Open Access Article
Open Access ArticleNano- and biosensors for the detection of SARS-CoV-2: challenges and opportunities
Siavash
Iravani

Faculty of Pharmacy and Pharmaceutical Sciences, Isfahan University of Medical Sciences, Iran. E-mail: siavashira@gmail.com
First published on 2nd November 2020
Abstract
Nanotechnology and biotechnology are currently being focused on pathogenic viruses, and researchers are ready to use these approaches to detect viral infections. Indeed, during pandemics, innovative nano-based structures and nanobiotechnology can be employed for the rapid, sensitive, and reliable detection of pathogenic viruses to control and prevent/reduce their spread, which is important in the case of the COVID-19 pandemic. Generally, the currently employed detection technique for COVID-19 is quantitative real-time polymerase chain reaction (qRT-PCR) technology, but it is labor-intensive, time-consuming, and cannot be promptly used in remote or resource-limited settings. This may lead to obstacles in obtaining actual data on the infectivity and transmission of SARS-CoV-2. Accordingly, nano- and biosensors should have sufficient sensitivity, selectivity, user-friendliness, scalability, authenticity, portability, specificity, and rapid/robust properties, with the potential for highly qualified and reliable screening, and great sensitivity, with minimal false positive/negative responses. This paper summarizes important alternative nano- and biosensor-based diagnostics approaches in comparison with the conventional methods used for detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Additionally, current important challenges and future perspectives related to the development of these innovative sensors for the detection of SARS-CoV-2 are discussed.
1. Introduction
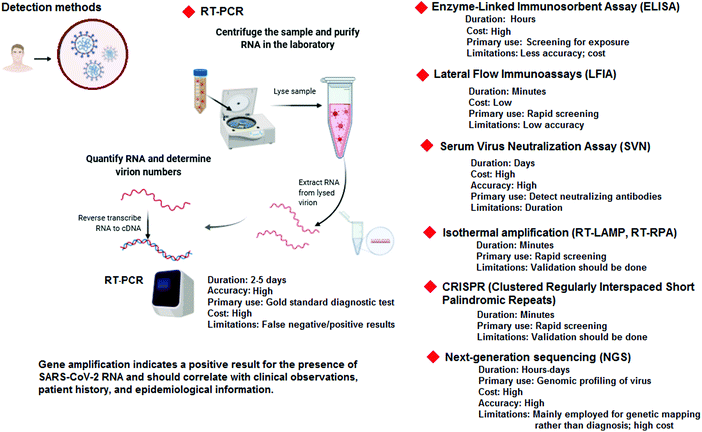
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an enveloped, single-stranded, and positive (+)-sense RNA virus, belonging to the beta-coronavirus (CoV) genera in the family Coronaviridae. The genome of this and other emerging pathogenic human coronaviruses (CoVs) encodes four major structural proteins, including spike (S), envelope (E), membrane (M), and nucleocapsid (N), about 16 nonstructural proteins (nsp1–16), and five to eight accessory proteins; importantly, the S protein plays a critical role in viral attachment, fusion, entry, and transmission.1–3 It includes an N-terminal S1 subunit responsible for virus-receptor binding and a C-terminal S2 subunit responsible for virus-cell membrane fusion. S1 is further divided into an N-terminal domain and a receptor-binding domain. During the infection, CoV first binds to the host cell through an interaction between its S1-receptor-binding domain and the cell membrane receptor, causing conformational alterations in the S2 subunit that result in virus fusion and entry into the target cell.1,4–6Considering the urgency of global healthcare, various technologies, including whole-genome sequencing and computed tomography (CT) imaging have been employed for the diagnosis of infected humans. Importantly, rapid and sensitive diagnosis and detection are urgently needed for epidemiological measurement, infection control, antiviral treatment and vaccine research.2,7 Considerable efforts have also been made to detect and prevent various patterns of community transmission (Fig. 1). Generally, diverse diagnostic and testing kits/assays, such as real-time reverse transcriptase polymerase chain reaction (PCR), enzyme-linked immunosorbent assay (ELISA)-based immunoassays, thermal screening guns, and point-of-care (POC) tests, have been employed or are under further investigation for the detection of SARS-CoV-2 and characterizing the cellular and antibody responses to viral infections. However, these methods have some drawbacks and restrictions/limitations, including high costs, non-specificity, false positive/negative responses, long duration of testing, and they are labor intensive.2,8
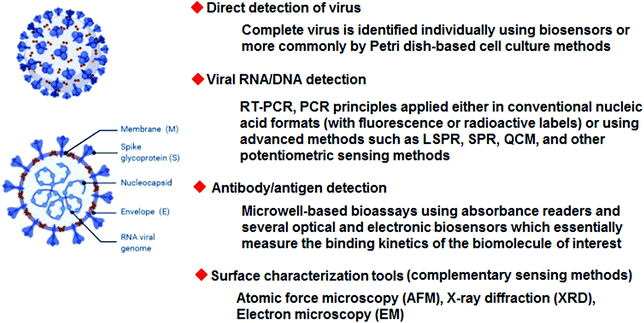 | ||
| Fig. 1 Some important biosensing methods and surface analysis techniques with the potential for detecting SARS-CoV-2. | ||
Typically, methods for the detection of viral infections are based on molecular diagnostics, with the potential to detect the presence of the pathogen, either by recognizing its genetic material or the unique markers of the pathogen itself. In the case of COVID-19, molecular diagnosis mainly relies on the detection of RNA of the SARS-CoV-2 virus. However, detection based on viral proteins can be considered, but unlike nucleic acids, proteins cannot be directly amplified; with no amplification, direct detection of trace amounts of viral proteins is very challenging, and may have some limitations for detection.9 The molecular-based methods generally require samples (containing viruses) from patients, including nasopharyngeal swabs (most reliable) or sputum samples. Moreover, these molecular-based techniques are comparatively more sensitive and are quicker than immunoassays, and could be utilized not only in a facile form towards the manual detection of a virus, but also as an embedded component of more complex systems. Despite these promising applications, most of them still have diverse potential limitations in accuracy, repeatability, specificity, and sensitivity, often caused by the high genetic variability of several viruses. On the other hand, real-time polymerase chain reaction technology (RT-PCR) has been generally deployed as a routine diagnosis method for the detection of CoVs.10 However, some false positive/negative responses are observed, especially in the case of COVID-19. Although, RT-PCR analyses are broadly applied, the testing capacity and availability cannot meet the unprecedented global demands for rapid, low-cost, reliable, and broadly accessible molecular diagnosis. Some challenging issues regarding the collection/treatment of specimens and the amplification and identification of viral RNA, as well as the validation procedure of clinical sensitivity/specificity, still remain. Some important diagnostic methods for the detection of SARS-CoV-2 are summarized in Fig. 2.11–13 Currently, COVID-19 detection is mainly based on the combination of some techniques which include RT-PCR, chest X-ray, CT scans, and identification of some main biomarkers in the blood; the detection of the level of biomarkers, including procalcitonin (low level), C-reactive protein (elevated level), lymphocyte counts (low level), and interleukins 6 and 10 (high concentrations) are important.11,14 Additionally, ELISA and nucleic acid mediated assays provide considerable amplification of detection signals, permitting indirect detection of specific proteins; they require affinity ligands (e.g., antibodies, receptors, aptamers, and peptides) for binding with the precise viral proteins (the binding affinity/specificity are important issues).9
Detection based on non-contact optical approaches is important for controlling the spread of viruses and surface disinfection. In this regard, strategies based on nanomaterials and nanotechnology are efficiently applied for the rapid detection of viruses.15 Indeed, there are various nano-based structures, including metallic nanoparticles (NPs), graphene oxide (GO), graphene, quantum dots (QDs), polymeric nanomaterials, nanocomposites and carbon nanotubes for the diagnosis of viral infectious diseases and the detection of pathogenic viruses.3 For instance, gold NPs combined with silver staining have been applied for the identification of HPV (human papillomavirus) in cervical carcinoma cells.16 These metallic nanoparticle (NP)-based detection methods are reported for various types of clinically relevant viruses with a particular focus on NP bio-hybrid systems, virus detection targets, and assay modalities.17 DNA, RNA, antibodies and antigens conjugated to the surface of various NPs can be utilized for rapid, sensitive, specific, direct and facile detection with extraordinary multiplexing potentials.18–20 Furthermore, the functionalization of nanomaterials with antibodies or nucleic acids can be employed for nano-based detection techniques, through antigen-binding/colorimetric assays and light/photothermal systems and platforms (Fig. 3). It should be noted that because of the superior surface-to-volume ratios, nanostructure materials can take part in remarkably suitable surface interactions between sensors and analytes, or any chemical constituents that should be measured or evaluated, permitting highly-qualified and sensitive viral detection with a good reliability and selectivity.3,21 On the other hand, biosensors are suitable for viral detection with a good sensitivity/selectivity, and can be employed for immediate measurements with robust and easy to perform processes. They have remarkable potential for use in on-site field detection and POC analyses (Tables 1 and 2).22–25 This paper provides recent advances in the design of nano- and biosensors, as well as their important challenges and opportunities for SARS-CoV-2 detection and COVID-19 diagnosis.
