, 6620–6625 CrossRef CAS PubMed.
- V. Sanna, N. Pala, G. DessÌ, P. Manconi, A. Mariani, S. Dedola, M. Rassu, C. Crosio, C. Iaccarino and M. Sechi, Int. J. Nanomed., 2014, 9, 4935–4951 Search PubMed.
- S. Goy-López, J. Juárez, M. Alatorre-Meda, E. Casals, V. F. Puntes, P. Taboada and V. Mosquera, Langmuir, 2012, 28, 9113–9126 CrossRef PubMed.
- A. Basu, S. Bhayye, S. Kundu, A. Das and A. Mukherjee, RSC Adv., 2018, 8, 30717–30724 RSC.
- S. Sen, S. Konar, B. Das, A. Pathak, S. Dhara, S. Dasgupta and S. DasGupta, RSC Adv., 2016, 6, 43104–43115 RSC.
- A. Micsonai, F. Wien, L. Kernya, Y.-H. Lee, Y. Goto, M. Réfrégiers and J. Kardos, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E3095–E3103 CrossRef CAS PubMed.
- C. Basu and R. Sur, BioMed Res. Int., 2018, 2018, 1–14 CrossRef PubMed.
- L. L. C. Schrödinger, Induced Fit Docking, Glide, New York, NY, 2013 Search PubMed.
- P. Biswas, A. Dutta, S. Dasgupta, A. K. Das, S. Chaudhury and S. Bag, Spectrochim. Acta, Part A, 2017, 192, 318–327 Search PubMed.
- R. Erler, A. F. Thu, S. Sokolov, T. T. Ahner, K. Rademann, F. Emmerling and R. Kraehnert, ACS Nano, 2010, 4, 1076–1082 CrossRef PubMed.
- T. C. Prathna, N. Chandrasekaran, A. M. Raichur and A. Mukherjee, Colloids Surf., B, 2011, 82, 152–159 CrossRef CAS PubMed.
- S. Das, P. Roy, S. Mondal, T. Bera and A. Mukherjee, Colloids Surf., B, 2013, 107, 27–34 CrossRef CAS PubMed.
- T. Kongpichitchoke, J.-L. Hsu and T.-C. Huang, J. Agric. Food Chem., 2015, 63, 4580–4586 CrossRef CAS PubMed.
- K. Pyrzynska and A. Pekal, Anal. Methods, 2013, 5, 4288–4295 RSC.
- I. M. C. Brighente, M. Dias, L. G. Verdi and M. G. Pizzolatti, Pharm. Biol., 2007, 45, 156–161 CrossRef CAS.
- F. G. Milanezi, L. M. Meireles, M. M. de Christo Scherer, J. P. de Oliveira, A. R. da Silva, M. L. de Araujo, D. C. Endringer, M. Fronza, M. C. C. Guimarães and R. Scherer, Saudi Pharm. J., 2019, 27, 968–974 CrossRef CAS PubMed.
- H. LeVine, Methods in Enzymology, 1999, vol. 309, pp. 274–284 Search PubMed.
- M. Calamai, F. Chiti and C. M. Dobson, Biophys. J., 2005, 89, 4201–4210 CrossRef CAS PubMed.
- J. Juárez, P. Taboada and V. Mosquera, Biophys. J., 2009, 96, 2353–2370 CrossRef PubMed.
- M. K. Siddiqi, P. Alam, S. K. Chaturvedi and R. H. Khan, Int. J. Biol. Macromol., 2016, 92, 1220–1228 CrossRef CAS PubMed.
- T. Shirahama and A. S. Cohen, J. Cell Biol., 1967, 33, 679–708 CrossRef CAS PubMed.
- S. Sen, S. Konar, A. Pathak, S. Dasgupta and S. Dasgupta, J. Phys. Chem. B, 2014, 118, 11667–11676 CrossRef CAS PubMed.
- N. Xiong, X. Y. Dong, J. Zheng, F. F. Liu and Y. Sun, ACS Appl. Mater. Interfaces, 2015, 7, 5650–5662 CrossRef CAS PubMed.
- N. K. Holm, S. K. Jespersen, L. V. Thomassen, T. Y. Wolff, P. Sehgal, L. A. Thomsen, G. Christiansen, C. B. Andersen, A. D. Knudsen and D. E. Otzen, Biochim. Biophys. Acta, Proteins Proteomics, 2007, 1774, 1128–1138 CrossRef CAS PubMed.
- M. Jafari, F. Rokhbakhsh-Zamin, M. Shakibaie, M. H. Moshafi, A. Ameri, H. R. Rahimi and H. Forootanfar, Biocatal. Agric. Biotechnol., 2018, 15, 245–253 CrossRef.
- N. M. Schaeublin, L. K. Braydich-Stolle, A. M. Schrand, J. M. Miller, J. Hutchison, J. J. Schlager and S. M. Hussain, Nanoscale, 2011, 3, 410–420 RSC.
- X. Meng, L. A. Munishkina, A. L. Fink and V. N. Uversky, Biochemistry, 2009, 48, 8206–8224 CrossRef CAS PubMed.
- N. K. Ramesh, S. Sudhakar and E. Mani, Langmuir, 2018, 34, 4004–4012 CrossRef CAS PubMed.
- R. K. Gangwar, V. A. Dhumale, D. Kumari, U. T. Nakate, S. W. Gosavi, R. B. Sharma, S. N. Kale and S. Datar, Mater. Sci. Eng., C, 2012, 32, 2659–2663 CrossRef CAS.
- S. Guedes, R. Vitorino, R. Domingues, F. Amado and P. Domingues, Rapid Commun. Mass Spectrom., 2009, 23, 2307–2315 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ma00274g |
| This journal is © The Royal Society of Chemistry 2020 |