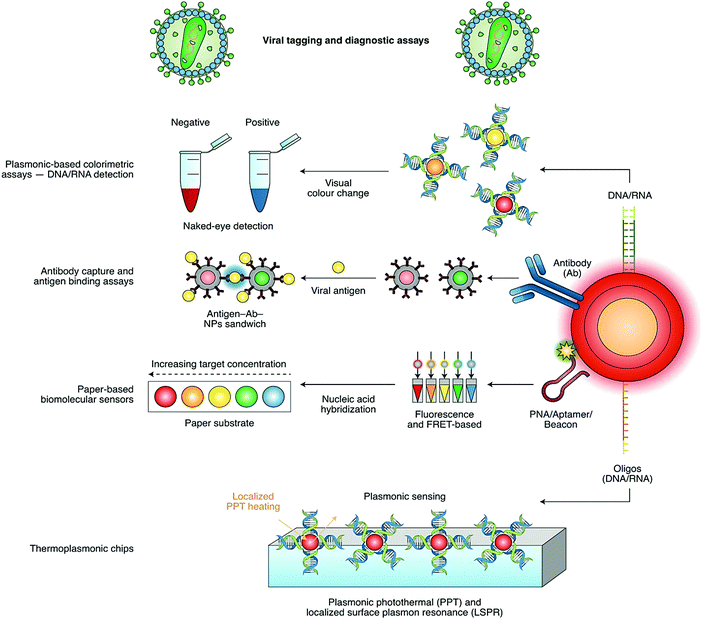 | ||
| Fig. 3 Nano-based sensors for the detection of SARS-CoV-2 based on viral tagging and diagnostic assays: Ab, antibody; FRET, Förster resonance energy transfer; LSPR, localized surface plasmon resonance; NPs, nanoparticles; PNA, peptide nucleic acid; and PPT, photothermal therapy. Reproduced with permission from ref. 21. | ||
| Recognition element/analyte | Detection method | Limit of detection (LOD) | Range of detection | Ref. |
|---|---|---|---|---|
| Anti-SARS-CoV N protein/SARS-CoV N protein | Optical-LSPR | 1.00 pg mL−1 | 0.100 pg mL−1–1.00 ng mL−1 | 26 |
| SARS-CoV N protein/quantum dot-conjugated RNA aptamer | Optical-confocal laser scanning microscopy | 0.100 pg mL−1 | 0.1–50 pg mL−1 | 27 |
| SARS-CoV 2 cDNA/SARS-CoV 2 nucleic acid | Optical-LSPR | 0.220 pM | 0.100 pM–1.00 μM | 10 |
| SARS-CoV oligonucleotide probe | Optical-LSPR | 2.00 nM | 1.00 nM–1 μM | 28 |
| SARS-CoV NG-8 aptamer/SARS-CoV helicase protein | Piezoelectric immunosensor | 3.50 ng mL−1 | 0.050–1.00 μg mL−1 | 29 and 30 |
| MERS-CoV and human CoV proteins/antibody for each virus | Electrochemical square wave voltammetry (SWV) | 0.400 pg mL−1 | 0.010–1.00 × 104 ng mL−1 | 31 |
| Biosensor type | Properties | Ref. |
|---|---|---|
| Plasmonic biosensors | These biosensors are label-free and remarkably sensitive, and they can be employed for various types of clinically interesting target analytes; human serum samples can be utilized without dilution for detecting nucleocapsid antibodies (specific against SARS-CoV-2) via applying a surface plasmonic resonance (SPR) biosensor. | 10 |
| Field-effect transistor (FET)-based biosensing | FET-based biosensing platforms have various promising advantages, including the capability of being highly sensitive and to detect small volumes of target analyte instantaneously. These types of biosensors have the potential to be employed in clinical analysis, POC tests, and on-site diagnostics. | 32 |
| Electrochemical biosensors | Electrochemical biosensors are used by researchers owing to their simplicity, high sensitivity/specificity, ease of operation, cost-effectiveness, and ease of miniaturization and bulk fabrication. These biosensors also have POC usability in homes/clinics. | 33 |
| Surface-enhanced Raman scattering (SERS)-based biosensors | These biosensors are used by researchers owing to their remarkably sensitive and quantitative determination of analytes using SERS-encoded NPs (SERS tags) as an alternative to colloidal gold to report a signal. There are three basic parts of SERS tags: gold/silver NPs as Raman enhanced substrates, adsorbed Raman reporter dyes to generate characteristic SERS signals, and precise antibodies for binding the targets. | 34 |
2. Nano- and biosensors for SARS-CoV-2 detection
Owing to the possibility of such pandemics in the future and in order to control and prevent the further spread of viral diseases, there is an urgent and vital demand to develop rapid, sensitive and low cost methods for the detection of viruses and diagnosis of related viral infections. Nano- and biosensors can help provide rapid and sensitive detection of SARS-CoV-2 virus particles, helping to better control, diagnose and treat COVID-19.22,24,35 Generally, biosensor platforms for the detection of SARS-CoV-2 are based on three important aspects, including the target for identification (e.g., viral RNA and proteins, or human immunoglobulins), identification methods (based on aptamers, antibodies, nucleic acid probes, receptors), and the amplification of signals and transduction systems (based on electrical, surface plasmon resonance, electrochemical, optical, mechanical systems, and fluorescent signals).36 The interaction between the antibody and antigen or the receptor and related ligand can be identified through the conformational alterations of sensor proteins. Additionally, the detection based on enzymatic reactions is one of the most important recognition methods, as an example, the detection of proteolytic cleavage by precise protease.36 Overall, an ideal biosensor should have some important features, including being mass producible, autonomous, possessing a remarkable sensitivity/selectivity, quick response time, multiplexing capabilities, multiple sensing modes, disposable, long shelf-life, cost effective, and easy to use.11,232.1. For the detection of surface antigens and/or the whole virus
The detection of SARS-CoV-2 in medical samples was performed using a field-effect transistor (FET)-based biosensing gadget, and its performance/efficacy was evaluated by applying a cultured virus, antigen protein and nasopharyngeal swab samples from COVID-19 patients.32 For fabrication of the biosensor, the graphene sheets of the FET were coated by precise Ab against the SARS-CoV-2 spike protein, and these sheets were decorated with the SARS-CoV-2 spike Ab via 1-pyrenebutyric acid N-hydroxysuccinimide ester as a probe linker. Thus, the produced FET tool could detect the SARS-CoV-2 spike protein at concentrations of 1 fg mL−1 in phosphate-buffered saline and a 100 fg mL−1 medical transfer vehicle. In addition, this sensor successfully detected SARS-CoV-2 in a culture with a limit of detection (LOD) of about 1.6 × 101 pfu mL−1 and medical tests with a LOD of about 2.42 × 102 copies mL−1. The FET biosensor exhibited a suitable sensitivity for the identification of COVID-19 with no sample pretreatment or labeling, but different materials may be considered for improvement of the signal-to-noise ratio.32 Importantly, the device showed no measurable cross-reactivity with the Middle East respiratory syndrome coronavirus (MERS-CoV) antigen, demonstrating the remarkable potential of this sensor to identify the SARS-CoV-2 antigen protein from that of MERS-CoV.32A biosensor for detection of the SARS-CoV-2 S1 spike protein expressed on the surface of the virus has been introduced.37 This biosensor was based on membrane-engineered mammalian cells bearing the human chimeric spike S1 antibody. It was revealed that the protein attachment to the membrane-bound antibodies caused the selective and remarkable alteration in the cellular bioelectric characteristics evaluated by applying an assay based on the bioelectric recognition approach. The prepared biosensor demonstrated an ultra-rapid behavior (∼3 min) with a LOD of about 1 fg mL−1, and a semi-linear range of response between 10 fg and 1 μg mL−1. Additionally, no cross-reactivity could be detected against the SARS-CoV-2 nucleocapsid protein. The biosensor was designed as a ready-to-use platform, including a portable read-out device that functioned through a smartphone or tablet. It was revealed that this biosensor may be applicable for the detection of SARS-CoV-2 surface antigens with no further sample processing/managing, thus suggesting a promising approach for widespread screening/monitoring, and subsequently controlling the global CoV pandemic.37 In another investigation, rapid and direct optical measurement of SARS-CoV-2 viruses was introduced in one step, almost without any sample preparation by applying a spike protein specific nanoplasmonic resonance sensor.38 It was reported that the sensor detected as few as 30 virus particles in one step within 15 min (LOD = 370 vp mL−1) (Fig. 4). Analysis of both the generic microplate reader and a handheld smartphone connected device revealed that this detection technique may be adopted rapidly under both a regular clinical environment and resource-limited settings.38
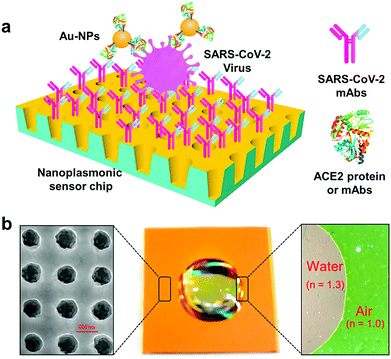 | ||
| Fig. 4 The one-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors: (a) the designed nanoplasmonic resonance sensor for evaluating the SARS-CoV-2 pseudovirus concentration; and (b) a photograph (middle) of one piece of the Au nanocup array chip with a drop of water on top. The scanning electron microscopy image (left) demonstrates the replicated nanocup array. The transmission microscopy image (right) demonstrates that air and water on the device surface have different colors, green and red/pink, respectively. Reproduced with permission from ref. 38. | ||
The decoration of nano-based particles with magnetic characteristics with precise receptors for viruses means that the attachment of virus molecules to the NPs can lead to magnetic extraction/separation by applying external magnetic fields.21 The viral detection system based on the viral genome, such as +ssRNA and the S (spike)-protein contained in SARS-CoV-2, was designed by applying magnetic NPs. For the detection of SARS-CoV-2, a giant magneto-resistive (GMR) biosensor along with magnetic NPs showed promising and sensitive biosensing potential, but additional evaluations should be systematically managed.39 Additionally, the G-quadruplex-based biosensor showed remarkable potential for the detection of SARS-CoV-2.40 The surface protein of SARS-CoV-2 is a potential target for detecting and treating this virus, which could be detected by the exclusive spatial structure of the G-quadruplex. The selected G-quadruplex sequences, as recognition elements, could be combined with other signal molecules to engineer different types of biosensors, in which SARS-CoV-2 was identified by reading electrochemical, florescent or colorimetry signal changes. G-quadruplex based biosensors have the potential to replace antibody-based detection and to develop the detection of SARS-CoV-2 and other pathogens.40
Molecular imprinted polymer based sensors with a unique selectivity and sensitivity can be employed for the detection of SARS-CoV-2.41 Previously, the detection of the Zika virus, Ebola virus and Norovirus have been successfully performed by selecting their DNA or virus specific aptamers as the recognition element, respectively. Thus, the detection of SARS-CoV-2 can be suggested by choosing a specific and suitable polymer and applying the CoV specific aptamer as the recognition element. To develop the sensing materials, polymers belonging to the acrylic group, such as acrylamide, acrylic acid, methyl acrylate, methyl methacrylate, ethyl acrylate and their mixtures are preferred. They can be additionally reinforced on graphene or conducting metal oxides to offer improved and enhanced conductivity.41 Indeed, molecular imprinting technology can be considered as a suitable strategy to apply molecularly imprinted NPs with a unique capability for selective recognition and binding of the spike protein receptor-binding domain of SARS-CoV-2.42 In one study, the potential application of the biocompatible polymeric material as molecular imprinted polymer-based monoclonal-type plastic antibodies for blocking the function of the virus spike protein was proposed.42 Additionally, these designed NPs could be potentially utilized as free-drug therapeutics for treating COVID-19, and when also loaded with antiviral agents, could be employed as a strong multimodal system combining their ability to block the virus spike protein with the targeted delivery of the loaded drug. Furthermore, they can be designed to become an immuno-protective vaccine or a molecular imprinted polymer-based sensor for the detection of SARS-CoV-2.42
An electrochemical immunosensor fabricated using a gold NPs modified carbon electrode and the recombinant spike protein S1 (as the biomarker) was reported for the detection of MERS-CoVs.31 This immunosensor holds promise for the simultaneous detection of various CoVs (20 min), and demonstrated a linear response of 0.001 to 100 ng mL−1 and 0.01 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ng mL−1 for detecting MERS-CoVs and human CoVs, respectively. Additionally, the resulting LOD values were 1.0 and 0.4 pg mL−1 for MERS-CoVs and human CoVs, respectively.31 In another study, a gold-NPs based electrochemical biosensor was constructed for detection of the SARS-CoV-2 spikeS1 protein antigen. The biosensor was fabricated by applying a fluorine-doped tin oxide based substrate, and gold NPs were utilized as a signal amplifier owing to the significant electrical conductivity.33 To produce the biosensing platform, gold NPs (∼29 nm) were drop cast, and then the monoclonal antibodies against SARS-CoV-2 were immobilized to prepare the immunosensor. The LOD of the immunosensor was about 10 fM for detection of the COVID-19 antigen (spike protein), and it could be employed for detection of the COVID-19 antigen (spike protein) at a concentration up to 120 fM, and more than three times.33 Additionally, a cheap and sensitive cobalt-functionalized TiO2 nanotubes-based electrochemical sensor was designed for the detection of SARS-CoV-2 via sensing of the spike present on the surface of the virus within about 30 s. The sensor specifically detected the S-receptor binding domain protein of SARS-CoV-2, even at a very low concentration (14–1400 nM); the sensor exhibited a linear response for the detection of the viral protein over the concentration range.43
000 ng mL−1 for detecting MERS-CoVs and human CoVs, respectively. Additionally, the resulting LOD values were 1.0 and 0.4 pg mL−1 for MERS-CoVs and human CoVs, respectively.31 In another study, a gold-NPs based electrochemical biosensor was constructed for detection of the SARS-CoV-2 spikeS1 protein antigen. The biosensor was fabricated by applying a fluorine-doped tin oxide based substrate, and gold NPs were utilized as a signal amplifier owing to the significant electrical conductivity.33 To produce the biosensing platform, gold NPs (∼29 nm) were drop cast, and then the monoclonal antibodies against SARS-CoV-2 were immobilized to prepare the immunosensor. The LOD of the immunosensor was about 10 fM for detection of the COVID-19 antigen (spike protein), and it could be employed for detection of the COVID-19 antigen (spike protein) at a concentration up to 120 fM, and more than three times.33 Additionally, a cheap and sensitive cobalt-functionalized TiO2 nanotubes-based electrochemical sensor was designed for the detection of SARS-CoV-2 via sensing of the spike present on the surface of the virus within about 30 s. The sensor specifically detected the S-receptor binding domain protein of SARS-CoV-2, even at a very low concentration (14–1400 nM); the sensor exhibited a linear response for the detection of the viral protein over the concentration range.43
For rapid and sensitive detection of the SARS-CoV-2 spike 1 (S1) protein, an innovative method was introduced by applying the SARS-CoV-2 receptor ACE2, which can produce matched pairs with commercially available antibodies (Fig. 5).44 ACE2 and S1-mAb were paired with each other for capture and detection in a lateral flow immunoassay (LFIA) that did not cross-react with the SARS-CoV Spike 1 or MERS-CoV Spike 1 protein. As a result, the SARS-CoV-2 S1 (<5 ng of recombinant proteins/reaction) was identified by the ACE2-based LFIA. The LOD was about 1.86 × 105 copies mL−1 in clinical specimens from COVID-19 patients, without any cross-reactivity for nasal swabs from healthy subjects.44
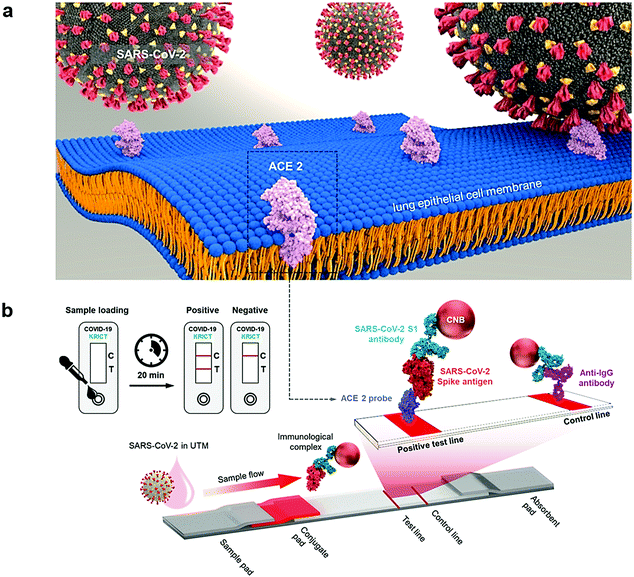 | ||
| Fig. 5 (a) The detection of SARS-CoV-2 spike 1 antigens using a cellular receptor (ACE2)-based LFIA. (b) The designed ACE2-based LFIA included a sample pad, conjugate pad, nitrocellulose membrane, and absorbent pad. The test line placed on the nitrocellulose membrane contains ACE2 for the detection of the SARS-CoV-2 spike antigen. The anti-IgG antibody was applied in the control line; and the detection result was achieved within 20 min. Reproduced with permission from ref. 44. | ||
2.2. For the detection of antibodies
A rapid and sensitive LFIA was introduced by applying lanthanide-doped polystyrene NPs to detect anti-SARV-CoV-2 IgG in human serum.45 A recombinant nucleocapsid phosphoprotein of SARS-CoV-2 was dispensed onto a nitrocellulose membrane to capture the specific IgG. Mouse anti-human IgG antibody was labeled with self-assembled lanthanide-doped polystyrene NPs, which served as a fluorescent reporter. This assay can be employed as a rapid and sensitive method for the detection of anti-SARS-CoV-2 IgG in human serum and can permit positive detection in suspicious cases; additionally, it can be promising for evaluating COVID-19 progression and analyzing a patients’ response to treatment.45 Additionally, a lateral flow combined IgG–IgM immunochromatographic assay was reported for rapid and simultaneous identification of IgM and IgG antibodies against SARS-CoV-2 in clinical blood samples within 15 min.46 After clinical evaluations, it was revealed that the sensitivity and specificity of this assay were 85.29% and 100.00%, respectively. It should be noted that the detection of SARS-CoV-2 related IgM tends to indicate a recent exposure to the virus, while detection based on IgG demonstrates exposure to the virus some time ago. Thus, compared with a single IgG and IgM evaluation, the combined IgG–IgM immunochromatographic strip test had a better sensitivity, and it showed potential for monitoring COVID-19 patients.46 For detection of IgM and IgG antibodies concurrently against SARS-CoV-2 (∼15 min), researchers developed a rapid and simple POC lateral flow immunoassay that is applicable at different stages of viral infection. Accordingly, the sensitivity and specificity were 88.66% and 90.63%, respectively. This colloidal gold-based test kit was based on the conjugation of gold NPs to IgM/IgG antibodies in human serum, plasma and whole blood samples, but it appears that the specificity is not sufficient for COVID-19. Thus, it can cause some false responses for patients with irrelevant infections.47 Furthermore, a multiplexed grating-coupled fluorescent plasmonics biosensor platform was introduced for rapid and accurate measurement of antibodies against SARS-CoV-2 in human blood serum and dried blood spot samples.48 This technique could be employed for the successful detection of IgM, IgG and IgA antibody–antigen interactions. Accordingly, the biosensor could evaluate antibody–antigen binding interactions for multiple targets in a single sample, and showed a high selectivity and sensitivity when measuring serum IgG levels against three SARS-CoV-2 antigens (spike S1, spike S1S2, and the nucleocapsid protein); the platform could result in a quantitative, linear response for serum samples diluted to as low as 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1600 dilution. For evaluation of the test efficacy with other sample matrices, dried blood spot samples were utilized and analyzed using the biosensor, yielding 100% selectivity and 86.7% sensitivity for diagnosing prior COVID-19.48
1600 dilution. For evaluation of the test efficacy with other sample matrices, dried blood spot samples were utilized and analyzed using the biosensor, yielding 100% selectivity and 86.7% sensitivity for diagnosing prior COVID-19.48
The properties of nanomaterials, such as a high surface-to-volume ratio, quantum size effects, remarkable adsorption and reactive capacity, as compared to their bulk form, are imperative to the design of biosensing techniques.3,49,50 Additionally, the size and shape of nanomaterials and nanoarchitectures can be optimized and designed, and thus surface modification/immobilization with various biological species through covalent or non-covalent bonding is possible to improve the biosensing features in terms of the LOD (increased up to several order of magnitudes), significant sensitivity/selectivity and rapid response towards the sample analytes.49,50 In one study, an opto-microfluidic sensing platform with gold nanospikes was developed for detection of the presence and amount of antibodies specific to the SARS-CoV-2 spike protein in 1 μL of human plasma diluted in 1 mL of buffer solution, within![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 min (Fig. 6).51 The target antibody concentration can be correlated with the LSPR wavelength peak shift of the gold nanospikes caused by the local refractive index alteration because of the antigen–antibody binding. The label-free microfluidic platform demonstrated a LOD of
30 min (Fig. 6).51 The target antibody concentration can be correlated with the LSPR wavelength peak shift of the gold nanospikes caused by the local refractive index alteration because of the antigen–antibody binding. The label-free microfluidic platform demonstrated a LOD of![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.08 ng mL−1, falling under the clinically relevant concentration range.51 This platform provided a promising POC testing tool to complement standard serological assays and can make SARS-CoV-2 quantitative diagnostics easier, cheaper, and faster. However, it is critical to validate it for antibody tests for the COVID-19 pandemic, and also the electrodeposition process should be optimized to produce gold nanostructures with a smaller spacing and a higher aspect ratio, thus the antibody–antigen binding can generate a larger shift in the LSPR peak and improve the signal-to-noise ratio of the sensor.51
0.08 ng mL−1, falling under the clinically relevant concentration range.51 This platform provided a promising POC testing tool to complement standard serological assays and can make SARS-CoV-2 quantitative diagnostics easier, cheaper, and faster. However, it is critical to validate it for antibody tests for the COVID-19 pandemic, and also the electrodeposition process should be optimized to produce gold nanostructures with a smaller spacing and a higher aspect ratio, thus the antibody–antigen binding can generate a larger shift in the LSPR peak and improve the signal-to-noise ratio of the sensor.51
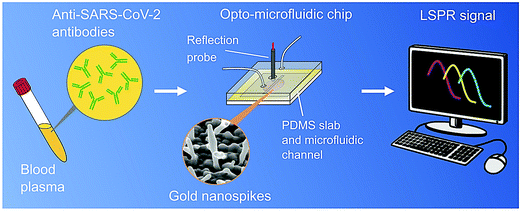 | ||
| Fig. 6 An opto-microfluidic sensing platform prepared for the rapid detection of antibodies against the SARS-CoV-2 spike protein in diluted human plasma with remarkable sensitivity. The sensing principle is based on LSPR, involving gold nanospikes in a microfluidic device, coupled with an optical probe. Reproduced with permission from ref. 51. | ||
2.3. For the detection of cytokines
Application of biosensors for the constant evaluation of cytokine levels have demonstrated remarkable potentials for the diagnosis of COVID-19 progression/severity stages and analysis of the efficacy of anti-inflammatory treatments.13 For instance, needle-shaped microelectrodes were suggested for the identification of changes in Interleukin-6 (IL-6) levels in real time.12 Additionally, by applying aptamer-modified graphene as the conducting channel, a wearable and deformable graphene-based field-effect transistor biosensor was designed for sensitive, consistent and time-resolved detection of cytokines in human biofluids (Fig. 7).52 The designed biosensor showed advantages, including a remarkable mechanical durability and consistent sensing responses, while conforming to non-planar surfaces such as the human body and withstanding large deformations. Additionally, a nonionic surfactant was deployed to reduce the nonspecific adsorption of the biosensor, hence, enabling cytokine detection (inflammatory cytokines such as TNF-α and IFN-γ) in artificial tears (as a biofluid). As a result, this biosensor demonstrated a high potential for use in the consistent and sensitive detection of TNF-α and IFN-γ (with the LOD down to 2.75 and 2.89 pM, respectively).52 It appears that biosensors should be further evaluated and developed for clinical applications; wearable biosensors are promising candidates for continuous monitoring and follow up of patients with COVID-19.13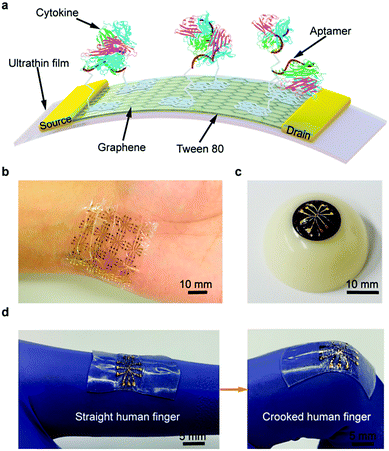 | ||
| Fig. 7 An ultra-flexible graphene field-effect transistor biosensor: (a) a schematic diagram of the biosensor produced on an ultrathin film. A photograph of the flexible device conformably attached onto (b) a human wrist and (c) an artificial eyeball. (d) The stretchable biosensor can be stretched with the activity of the human body. Reproduced from ref. 52, (CC BY 4.0). | ||
2.4. For the detection of nucleic acids
A colorimetric assay based on gold NPs was developed, and capped by appropriately constructed thiol-modified antisense oligonucleotides with sufficient specificity for the N-gene (nucleocapsid phosphoprotein) of SARS-CoV-2; accordingly it showed potential for the selective and visual naked-eye diagnosis of COVID-19 (∼10 min), but its performance can be influenced by the quantity of loaded virus (Fig. 8).53 The thiol-modified antisense oligonucleotide-capped gold NPs agglomerated selectively in the presence of the target RNA sequence of SARS-CoV-2 and showed an alteration in its surface plasmon resonance (SPR).53 Additionally, the utilization of RNaseH could cleave the RNA strand from the RNA-DNA hybrid, causing a visually detectable precipitate from the solution mediated by the additional agglomeration among the gold NPs. The selectivity of the assay has been checked in the presence of MERS-CoV viral RNA with a LOD of about 0.18 ng μL−1 of RNA having a SARS-CoV-2 viral load. Thus, this study reported selective and visual naked-eye detection of SARS-CoV-2, with no requirement for any sophisticated instrumental techniques.53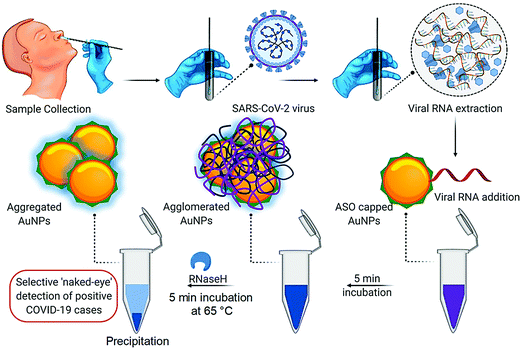 | ||
| Fig. 8 The process of detecting SARS-CoV-2 RNA mediated using appropriately constructed thiol-modified antisense oligonucleotide (ASO)-capped gold (Au) NPs with remarkable selectivity. Reproduced with permission from ref. 53, copyright© 2020 American Chemical Society. | ||
Interestingly, a double-operational plasmonic biosensor combining the LSPR sensing transduction and the PPT influence, offered a promising substitute for the diagnosis of COVID-19, clinically.10 The delicate recognition of certain arrangements from SARS-CoV-2 via nucleic acid hybridization was performed by using the 2-D gold nano-islands decorated with complementary DNA receptors.10 The thermos-plasmonic heat was produced on the chip for an improved sensing performance, which upon illumination at their plasmonic resonance frequency and under local PPT heat, could improve the in situ hybridization temperature and enable the precise discernment of two similar gene sequences. This biosensor showed remarkable sensitivity for the examined SARS-CoV-2 sequences with a much lower detection limit at a concentration of 0.22 pM, and permitted specific target recognition in a multigene mixture.10 In this study, Qui et al.,10 reported that by using LSPR excited at two different wavelengths and the plasmonic resonances of PPT, remarkable stability, reliability and sensitivity properties in diagnosis are achievable.
A paper-based colorimetric assay for DNA detection based on pyrrolidinyl peptide nucleic acid (acpcPNA)-induced NP aggregation has been proposed as an alternative to conventional techniques for the detection of SARS-CoV-2.54 These PNA probes are an attractive alternative to DNA and RNA probes owing to the chemical and biological stability and simple related synthetic methods; they can also hybridize successfully with the complementary DNA strands. Indeed, the acpcPNA probe comprises a single positive charge from the lysine at the C-terminus and results in aggregation of the citrate anion-stabilized silver NPs in the absence of complementary DNA.54 In the presence of the target DNA, the anionic DNA-acpcPNA duplex was generated, which can cause the dispersion of silver NPs as a result of the electrostatic repulsion, providing an enhancement in the detectable color change. This technique was introduced for detecting and analyzing synthetic MERS-CoV, Mycobacterium tuberculosis, and human papillomavirus DNA; however, it may also be suitable for the detection of SARS-CoV-2. The acpcPNA probe showed significant selectivity for the complementary oligonucleotides over single-base-mismatch, two-base-mismatch, and noncomplementary DNA targets. Importantly, some parameters for controlling the sensitivity/selectivity issues of this assay should be noted, such as the silver NP concentration, DNA strand mismatches, ionic strength, and PNA concentration.54 Additionally, aptamers and molecular beacons have been suggested instead of PNA.21
3. Challenges and opportunities
One of the important challenges for constructing suitable biosensors is to capture a signal of very low magnitude that takes place between the biological species (bio-receptor and analyte).24 To solve the problem, nanomaterials can be employed as labels to obtain a remarkable improvement in the signal, high enough to be easily detectable.50,55,56 As an example, metallic nanomaterials (e.g., gold or silver NPs) and quantum dots can be analyzed and employed for labeling by attaching them onto the targeted DNA/bio-recognizing probe.55,56 This can cause a synergetic effect owing to the nano-labeling consequences to significantly amplify the electrochemical signal, and make it possible to design ultrasensitive and selective labeled-biosensing strategies.56 In this regard, future studies should be performed based on taking advantage of the various attractive physicochemical characteristics of nanoscale materials (especially optical, electrical, magnetic, and opto-magnetic properties) for the development of nano-enabled biosensing approaches for the specific detection of viruses, especially MERS-CoV, SARS-CoV and SARS-CoV-2 (Table 3).3,57 However, limited analyses have been performed to develop these nanomaterials-based biosensors for SARS-CoV-2 detection. These techniques can be applied instead of PCR-based testing for COVID-19, with the advantages of simplicity, cost-effectiveness, fast response and real-time diagnostic procedures. These nanomaterial-enabled biosensors primarily rely on the nucleic acid and protein (antigen/antibody) mediated detection of SARS-CoV-2, although they have not yet yielded 100% accuracy owing to the contamination of these highly sensitive bio-receptors, and ultrasensitive, rapid, and portable SARS-CoV-2 sequence detection methods are highly demanded. For instance, the technique based on CRISPR-Cas12,58 or in the case of nanomaterial-based biosensors based on the aerosol mediated diagnosis method, have the advantages of a fast response, sensitivity, and a lack of sample perturbation.59 Importantly, the scale up of these detection methods is very important; in the lab scale, these biosensors have exhibited an approved stability, fast response time, and high sensitivity/selectivity.50 However, this may change in the case of real sample evaluations, and several factors, such as the characteristics of the target antigen/protein/antibody, nanomaterial features, and other important biomolecules may influence the obtained results, as well as the performance of the nanomaterial based biosensing devices.50| Nanomaterial | Biomarker | Findings | Target | Ref. |
|---|---|---|---|---|
| Magnetic NPs | Poly(amino ester) with carboxyl group-coated magnetic NPs | Linear correlation: 10 to 105 copies of SARS-CoV-2 pseudovirus particles; 10-copy sensitive; NPs enabled improved RNA extraction | SARS-CoV-2 virus RNA | 62 |
| Biotinylated probe | LOD was about 0.4 fM; total assay time was about 100 min; dynamic detection range covers three orders of magnitude; the process was based on circle to circle amplification based optomagnetic biosensing | RdRp sequence (synthetic complementary DNA of SARS-CoV-2) | 63 | |
| Gold NPs | Thiol-c-DNA receptors/nucleic acid | LOD of about 0.22 ± 0.08 pM; detection range was about 0.1 pM to 1 μM; the process was based on the combined effects of PPT and LSPR mediated biosensing | ORF1ab-COVID, and E genes from SARS-CoV-2; viral sequences including RdRp-COVID | 10 |
| Thiol-modified antisense oligonucleotides specific for the N-gene (nucleocapsid phosphoprotein) of SARS-CoV-2 | Detection limit of about 0.18 ng μL−1; detection range: 0.2–3 ng μL−1; assay time was about 10 min; the process was based on plasmonic effect based colorimetric biosensing | RNA [N-gene of SARS-CoV-2] | 53 | |
| Oligo probe | Detection limit of about 0.5 ng; total assay time was about 30 min; the process was based on plasmonic effect based colorimetric biosensing | RNA [RdRp gene of SARS-CoV-2] | 64 | |
| Recombinant antigen of SARS-CoV-2 and rabbit-IgG | Assay time was about 15 min; the process was based on colorimetric dependent lateral flow immunoassay based biosensing | IgM and IgG antibodies | 47 | |
| Monoclonal antibodies against SARS-CoV-2 | LOD of about 10 fM; sensitive for detecting SARS-CoV-2 antigens; amperometric biosensing technique | SARS-CoV-2 spike antigen in saliva samples | 33 | |
| Screen printed carbon electrode (SPCE)-based sensing device | Monoclonal antibodies against SARS-CoV-2 | Detection of viral antigen at 10 fM concentration in standard buffer sample; LOD was about 90 fM in the case of spiked saliva samples using an SPCE mediated homemade biosensor device; detection of antigen traces in patient saliva within one min; stability for up to 28 d; amperometric biosensing technique | SARS-CoV-2 spike antigen in saliva samples | 33 |
| Polymer NPs coated with dye streptavidin (Crimson red) | Rabbit anti-fluorescein antibody, sheep anti-digoxigenin antibody, and biotinylated bovine serum albumin | LOD was about 12 copies (for each detection target) per reaction; total diagnostic test was completed within 1 h from sample collection to result interpretation; analytical sensitivity was 100%; specificity was 100%; the assay required simple heating equipment to maintain a constant temperature of 63 °C for 40 min; the process was based on multiplex reverse transcription loop mediated isothermal amplification coupled with a NP-based lateral flow biosensor | ORF1ab and N genes of SARS-CoV-2 | 65 |
| Graphene | Spike protein antibody | FET-based biosensing method for the detection of the SARS-CoV-2 spike protein; LOD was about 1.6 × 101 pfu mL−1 in culture medium, whereas it was 2.42 × 102 copies mL−1 in clinical samples. | SARS-CoV-2 antigen protein | 32 |
Nano and biosensors should be technically developed for highly efficient detection of SARS-CoV-2 in various clinical samples, including urine, blood, saliva, and nasopharyngeal swabs.50,60 One of the important limitations for developing biosensors based on biological reagents (such as antibodies, DNA, and antigens) is the time consuming and costly procedure of extraction and/or preparation of the target biological metrics/reagents.61 Thus, designing nanomaterials-enabled biosensors that do not need extra extraction procedures is very necessary. For instance, a rapid electrochemical diagnostic kit composed of fixed/screen printed electrodes was constructed for the detection of SARS-CoV-2 via the differentiable fingerprint of the viral glycoproteins at various voltage positions.61 The sensor was activated upon coating a layer of coupled graphene oxide with sensitive chemical compounds along with gold nanostars; this technique, without the need for extraction procedures and biomarkers, could identify trace amounts of the virus in about 1 min with a high sensitivity. The electrochemical detection mechanistic evaluations showed the surface-confined mechanism via the adsorption procedure, providing differentiable detection of pathogenic viruses within selected aquatic biological media.61
The detection limit is very important because of the relatively low viral concentration in patient samples; it appears that magnetic, plasmonic, and electrochemical devices have exhibited the lowest LOD, indicating they are promising techniques.66 However, an important drawback regarding these magnetic and plasmonic methods is that they typically require specialized instrumentation both for production and operation, creating challenging portability issues. Therefore, improved procedures are highly necessary to make them more portable, and also improve their sensitivity to be applicable to POC use.66 Furthermore, some techniques based on colorimetric, electrochemical, and lateral flow assays have demonstrated greater portability, permitting better operation in the field as they do not need a laboratory infrastructure and instrumentation to obtain the results.67,68 On the other hand, the plasmonic and magnetic techniques require a laboratory infrastructure, and also have the advantage of a higher throughput, allowing more samples to be evaluated in one step; the electrochemical and plasmonic techniques allow a better and fastest readout compared with the colorimetric and lateral flow methods.24,58,66 Additionally, to design well-organized biosensors, one of the most important parts is transporting the targeted molecules to the functionalized surface of them.69 In this regard, robust simulation (reliable, fast, and stable numerical modelling) of these systems and their related chemical reactions can help improve the designed biosensors. For instance, computational fluid dynamics (CFD) have been employed for various related biomedical studies involving design, validation and proof-of-concept.69
4. Conclusions and future outlooks
Rapid, convenient, and large-scale diagnosis is vital for the treatment and management of COVID-19 (especially asymptomatic and/or early-stage patients) resulting from SARS-CoV-2, especially for reducing and controlling its spread. Traditional techniques for the detection of respiratory viruses are generally costly, time consuming, and labor intensive, and they require specialized laboratory equipment and expertise. Nano- and biosensors can be considered as sensitive, selective, and affordable analytical diagnostic systems for the detection of SARS-CoV-2 and related viral infections. Indeed, the low-level detection of a targeted disease biomarker can be highly suitable for the better evaluation, management, and treatment of viral disease progression. The surface functionalization and modification of NPs can facilitate the faster and more specific detection of SARS-CoV-2, even in the early stages of infection. Moreover, to improve the diagnostic performance and accuracy, combinational diagnosis/detection procedures for various kinds of biomarkers, via applying multiplex nano- and biosensors, could be a useful strategy. On the other hand, to improve the reliability and reproducibility of these sensors, machine learning-based signal procedures and the related obtained details are an essential area of focus for researchers.It appears that more elaborate academic analyses and studies should be systematically carried out to solve these challenging issues, especially with regards to false negative/positive results, validation processes, detection speed, simplicity, selectivity, sensitivity, cost-effectiveness, and public usability. Nanotechnology can enable extreme progress regarding the fabrication of innovative diagnostic sensors, the integration of novel devices, better optimization/validation, and improvements in sensing performances at the POC. Future studies should be planned to find and design innovative and next-generation non-invasive, specific, affordable, and fast biosensing methodologies and technologies for diagnostic applications, to manage pandemics and life-threatening infectious diseases in particular. Remarkably, sensing devices with suitable cost-effectiveness, miniaturization, and user-friendly features have received considerable academic attention, and companies and industry in particular are focused on these smart nano- and biosensors (with high sensitivity/selectivity features) for use as highly qualified and intelligent diagnostic systems/platforms. Importantly, owing to the occurrence of asymptomatic SARS-CoV-2 patients, public and home-use nano- and biosensors may enable immediate detection of this virus and accordingly provide better control and management of COVID-19. In this regard, novel smartphone-based biosensors and colorimetric strips targeting antibodies/antigens have shown potential for use in the home and for simple POC testing.
Conflicts of interest
There are no conflicts to declare.References
- S. Jiang, C. Hillyer and L. Du, Trends Immunol., 2020, 41, 355–359 CrossRef CAS.
- G. Jamalipour Soufi, A. Hekmatnia, M. Nasrollahzadeh, N. Shafiei, M. Sajjadi, P. Iravani, S. Fallah, S. Iravani and R. S. Varma, Appl. Sci., 2020, 10, 3641 CrossRef.
- M. Nasrollahzadeh, M. Sajjadi, G. Jamalipour Soufi, S. Iravani and R. S. Varma, Nanomaterials, 2020, 10, 1072 CrossRef CAS.
- H. F. Florindo, R. Kleiner, D. Vaskovich-Koubi, R. C. Acúrcio, B. Carreira, E. Yeini, G. Tiram, Y. Liubomirski and R. Satchi-Fainaro, Nat. Nanotechnol., 2020, 15, 630–645 CrossRef CAS.
- S. Talebian and J. Conde, Matter, 2020, 3, 598–601 CrossRef.
- M. D. Shin, S. Shukla, Y. H. Chung, V. Beiss, S. K. Chan, O. A. Ortega-Rivera, D. M. Wirth, A. Chen, M. Sack, J. K. Pokorski and N. F. Steinmetz, Nat. Nanotechnol., 2020, 15, 646–655 CrossRef CAS.
- F. Cui and H. S. Zhou, Biosens. Bioelectron., 2020, 165, 112349 CrossRef CAS.
- B. Udugama, P. Kadhiresan, H. N. Kozlowski, A. Malekjahani, M. Osborne, V. Y. C. Li, H. Chen, S. Mubareka, J. B. Gubbay and W. C. W. Chan, ACS Nano, 2020, 14, 3822–3835 CrossRef CAS.
- W. Feng, A. M. Newbigging, C. Le, B. Pang, H. Peng, Y. Cao, J. Wu, G. Abbas, J. Song, D.-B. Wang, M. Cui, J. Tao, D. L. Tyrrell, X.-E. Zhang, H. Zhang and X. C. Le, Anal. Chem., 2020, 92, 10196–10209 CrossRef CAS.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS.
- N. Bhalla, Y. Pan, Z. Yang and A. F. Payam, ACS Nano, 2020, 14, 7783–7807 CrossRef CAS.
- C. Russell, A. C. Ward, V. Vezza, P. Hoskisson, D. Alcorn, D. P. Steenson and D. K. Corrigan, Biosens. Bioelectron., 2019, 126, 806–814 CrossRef CAS.
- S. M. Russell, A. Alba-Patiño, E. Baron, M. Borges, M. Gonzalez-Freire and R. de la Rica, ACS Sens., 2020, 5, 1506–1513 CrossRef CAS.
- T. Ai, Z. Yang, H. Hou, C. Zhan, C. Chen, W. Lv, Q. Tao, Z. Sun and L. Xia, Radiology, 2020, 200642 Search PubMed.
- N. Rabiee, M. Bagherzadeh, A. Ghasemi, H. Zare, S. Ahmadi, Y. Fatahi, R. Dinarvand, M. Rabiee, S. Ramakrishna, M. Shokouhimehr and R. S. Varma, Int. J. Mol. Sci., 2020, 21, 5126 CrossRef CAS.
- I. Zehbe, G. W. Hacker, H. Su, C. Hauser-Kronberger, J. F. Hainfeld and R. Tubbs, Am. J. Pathol., 1997, 150, 1553 CAS.
- M. S. Draz and H. Shafiee, Theranostics, 2018, 8, 1985 CrossRef CAS.
- X. Wang, L.-H. Liu, O. Ramstroem and M. Yan, Exp. Biol. Med., 2009, 234, 1128–1139 CrossRef CAS.
- P. Alivisatos, Nat. Biotechnol., 2004, 22, 47–52 CrossRef CAS.
- N. L. Rosi and C. A. Mirkin, Chem. Rev., 2005, 105, 1547–1562 CrossRef CAS.
- S. Talebian, G. G. Wallace, A. Schroeder, F. Stellacci and J. Conde, Nat. Nanotechnol., 2020, 15, 618–621 CrossRef CAS.
- H. A. Hussein, R. Y. A. Hassan, M. Chino and F. Febbraio, Sensors, 2020, 20, 4289 CrossRef CAS.
- R. Samson, G. R. Navale and M. S. Dharne, 3 Biotech, 2020, 10, 385 CrossRef.
- M. Asif, M. Ajmal, G. Ashraf, N. Muhammad, A. Aziz, T. Iftikhar, J. Wang and H. Liu, Curr. Opin. Electrochem., 2020, 23, 174–184 CrossRef.
- I. Santiago, ChemBioChem, 2020, 21, 2880–2889 CrossRef CAS.
- J. C. Huang, Y.-F. Chang, K.-H. Chen, L.-C. Su, C.-W. Lee, C.-C. Chen, Y.-M. A. Chen and C. Chou, Biosens. Bioelectron., 2009, 25, 320–325 CrossRef CAS.
- C. Roh and S. K. Jo, J. Chem. Technol. Biotechnol., 2011, 86, 1475–1479 CrossRef CAS.
- L. Shi, Q. Sun, J. He, H. Xu, C. Liu, C. Zhao, Y. Xu, C. Wu, J. Xiang, D. Gu, J. Long and H. Lan, Biomed. Mater. Eng., 2015, 26, S2207–S2216 Search PubMed.
- T. Lee, J.-H. Ahn, S. Y. Park, G.-H. Kim, J. Kim, T.-H. Kim, I. Nam, C. Park and M.-H. Lee, Micromachines, 2018, 9, 651 CrossRef.
- D. R. B. Albano, K. Shum, J. A. Tanner and Y. S. Fung, 17th Int. Meet. Chem. Sensors - IMCS, AMA, Viena, Austria, 2018, 211–213.
- L. A. Layqah and S. Eissa, Mikrochim. Acta, 2019, 186, 224 CrossRef.
- G. Seo, G. Lee, M. J. Kim, S. H. Baek, M. Choi, K. B. Ku, C. S. Lee, S. Jun, D. Park, H. G. Kim, S. J. Kim, J. O. Lee, B. T. Kim, E. C. Park and S. I. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS.
- S. Mahari, A. Roberts, D. Shahdeo and S. Gandhi, bioRxiv, 2020 DOI:10.1101/2020.04.24.059204.
- C. Wang, C. Wang, X. Wang, K. Wang, Y. Zhu, Z. Rong, W. Wang, R. Xiao and S. Wang, ACS Appl. Mater. Interfaces, 2019, 11, 19495–19505 CrossRef CAS.
- L. Krejcova, P. Michalek, M. Merlos Rodrigo, Z. Heger, S. Krizkova, M. Vaculovicova, D. Hynek, V. Adam and R. Kizek, Nanobiosens. Dis. Diagn., 2015, 4, 47–66 Search PubMed.
- K.-H. Liang, T.-J. Chang, M.-L. Wang, P.-H. Tsai, T.-H. Lin, C.-T. Wang and D.-M. Yang, J. Chin. Med. Assoc., 2020, 83, 701–703 CAS.
- S. Mavrikou, G. Moschopoulou, V. Tsekouras and S. Kintzios, Sensors, 2020, 20, 3121 CrossRef CAS.
- L. Huang, L. Ding, J. Zhou, S. Chen, F. Chen, C. Zhao, J. Xu, W. Hu, J. Ji, H. Xu and G. L. Liu, Biosens. Bioelectron., 2020, 112685 Search PubMed.
- A. Islam and Z. Ahsan, Am. J. Nanosci., 2020, 6, 6–13 CrossRef.
- H. Xi, M. Juhas and Y. Zhang, Biosens. Bioelectron., 2020, 167, 112494 CrossRef CAS.
- T. N. Chatterjee and R. Bandyopadhyay, Trans. Indian Natl. Acad. Eng., 2020, 1–4, DOI:10.1007/s41403-41020-00125-41407.
- O. I. Parisi, M. Dattilo, F. Patitucci, R. Malivindi, V. Pezzi, I. Perrotta, M. Ruffo, F. Amone and F. Puoci, BioRxiv, 2020 DOI:10.1101/2020.05.28.120709.
- B. S. Vadlamani, T. Uppal, S. C. Verma and M. Misra, Sensors, 2020, 20, 5871 CrossRef.
- J.-H. Lee, M. Choi, Y. Jung, S. K. Lee, C.-S. Lee, J. Kim, J. Kim, N. H. Kim, B.-T. Kim and H. G. Kim, Biosens. Bioelectron., 2021, 171, 112715 CrossRef CAS.
- Z. Chen, Z. Zhang, X. Zhai, Y. Li, L. Lin, H. Zhao, L. Bian, P. Li, L. Yu, Y. Wu and G. Lin, Anal. Chem., 2020, 92, 7226–7231 CrossRef CAS.
- L. Zeng, Y. Li, J. Liu, L. Guo, Z. Wang, X. Xu, S. Song, C. Hao, L. Liu, M. Xin and C. Xu, Mater. Chem. Front., 2020, 4, 2000–2005 RSC.
- Z. Li, Y. Yi, X. Luo, N. Xiong, Y. Liu, S. Li, R. Sun, Y. Wang, B. Hu, W. Chen, Y. Zhang, J. Wang, B. Huang, Y. Lin, J. Yang, W. Cai, X. Wang, J. Cheng, Z. Chen, K. Sun, W. Pan, Z. Zhan, L. Chen and F. Ye, J. Med. Virol., 2020, 92, 1518–1524 CrossRef CAS.
- N. C. Cady, N. Tokranova, A. Minor, N. Nikvand, K. Strle, W. T. Lee, W. Page, E. Guignon, A. Pilar and G. N. Gibson, Biosens. Bioelectron., 2021, 171, 112679 CrossRef CAS.
- G. Maduraiveeran, M. Sasidharan and V. Ganesan, Biosens. Bioelectron., 2018, 103, 113–129 CrossRef CAS.
- M. Srivastava, N. Srivastava, P. K. Mishra and B. D. Malhotra, Sci. Total Environ., 2021, 754, 142363 CrossRef CAS.
- R. Funari, K.-Y. Chu and A. Q. Shen, Biosens. Bioelectron., 2020, 169, 112578 CrossRef CAS.
- Z. Wang, Z. Hao, S. Yu, C. Huang, Y. Pan and X. Zhao, Nanomaterials, 2020, 10, 1503 CrossRef CAS.
- P. Moitra, M. Alafeef, K. Dighe, M. B. Frieman and D. Pan, ACS Nano, 2020, 14, 7617–7627 CrossRef CAS.
- P. Teengam, W. Siangproh, A. Tuantranont, T. Vilaivan, O. Chailapakul and C. S. Henry, Anal. Chem., 2017, 89, 5428–5435 CrossRef CAS.
- C. Kokkinos, Nanomaterials, 2019, 9, 1361 CrossRef CAS.
- Y. Saylan, Ö. Erdem, S. Ünal and A. Denizli, Biosensors, 2019, 9, 65 CrossRef CAS.
- T. Ozer, B. J. Geiss and C. S. Henry, J. Electrochem. Soc., 2020, 167, 037523 CrossRef CAS.
- C. Lucia, P.-B. Federico and G. C. Alejandra, bioRxiv, 2020 DOI:10.1101/2020.02.29.971127.
- J. A. Huffman, A. E. Perring, N. J. Savage, B. Clot, B. Crouzy, F. Tummon, O. Shoshanim, B. Damit, J. Schneider, V. Sivaprakasam, M. A. Zawadowicz, I. Crawford, M. Gallagher, D. Topping, D. Doughty, S. C. Hill and Y. Pan, Aerosol Sci. Technol., 2020, 54, 465–495 CrossRef CAS.
- M. Fani, M. Zandi, S. Soltani and S. Abbasi, Biotechnol. Appl. Biochem., 2020 DOI:10.1002/bab.2033.
- S. A. Hashemi, N. G. G. Behbahan, S. Bahrani, S. M. Mousavi, A. Gholami, S. Ramakrishna, M. Firoozsani, M. Moghadami, K. B. Lankarani and N. Omidifar, Biosens. Bioelectron., 2021, 171, 112731 CrossRef CAS.
- Z. Zhao, H. Cui, W. Song, X. Ru, W. Zhou and X. Yu, bioRxiv, 2020 DOI:10.1101/2020.02.22.961268.
- B. Tian, F. Gao, J. Fock, M. Dufva and M. F. Hansen, Biosens. Bioelectron., 2020, 165, 112356, DOI:10.1016/j.bios.2020.112349.
- V. Kumar, S. Mishra, R. Sharma, J. Agarwal, U. Ghoshal, T. Khanna, L. K. Sharma, S. K. Verma and S. Tiwari, bioRxiv, 2020 DOI:10.1101/2020.06.30.172833.
- X. Zhu, X. Wang, L. Han, T. Chen, L. Wang, H. Li, S. Li, L. He, X. Fu, S. Chen, M. Xing, H. Chen and Y. Wang, Biosens. Bioelectron., 2020, 166, 112437 CrossRef CAS.
- C. Tymm, J. Zhou, A. Tadimety, A. Burklund and J. X. J. Zhang, Cell. Mol. Bioeng., 2020, 13, 313–329 CrossRef CAS.
- V. N. Goral, N. V. Zaytseva and A. J. Baeumner, Lab Chip, 2006, 6, 414–421 RSC.
- A. Parihar, P. Ranjan, S. K. Sanghi, A. K. Srivastava and R. Khan, ACS Appl. Bio Mater., 2020 DOI:10.1021/acsabm.0c01083.
- F. Shahbazi, M. Jabbari, M. Nasr Esfahani and A. Keshmiri, Biosens. Bioelectron., 2021, 171, 112716 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |