Open Access Article
Open Access ArticleGraphene oxide-functionalized nanofibre composite matrices to enhance differentiation of hippocampal neuronal cells†
Moon Sung
Kang‡
a,
Jong Ho
Lee‡
b,
Su-Jin
Song
a,
Dong-Myeong
Shin
 c,
Jun-Hyeog
Jang
d,
Suong-Hyu
Hyon
e,
Suck Won
Hong
*a,
Jong Hun
Lee
c,
Jun-Hyeog
Jang
d,
Suong-Hyu
Hyon
e,
Suck Won
Hong
*a,
Jong Hun
Lee
 *f and
Dong-Wook
Han
*f and
Dong-Wook
Han
 *a
*a
aDepartment of Cogno-Mechatronics Engineering, College of Nanoscience & Nanotechnology, Pusan National University, Busan 46241, South Korea. E-mail: swhong@pusan.ac.kr; nanohan@pusan.ac.kr
bDaan Korea Corporation, Seoul 06252, South Korea
cDepartment of Mechanical Engineering, The University of Hong Kong, Hong Kong 999077, China
dDepartment of Biochemistry & IRIMS, Inha University School of Medicine, Incheon 22212, South Korea
eThe Joint Graduate School of Veterinary Medicine, Kagoshima University, Kagoshima 890-8580, Japan
fDepartment of Food Science and Biotechnology, Gachon University, Seongnam 13120, South Korea. E-mail: foodguy@gachon.ac.kr
First published on 2nd November 2020
Abstract
During the last decade, graphene-family nanomaterials have been widely utilized, forming a composite with various polymers for potential applications in tissue engineering and regeneration due to their excellent physicochemical characteristics and biocompatibility. Above all, these graphene-family nanomaterials allow polymers to have novel biofunctional properties, while maintaining their intrinsic character. By virtue of this bioactivity of nanocomposites, they are able to modulate cellular behaviours such as cell attachment, proliferation and differentiation. In this study, the nanofibre composite matrices of poly(L-lactide-co-ε-caprolactone) (PLCL) and laminin (Lam) functionalized with graphene oxide (GO) (PLCL/Lam/GO) were prepared via electrospinning to explore whether they have the potential to enhance the differentiation of hippocampal neuronal cells. The PLCL/Lam/GO nanofibre matrices could support the adhesion and proliferation of hippocampal neuronal cells by providing the extracellular matrix-like structure. Furthermore, these PLCL/Lam/GO composite matrices were found to promote neurite outgrowth and alignment from hippocampal neuronal cells through providing optimal microenvironments for neuronal differentiation. The underlying mechanism of this phenomenon was proposed according to the scenario in which neurite outgrowth was guided by biofunctional cues, such as Lam and GO, with nanofibre-mediated topographical cues, which upregulated the expression of some specific genes (i.e., DCX, MAP2 and Nestin) closely related to cytoskeletal interactions. In conclusion, it is suggested that these nanofibre composite matrices can be exploited to craft a range of strategies for the development of novel scaffolds to accelerate neural regeneration.
1. Introduction
Due to the sophisticated system of the nerve tissue and its still-veiled mechanism of action, repairing damaged nerves to recover the function of the nervous system has been hard work compared to other tissues.1 Conventionally, surgical intervention and tubulation have been utilized. However, these methods are not so reliable due to the complexity of the operation and the risk of secondary surgery.2The fabrication of artificial scaffolds is extensively applied for nerve tissue regeneration since they can mimic the natural extracellular matrix (ECM) of nerve tissues to encourage neural cells to appropriately grow and differentiate.3–5 Various types of scaffolds have been introduced for neural tissue engineering. Among them, electrospun nanofibre matrices have been mostly highlighted owing to their several advantages such as suitable mechanical property, surface modification availability, precisely controllable fibrous structure, large surface–volume ratio, and ability to nano-topographically control the cells.6,7 In particular, the aligned nanofibre scaffolds can provide oriented topographical guidance to neural cells, which play an important role in the neurite outgrowth and nerve functions recovery.
Aligned nanofibre matrices based on synthetic polymers, such as poly(L-lactide-co-ε-caprolactone) (PLCL),8 polycaprolactone (PCL),9 polylactic acid (PLA),10 and poly(lactic-co-glycolic acid) (PLGA),11 are the most highlighted platform for neural tissue engineering because of their controllable degeneration rate, excellent mechanical properties, and biocompatibility. These synthetic polymer-based aligned nanofibre matrices have been utilized to promote the growth and differentiation of neuronal cells, including Schwann cells,12,13 PC12 pheochromocytoma cells,14,15 and HT22 hippocampal cells.16 These studies also indicated that the topographical control of aligned nanofibres supported cell growth and neonatal neuritogenesis. Among those synthetic polymers, PLCL polymers have the potential for nerve tissue scaffolds due to their superior mechanical and physicochemical properties, whereas their lack of surface cell recognition site and biofunctionality mainly hinders their application to tissue engineering scaffolds. To improve the functionality of the aligned polymer-based nanofibre matrices, cell adhesive ECM components, biomolecules, and nanomaterials were incorporated into the matrices.8,17–20 By developing the composite nanofibre scaffolds, more cytocompatible properties were achieved, while maintaining the intrinsic nature of a single material. Among various components of ECM, laminin (Lam) is a plentiful glycoprotein in the basement membrane of the nerve tissues, which comprises a multi-domain, cross-link-shaped structure consisting of three polypeptide chains (α1, β1, and β2).21 Lam in the nerve tissue membrane gives signals for axonal guidance and neurite outgrowth, and controls cellular behaviours such as adhesion, migration, proliferation, and differentiation.21 Therefore, the incorporation of Lam was predicted to promote neuronal cell growth and neurite outgrowth.
Meanwhile, graphene and its derivatives belong to the novel nanomaterial family, which are two-dimensional (2D) monolayered sp2-bonded carbon atoms, and feature exceptional optical, physicochemical, electrical, and mechanical properties.22–24 Among the graphene derivatives, graphene oxide (GO), which is highly oxidative graphene, has specific characteristics beneficial to tissue engineering and regeneration, because of the presence of oxygen-containing moieties, such as hydroxyl, carbonyl and carboxylic groups, and an epoxide, which promotes protein adsorption and cell–matrix signalling.25–27 Previous studies have shown that neuronal cells exhibited better adhesion and proliferation on GO-containing nanofibre matrices.28,29 Moreover, GO has been proved to induce neurogenic differentiation of stem cells (SCs) and enhanced neuritogenesis of neuronal cells.29–31 For example, PCL/GO nanofibre matrices enhanced the neurogenic differentiation of PC12 cells by improving cell–matrix interaction and specific biochemical cues.32 Another study also reported that aligned PCL/GO promoted neuronal SCs into matured oligodendrocytes without any differentiation-inducing factors.19
During the last few decades, considerable research on the strategies of neural tissue engineering and regeneration has been performed to develop optimal composite scaffolds. Hence, complete neural regeneration remains a challenge, and the search for an ideal approach is still ongoing. However, the use of PLCL and Lam nanofibres functionalized with GO (PLCL/Lam/GO) is novel and challenging. To our knowledge, this study is the first to report the direct incorporation of PLCL nanofibre matrices with Lam and GO simultaneously. Herein, we explored the potential of the PLCL/Lam/GO nanofibre composites as alternative scaffolds to enhance the differentiation of the hippocampal neuronal cells (Fig. 1).
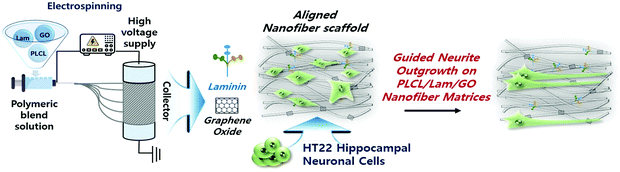 | ||
| Fig. 1 Schematic illustration of PLCL/Lam/GO nanofibre composite matrices fabricated by electrospinning for enhancement of differentiation of HT22 hippocampal neuronal cells. | ||
2. Results and discussion
2.1 Physicochemical characterizations of PLCL/Lam/GO nanofibre composite matrices
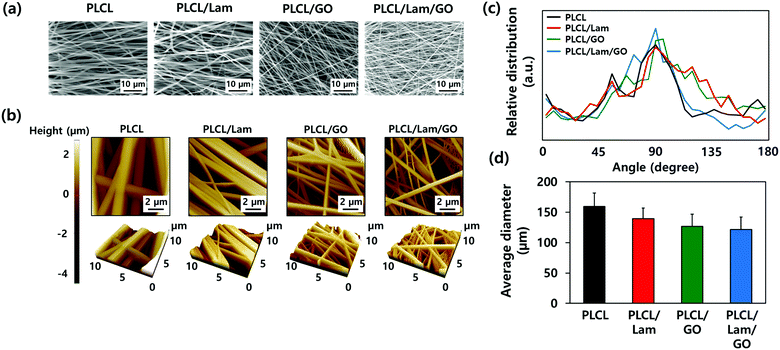
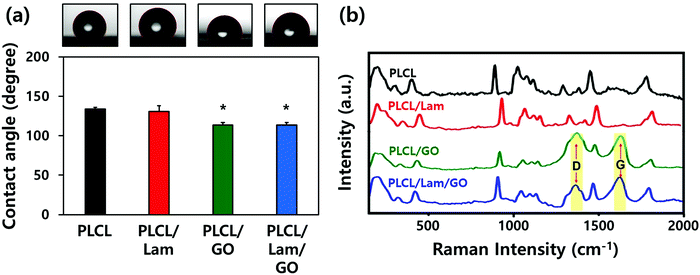
The morphological characteristics of prepared PLCL/Lam/GO nanofibre composites were observed by scanning electron microscopy (SEM) and atomic force microscopy (AFM) (Fig. 2(a) and (b), respectively). SEM images showed that all of the fabricated nanofibre composites (i.e., PLCL, PLCL/Lam, PLCL/GO, and PLCL/Lam/GO) have fine and uniform fibrous structure without any beads (Fig. 2(a)). 3D AFM images revealed that the surface roughness is lower than 2 μm (Fig. 2(b)). Every prepared nanofibre was found to be aligned at a fairly high rate (Fig. 2(c)). Moreover, as shown in Fig. 2(d), the diameter of the prepared nanofibres somewhat changed upon incorporation of Lam and GO. By containing Lam and GO, the diameter of the PLCL nanofibres was reduced from 159 nm to 139 (PLCL/Lam), 136 (PLCL/GO), and 131 (PLCL/Lam/GO) nm due to the higher conductivity of the polymer solution. It is suggested that PLCL/Lam/GO is the most suitable scaffold candidate in terms of the fibre diameter because as the fibre diameter decreases, the surface–volume-ratio increases, leading to the effective promotion of the cell–matrix interaction.33 These results indicated that the fabricated nanofibre matrices can mimic the natural ECM structure with their interconnected 3D network structure. The highly porous structure of the nanofibre matrices have advantages that facilitate the cell migration and transport of nutrients during tissue regeneration because the 3D structure can give a suitable microenvironment by providing an adhesion site and appropriate mechanical signals. Moreover, the highly aligned microstructure can enhance the growth of neuronal cells and their neurite outgrowth. This is because they can mimic the aligned nerve fibre structure to induce the neurite extension and outgrowth by mimicking the epineurium layer of the natural ECM.34The hydrophobic nature of PLCL is one of the hurdles for the tissue engineering application because it hinders the formation of a focal adhesion and integrin-mediated signalling.35,36 Therefore, we expected that the hydrophobicity of the PLCL matrices would be decreased through the hydrophilic properties of Lam and GO, making them suitable for cell adhesion and proliferation. The water contact angle measurements indicated that all of the prepared nanofibre matrices have a relatively hydrophobic surface (Fig. 3(a)). As Lam and GO were incorporated into the PLCL nanofibres, their contact angle was decreased from 133.9° to 130.5° and 113.4°, respectively. When Lam and GO were incorporated into the PLCL nanofibre matrices at the same time, the contact angle was significantly (p < 0.05) decreased to 113.7°. It is suggested that GO has the potential to increase the hydrophilicity, mainly due to their oxygen-containing moieties, such as epoxide, hydroxyl, carbonyl, and carboxylic groups.37
The Raman spectra of the PLCL, PLCL/Lam, and PLCL/Lam/GO nanofibres showed the characteristic peaks of each material (Fig. 3(b)). Because PLCL is a copolymer of PLA and PCL, the peaks for pristine PLA and PCL were observed.37,38 The peaks at 873 cm−1 (νC–COO), 1042 cm−1 (νC–CH2), 1092 cm−1 (νC–O–C), 1128 cm−1 (γCH3), and 1452 cm−1 (δCH3) denote the semicrystalline phase of pristine PLA.39 In the pristine PCL, the peaks at 1730 and 1736 cm−1 correspond to the amorphous phase, and the peak at 1720 cm−1 is assigned to the crystalline phase. Other peaks at 1470–1415 cm−1, 1303–1281 cm−1, 1107–1033 cm−1, and 912 cm−1 reveal the crystalline phase of the pristine PLCL.40,41 The C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration is assigned to a peak at around 1700 cm−1.42 Raman spectra of all nanofibre matrices indicate that the PLCL peaks are present in all groups. In particular, the Raman spectra of the PLCL/GO and PLCL/Lam/GO nanosheets exhibited the two characteristic peaks D-band (first-order scattering of the E2g mode) and G band (breathing mode of j-point phonons of A1g symmetry), which were marked with yellow column (Fig. 3(b)).42,43 These results suggest that all prepared nanofibre matrices were successfully fabricated.
O stretching vibration is assigned to a peak at around 1700 cm−1.42 Raman spectra of all nanofibre matrices indicate that the PLCL peaks are present in all groups. In particular, the Raman spectra of the PLCL/GO and PLCL/Lam/GO nanosheets exhibited the two characteristic peaks D-band (first-order scattering of the E2g mode) and G band (breathing mode of j-point phonons of A1g symmetry), which were marked with yellow column (Fig. 3(b)).42,43 These results suggest that all prepared nanofibre matrices were successfully fabricated.
2.2 Cell growth and morphology on the PLCL/Lam/GO nanofibre composite matrices
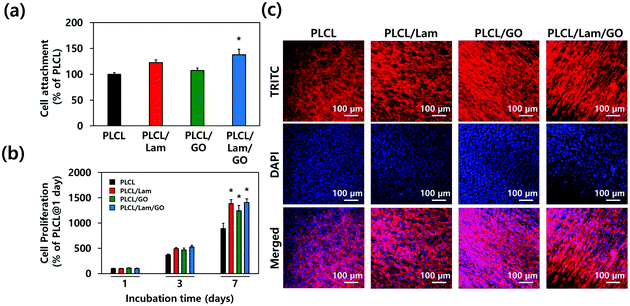
To assess the effectiveness of each nanofibre matrix as the tissue engineering scaffold, the cell adhesion and proliferation were determined. HT22 hippocampal neuronal cells were cultured on the PLCL, PLCL/Lam, PLCL/GO, and PLCL/Lam/GO nanofibre matrices. The initial attachment of the HT22 hippocampal neuronal cells was not hindered on PLCL/Lam and PLCL/GO. Rather, it significantly (p < 0.05) increased on PLCL/Lam/GO, suggesting the synergistic effects of Lam and GO on cell adhesion (Fig. 4(a)). On the other hand, cell proliferation was consistently increased from 1 to 3 days in every group, as shown in Fig. 4(b). After 7 days of incubation, the cell proliferation on PLCL/Lam, PLCL/GO, and PLCL/Lam/GO was significantly (p < 0.05) increased compared to the PLCL group. Moreover, the cells were fully grown on the nanofibre matrices, which were observed by confocal microscopy (Fig. 4(c)). These results suggest that Lam and GO can enhance the cell proliferation synergistically, which correlated to the other studies. Lam is a matrix component of the basement membrane of nerve tissue and plays an important role in the initiation of cell anchorage, and activates α3β1 integrin-related cell proliferation signalling.44 Meanwhile, GO is known to enhance cell adhesion, spreading, and proliferation by supporting the protein adsorption and intracellular protein delivery by the ionic bonding formation and the electrical conductivity of GO.45,46 The nanofibrous matrix of GO and silk fibroin was found to accelerate the early adhesion of human mesenchymal SCs by promoting F-actin assembly.47 Confocal microscopy of the HT22 hippocampal neuronal cells cultured on nanofibre matrices showed that the cells have been well spread and grown throughout the whole matrices with typical morphology, as reported elsewhere.48–50The cytotoxicity of the GO nanoparticles (GONPs) was evaluated to optimize the concentration of GONPs for their incorporation into nanofibres. We examined the cytotoxicity of GONPs against the HT22 hippocampal neuronal cells using a Cell Counting Kit-8 (CCK-8) and lactate dehydrogenase (LDH) assays. From the CCK-8 assay, based on the cell metabolic activity (Fig. S1(a), ESI†), it was found that the cell viability exhibited a decreasing tendency in a dose-dependent manner when the cells were treated with increasing concentrations (0.25–250 μg mL−1) of GONPs. At relatively low concentrations (<4 μg mL−1), over 90% of HT22 cells were viable, whereas the cell viability at 62.5 and 125 μg mL−1 decreased to about 77% and 60%, respectively. On the other hand, since the cytotoxic effects of GONPs can be ascribed to membrane disruption,51 the cytotoxicity of GONPs was also investigated using an LDH assay based on the cell membrane integrity. The extracellular release of LDH has been extensively used for investigating the cell membrane integrity, because LDH (a stable cytoplasmic enzyme) can only be released into the extracellular fluids upon plasma membrane disruption.52 As shown in Fig. S1(b) (ESI†), a significant (p < 0.05) LDH release (approximately 113% of the control) was detected only at the highest concentration of GONPs (125 μg mL−1). On the contrary, the LDH release was not detected at any other concentrations except for 125 μg mL−1 of the GONPs, indicating that relatively higher concentrations of GONPs did not induce a membrane disruption. The cytotoxicity of GONPs is known to vary, depending on their size and shape. It was revealed that the few layered GO nanosheets and graphene quantum dots exhibited different cytotoxicities on MCF-7 cells. The GO nanosheets exhibited around 78% of cell viability at a concentration of 100 μg mL−1, while the small sized graphene quantum dots retained 75% of cell viability until it reached the concentration of 400 μg mL−1, suggesting the size-dependent cytotoxicity of graphene NPs.53 Herein, the few layered sheet-like GONPs were used with dimensions of about 5 nm in height and several to tens of nm in lateral size (Fig. S2, ESI†). Therefore, it is expected that the cell viability was not hindered at concentrations lower than 31.3 μg mL−1. To determine whether the concentration of GONPs degraded from the PLCL nanofibre matrices is high enough to induce cytotoxicity, the concentration of GONPs incorporated into the nanofibre matrices should be considered. Herein, we added GONPs at the concentration of 10 μg mL−1. This means that even if all the GONPs were degraded in the media, it is too low compared to 62.5 μg mL−1. There is no need to consider the potential cytotoxicity from the degraded GONPs.
2.3 Neuritogenesis of the HT22 hippocampal neuronal cells on PLCL, PLCL/Lam, PLCL/GO, and PLCL/Lam/GO nanofibre matrices
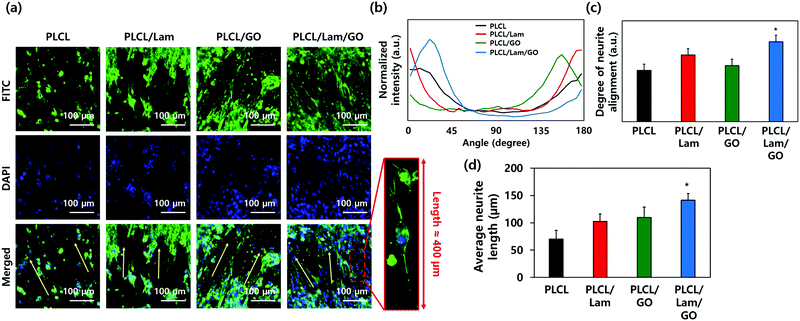
The morphologies of the HT22 hippocampal neuronal cells on each nanofibre matrix were identified using confocal microscopy (Fig. 5(a) and Fig. S3, ESI†). On all nanofibre matrices, the HT22 hippocampal neuronal cells were well-grown and aligned along the direction of the nanofibres (yellow arrows). The highly magnified image of the HT22 hippocampal neuronal cell on the PLCL/Lam/GO nanofibre matrices showed that the neurite was aligned along the fibre direction and elongated to around 400 μm. As shown in Fig. 5(b) and (c), the angle distribution of neurite and degree of neurite alignment were assessed. It is observed that the angle distribution of neurites of HT22 cells on the PLCL/Lam/GO matrices is the most aligned. The average neurite length of cells on PLCL, PLCL/Lam, PLCL/GO, and PLCL/Lam/GO was 70, 102, 109, and 141 nm, respectively (Fig. 5(d)). These results suggest that Lam and GO synergistically enhanced the neurite outgrowth of HT22 cells. Lam has been reported as an important neurite outgrowth-promoting factor throughout its heparin-binding domain.54,55 During the development of the central nervous system, haptotactic promoters such as Lam are known to stimulate the axonal guidance and extension of neurites.56 Haptotactic cues mediate contact-mediated signals, such as interaction with ECM proteins and guidance for the axon to its synaptic targets.55 Therefore, it plays critical roles in neuronal cell growth; for example, the behaviours of Schwann cells such as migration, proliferation, and differentiation for myelination could not achieved without the presence of Lam.57 Several in vitro studies also suggested that neurite outgrowth is enhanced, while covalently bound with Lam in the scaffolds such as microgrooves, hydrogels, and nanofibre matrices.58–62 These studies implied that the incorporation of Lam can physically and biochemically enhance the neurite outgrowth, and potentially improve the neuronal regeneration.Meanwhile, the high protein affinity of GO is thought to significantly enhance the neuritogenesis of the HT22 hippocampal neuronal cells. The oxygen-containing moieties of GO could enhance the protein sedimentation on the scaffolds. The exceptional high affinity contributes to the π–π stacking between the aromatic rings of several biomolecules and basal plane of graphene. Previous reports also implied that the GO in the scaffold surface exhibited good electrical coupling with the differentiated neurons; hence, the neural activity can be upregulated.63 Therefore, many studies incorporated GO in the artificial scaffolds for neuronal cell differentiation.64–68 Taken together, GO and Lam upregulated the cell–matrix interaction through several physical and biochemical ways synergistically. To elucidate how the enhanced neuritogenesis was originated by GO, the mRNA expression levels of neuronal markers were evaluated as below.
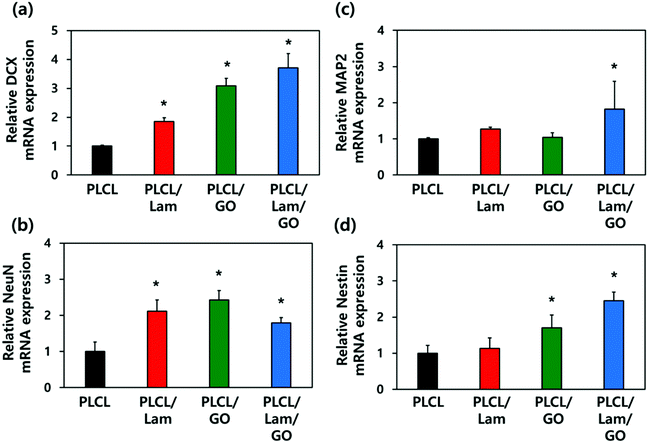
2.4 mRNA expression levels of neuronal markers on the PLCL/Lam/GO nanofibre composite matrices
To explore the underlying mechanism for the enhanced neuronal differentiation of the HT22 hippocampal neuronal cells on the PLCL/Lam/GO nanofibre matrices, we observed the expression of neuronal development-related genes such as DCX, NeuN, MAP2 and Nestin at the gene level using real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR). HT22 hippocampal neurons were seeded on 4 different types of nanofibre composite matrices, e.g., PLCL, PLCL/Lam, PLCL/GO, and PLCL/Lam/GO, and cultured in the growth medium for 3 days. Then, the medium was replaced with the differentiation medium, and further cultured for 7 days to induce neuronal differentiation. As shown in Fig. 6, regarding the mRNA expression levels of the four established neuronal differentiation markers, the presence of Lam or GO facilitated the mRNA expression, especially in the case of DCX and Nestin compared to the intact PLCL matrices without any incorporation of them.The expression of DCX, a marker for early stage nerve development, was shown to increase sequentially as the topographical cues. Conversely, HT22 cells are seeded, changing from PLCL nanofibre matrices to PLCL/Lam, PLCL/GO, and PLCL/Lam/GO nanofibre matrices. In particular, the PLCL/Lam/GO nanofibre matrix induced the expression of DCX mRNA about 3.7-fold higher than that of the PLCL matrix (Fig. 6(a). NeuN, the pre-mRNA alternative splicing regulator, also showed significantly (p < 0.05) increased expression on the nanofibre matrices incorporated with Lam, GO, or both by around 2.1-, 2.4-, and 1.8-fold, respectively, compared to the intact PLCL matrices (Fig. 6(b)). Interestingly, Fig. 6(c) showed that the mRNA expression level of MAP2, a representative neuron-specific microtubule-associated protein, significantly (p < 0.05) increased (about 1.8-fold to the intact PLCL matrices) only on the PLCL/Lam/GO matrices compared to the other groups. The expression of Nestin mRNA, encoding intermediate filament protein, significantly (p < 0.05) increased 1.7- and 2.5-fold as the biofunctional cue changed to GO and Lam/GO, showing a similar trend to that of DCX (Fig. 6(d)). As the increased expression of MAP2 that regulates microtubule and Nestin was induced by GO and Lam, it can be inferred that the outgrowth and alignment of neurite are increased by the protein expressed by these genes.69–72
Over the past years, graphene-family nanomaterials have been enormously studied for their applications in biomedical fields due to their extraordinary properties. Many studies reported that the GO particles, films or substrates could affect cell proliferation and morphology, as well as guide the differentiation of varying SCs into a specific lineage because they possess hydrophilic oxygen functional groups on the surface.73,74 The cell morphology is known as an early indicator of the SC fate. The chemical composition and functional moieties of the GO surface were found to regulate cell adhesion and morphology by promoting the adsorption of proteins and growth factors released from cells or contained in the culture medium.75,76 On the other hand, it has been reported that adding ECM to the neuronal culture matrix accelerates the development of mature neuronal networks.72 Additionally, the biofunctional hydrogel containing Lam showed a significant increase in DCX expression and neurite growth,77 which is consistent with our results.
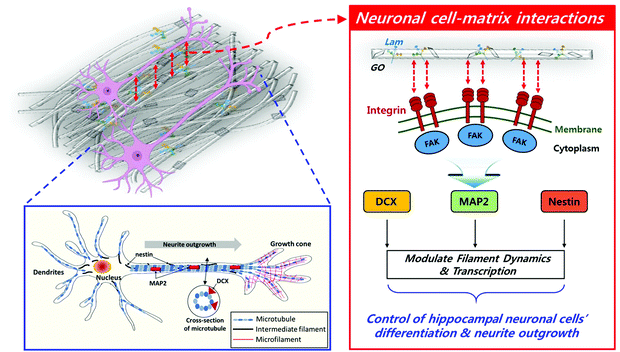
Taking all the results into consideration, the biochemically synergistic effects of Lam and GO in the PLCL nanofibre matrix on neurite outgrowth are described in Fig. 7. During the neuronal differentiation, neurons form neurites that extend from the globular cell body and elongate to form thin protrusions. This extensive remodelling for differentiation is regulated by cytoskeletal proteins, such as microtubules, intermediate filaments, and actin microfibers.78 In particular, microtubules play an important role in neurite formation by creating small bundles that invade lamellipodia in versatile directions.79 MAP2 can stabilize the microtubule bundle in the neuronal cells, which then move toward the cell periphery, prompting protrusions of the cell membrane.70 Moreover, several observations stated that the stability of the axonal microtubules, which is closely related to neurite outgrowth, is directly linked to intermediate filament levels,69 and the focal adhesion is modulated by intermediated filaments by various signalling molecules.71 DCX is able to stabilize the fully polymerised microtubule lattice (see the cross-section of microtubule in Fig. 7) against the disruption of the inter lateral contacts.80 On the other hand, Lam triggers axon formation through signalling with integrin. The increased expression of MAP2 and Nestin was observed during the axon formation.81 Therefore, it can be inferred that the enhanced outgrowth and alignment of neurites are promoted by these genes.
3. Experimental section
3.1 Preparation of PLCL/Lam/GO nanofibre composite matrices
GO (1 g) (Sigma Aldrich Co., St. Louis, MO, USA) was sonicated in 1 L deionized water for 2 h. Hydrazine hydrate (10 mL) was then added to the suspension, and the reaction proceeded at 100 °C for 24 h. After the reaction, the suspension was filtered and washed several times with ethanol and deionized water, respectively. Finally, the GONPs were prepared after drying in a vacuum oven at 80 °C for 12 h. PLCL (75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25, MW = 220 kDa, BMG Inc., Kyoto, Japan) and Laminin (MW = 850 kDa, Gibco, Singapore), isolated from Engelbreth-Holm-Swarm (EHS), were commercially purchased.
25, MW = 220 kDa, BMG Inc., Kyoto, Japan) and Laminin (MW = 850 kDa, Gibco, Singapore), isolated from Engelbreth-Holm-Swarm (EHS), were commercially purchased.
The nanofibres composed of PLCL, Lam and GONPs were fabricated by electrospinning. Briefly, PLCL with a concentration of 10 wt% was prepared by dissolving in 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP, Sigma-Aldrich) with stirring for 12 h. Lam was added to the completely dissolved PLCL solution with a weight ratio of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 = PLCL
6 = PLCL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Lam. Lastly, GONPs were added to the PLCL/Lam solution at 10 μg mL−1. Each solution of PLCL, PLCL/Lam, PLCL/GO, and PLCL/Lam/GO was loaded into a respective syringe (HSW, Tuttlingen, Germany) with a 25-gauge stainless-steel needle. The working distance between the needle and collector was set at 15 cm with a flow rate of 0.5 mL h−1, while a voltage of 18 kV (DC high voltage power supply, NanoNC, Seoul, Korea) and rotation of 2000 rpm were applied. Each type of nanofibre was fabricated, followed by slow-vacuum drying at room temperature overnight. Prior to use, the prepared nanofibres were cut into disks with 9 mm diameter and exposed under ultraviolet light for sterilization.
Lam. Lastly, GONPs were added to the PLCL/Lam solution at 10 μg mL−1. Each solution of PLCL, PLCL/Lam, PLCL/GO, and PLCL/Lam/GO was loaded into a respective syringe (HSW, Tuttlingen, Germany) with a 25-gauge stainless-steel needle. The working distance between the needle and collector was set at 15 cm with a flow rate of 0.5 mL h−1, while a voltage of 18 kV (DC high voltage power supply, NanoNC, Seoul, Korea) and rotation of 2000 rpm were applied. Each type of nanofibre was fabricated, followed by slow-vacuum drying at room temperature overnight. Prior to use, the prepared nanofibres were cut into disks with 9 mm diameter and exposed under ultraviolet light for sterilization.
3.2 Characterizations of PLCL/Lam/GO nanofibre composite matrices
To observe the surface morphology of the prepared nanofibres with SEM (TESCAN VEGA3, Brno, Czech Republic), each nanofibre was coated with an ultrathin layer of Pt prior to observation with the view field of 27.7 μm2 at an accelerated voltage of 30 kV. The nanofibres were also imaged with AFM (XE-100, Park Systems, Suwon, Korea) with a silicon cantilever in non-contact mode. The images with 10 μm × 10 μm or 45 μm × 45 μm scan sizes consisted of 512 × 512 points of height data and a scan rate of 1.0 Hz per line. Surface data from AFM were further processed with XEI software (Park Systems). Raman spectra of the prepared nanofibres were recorded using a Raman spectroscope (Ramboss 500i, Dong Woo Optron Co., Gwangju, Korea) equipped with a charge-coupled device camera (iDusDV420A-OE Andor Technology, Belfast, Ireland) and a precise motorized stage (SGSP 20–85, Sigma Koki Co., Ltd, Tokyo, Japan). A radially polarized solid-state laser of 532 nm (LasNova 50, LASOS, Jena, Germany) was focused onto the sample using a water immersion objective lens (×60 magnification, numerical aperture of 1.2 UPlanSApo, Olympus, Tokyo, Japan) and resolved with a monochromator (Monora500i, DongWoo Optron Co.). 50 mW laser power at 532 nm attenuated by using a neutral density filter of 50% transmittance at the objective was used for all measurements. Water contact angles of the nanofibres were measured with the sessile drop method using an OCA 10 goniometer (DataPhysics, Filderstadt, Germany). Then, 10 μL of distilled water was dropped onto the surface of the nanofibre and the images were captured and the contact angle was calculated by an optical system. All data quantification was assessed by ImageJ software (National Institutes of Health, Bethesda, MD).3.3 Cell culture
HT-22 mouse hippocampal neuronal cells were cultured in the growth medium (Dulbecco's modified Eagle's medium, DMEM, Welgene, Daegu, Korea) with high glucose concentration containing 10% fetal bovine serum (Welgene), and 1% antibiotic–antimycotic solution (Abs, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units of penicillin, 25 μg mL−1 of amphotericin B and 10 mg of streptomycin, Sigma-Aldrich) at 37 °C in a humid incubator with 5% CO2. Cells were seeded at a density of 1 × 104 cells on PLCL, PLCL/Lam, PLCL/GO, and PLCL/Lam/GO nanofibre matrices. To investigate the neuronal differentiation, cells were cultured on each nanofibre matrix for 3 days, and then the growth media were changed with the neurobasal media (Gibco) containing Abs, L-glutamine (Gibco), B-27 supplement (Gibco), N2 supplement (Gibco), epidermal growth factor (PeproTech, Houston, TX), and basic fibroblast growth factor (PeproTech).
000 units of penicillin, 25 μg mL−1 of amphotericin B and 10 mg of streptomycin, Sigma-Aldrich) at 37 °C in a humid incubator with 5% CO2. Cells were seeded at a density of 1 × 104 cells on PLCL, PLCL/Lam, PLCL/GO, and PLCL/Lam/GO nanofibre matrices. To investigate the neuronal differentiation, cells were cultured on each nanofibre matrix for 3 days, and then the growth media were changed with the neurobasal media (Gibco) containing Abs, L-glutamine (Gibco), B-27 supplement (Gibco), N2 supplement (Gibco), epidermal growth factor (PeproTech, Houston, TX), and basic fibroblast growth factor (PeproTech).
3.4 Cell adhesion and proliferation assays
Cell attachment and proliferation were assessed by a CCK-8 assay (Dojindo, Kumamoto, Japan). The cells on each nanofibre matrix were incubated in the growth media from 6 h (for initial attachment) up to 7 days (for proliferation). The CCK-8 assay was conducted according to the manufacturer's protocol. Briefly, the CCK-8 solution was dispensed into each matrix and incubated for 2 h at 37 °C in the dark. After the completion of the incubation period, the supernatant was collected, and then its absorbance was measured at 450 nm using a microplate reader (SpectraMax® 340, Molecular Device CO., Sunnyvale, CA).3.5 Immunocytochemical analysis
After 7 days of incubation, the cells were fixed with a 3.7% formaldehyde solution (Sigma-Aldrich) for 10 min and treated in 0.1% Triton X-100 (Sigma-Aldrich) for 5 min, followed by blocking with a 5% bovine serum albumin (BSA, GenDEPOT, Barker, TX) solution for 30 min. Anti-neurofilament heavy polypeptide antibody (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in PBS, Abcam, Cambridge, MA) was dispensed and reacted overnight in a 4 °C refrigerator. Subsequently, secondary goat anti-rabbit IgG heavy and light chains conjugated with fluorescein isothiocyanate (FITC) solution (diluted 1
1000 in PBS, Abcam, Cambridge, MA) was dispensed and reacted overnight in a 4 °C refrigerator. Subsequently, secondary goat anti-rabbit IgG heavy and light chains conjugated with fluorescein isothiocyanate (FITC) solution (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 in PBS, Abcam), 165 nM tetramethylrhodamine isothiocyanate (TRITC) – labelled phalloidin (Molecular Probes, Eugene, OR), and 300 nM 4′,6-diamidino-2-phenylindole (DAPI) solution (Sigma-Aldrich) were treated and reacted for 30 min at room temperature in the dark. The fluorescence images were obtained using a confocal laser scanning microscope (LSM 800, Zeiss, Oberkochen, Germany). The angle distribution of neurite on each nanofibre matrix was quantified by the FFT mode of ImageJ software (National Institutes of Health, Bethesda, MD) based on the confocal microscopic images. The degree of neurite alignment was determined by calculating the standard deviation (SD) of the angle distribution.
5000 in PBS, Abcam), 165 nM tetramethylrhodamine isothiocyanate (TRITC) – labelled phalloidin (Molecular Probes, Eugene, OR), and 300 nM 4′,6-diamidino-2-phenylindole (DAPI) solution (Sigma-Aldrich) were treated and reacted for 30 min at room temperature in the dark. The fluorescence images were obtained using a confocal laser scanning microscope (LSM 800, Zeiss, Oberkochen, Germany). The angle distribution of neurite on each nanofibre matrix was quantified by the FFT mode of ImageJ software (National Institutes of Health, Bethesda, MD) based on the confocal microscopic images. The degree of neurite alignment was determined by calculating the standard deviation (SD) of the angle distribution.
3.6 RNA isolation and real-time qRT-PCR
HT-22 hippocampal neuronal cells were seeded on each nanofiber matrix, and cultured with the growth medium for 3 days. Then, the medium was changed with the differentiation medium. After 7 days, the cells were dissociated with 0.05% trypsin-EDTA (Invitrogen, Carlsbad, CA) by mild pipetting. Total RNA was extracted from the detached cells using TRIzol reagent (Invitrogen) and an RNeasy Mini Kit (Qiagen, Grand Island, NY). SuperScript III First-Strand cDNA Synthesis System (Invitrogen) was further used to synthesize the first-strand cDNA from the total RNA, according to the manufacturer's instructions. The mRNA expression of the specific genes was then determined by real-time qRT-PCR using the total first-strand cDNA as the template and Power SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA). The following sequences of the primers for DCX, NeuN, MAP2, Nestin and 18S were used: DCX, 5′-ATGCAGTTGTCCCTCCATTC-3′ (sense) and 3′-ATGCCACCAAGTTGTCATCA-5′ (antisense); NeuN, 5′-CACCACTCTCTTGTCCGTTTGC-3′ (sense) and 3′-GGCTGAGCATATCTGTAAGCTGC-5′ (antisense); MAP2, 5′-GAGAAACGTTCTTCCCTCCC-3′ (sense) and 3′-GTGTGGAGGTGCCACTTTTT-5′ (antisense); Nestin, 5′-AGAGGACCAGGTGCTTGAGA-3′ (sense) and 3′-TCCTCTGCGTCTTCAAACCT-5′ (antisense); 18S, 5′-ATGCAGTTGTCCCTCCATTC-3′ (sense) and 3′-GAATCGAACCCTGATTCCCCGTC-5′ (antisense). The 18S rRNA expression level was used as an endogenous normalizer and the relative expression levels were calculated using the −ΔΔCt method.3.7 Statistical analysis
All variables were tested in three independent cultures for each experiment, which was repeated twice (n = 6). The quantitative data are expressed as the mean ± SD. The data were tested for the homogeneity of the variances using Levene's test, prior to statistical analysis. Statistical comparisons were carried out using a one-way analysis of variance (ANOVA; SAS Institute Inc., Cary, NC), followed by a Bonferroni test for multiple comparisons. A p value <0.05 was considered as statistically significant differences among the means.4. Conclusions
In the present study, GO-functionalized PLCL and Lam nanofibre composite matrices were prepared by an electrospinning process to explore whether they have the potential to enhance the differentiation of hippocampal neuronal cells. To prove this hypothesis, the physicochemical properties and biofunctionality of PLCL/Lam/GO nanofibre composite matrices were evaluated. Our fascinating results can be summarized as follows: first, the PLCL/Lam/GO nanofibre composite matrices were found to provide a three-dimensional structure similar to the natural ECM, leading to support of the attachment and proliferation of the hippocampal neuronal cells. Second, the PLCL/Lam/GO nanofibre matrices could promote neurite outgrowth and alignment from hippocampal neuronal cells through providing optimal microenvironments for neuronal differentiation. These results suggest that the PLCL/Lam/GO nanofibre matrices can be employed as potential scaffolds for neural tissue engineering and regeneration. Lastly, the underlying mechanism of this phenomenon could be proposed according to the scenario in which neurite outgrowth was guided by biofunctional cues (such as Lam and GO) with nanofibre-mediated topographical cues, which upregulated the expression of some specific genes (i.e., DCX, MAP2 and Nestin) closely related to cytoskeletal interactions. Based on this scenario, if biofunctional cues that regulate intracellular biochemical mechanisms are applied, the development of biomaterials with capability to induce more accurate and efficient cellular responses can be expected. Thus, these intriguing experimental results highlight the need for detailed studies on specific upstream signalling regulators that mediate cytoskeletal proteins.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Research Foundation (NRF) of Korea grant funded by the Korea government (MSIT) (No. 2019R1A4A1024116).References
- J. Faweett and R. J. Keynes, Annu. Rev. Neurosci., 1990, 13, 43–60 CrossRef.
- F. Ijpma, R. Van De Graaf and M. Meek, J. Hand Surg. Eur. Vol., 2008, 33, 581–586 CrossRef CAS.
- E. Kijeńska, M. P. Prabhakaran, W. Swieszkowski, K. J. Kurzydlowski and S. Ramakrishna, J. Biomed. Mater. Res., Part B, 2012, 100, 1093–1102 CrossRef.
- A. Subramanian, U. M. Krishnan and S. Sethuraman, J. Biomed. Sci., 2009, 16, 108 CrossRef.
- B. G. Zhang, A. F. Quigley, D. E. Myers, G. G. Wallace, R. M. Kapsa and P. F. Choong, Int. J. Artif. Organs, 2014, 37, 277–291 CrossRef.
- S. Kumbar, R. James, S. Nukavarapu and C. Laurencin, Biomed. Mater., 2008, 3, 034002 CrossRef CAS.
- M. P. Prabhakaran, J. Venugopal, C. K. Chan and S. Ramakrishna, Nanotechnology, 2008, 19, 455102 CrossRef.
- E. Kijeńska, M. P. Prabhakaran, W. Swieszkowski, K. J. Kurzydlowski and S. Ramakrishna, Eur. Polym. J., 2014, 50, 30–38 CrossRef.
- L. Ghasemi-Mobarakeh, M. P. Prabhakaran, M. Morshed, M.-H. Nasr-Esfahani and S. Ramakrishna, Biomaterials, 2008, 29, 4532–4539 CrossRef CAS.
- J. Xie, M. R. MacEwan, S. M. Willerth, X. Li, D. W. Moran, S. E. Sakiyama-Elbert and Y. Xia, Adv. Funct. Mater., 2009, 19, 2312–2318 CrossRef CAS.
- J. Wang, L. Tian, N. Chen, S. Ramakrishna and X. Mo, Mater. Sci. Eng., C, 2018, 91, 715–726 CrossRef CAS.
- J. Wang, L. Tian, B. Luo, S. Ramakrishna, D. Kai, X. J. Loh, I. H. Yang, G. R. Deen and X. Mo, Colloids Surf., B, 2018, 169, 356–365 CrossRef CAS.
- D. Gupta, J. Venugopal, M. P. Prabhakaran, V. G. Dev, S. Low, A. T. Choon and S. Ramakrishna, Acta Biomater., 2009, 5, 2560–2569 CrossRef CAS.
- L. Tian, M. P. Prabhakaran, J. Hu, M. Chen, F. Besenbacher and S. Ramakrishna, Colloids Surf., B, 2016, 145, 420–429 CrossRef CAS.
- Z. Zhang, M. L. Jorgensen, Z. Wang, J. Amaget, Y. Wang, Q. Li, M. Dong and M. Chen, Biomaterials, 2020, 253, 120108 CrossRef CAS.
- S. J. Song, Y. C. Shin, S. E. Kim, I. K. Kwon, J. H. Lee, S. H. Hyon, D. W. Han and B. Kim, Sci. Rep., 2018, 8, 1–11 CrossRef.
- E. Schnell, K. Klinkhammer, S. Balzer, G. Brook, D. Klee, P. Dalton and J. Mey, Biomaterials, 2007, 28, 3012–3025 CrossRef CAS.
- Y. Qian, X. Zhao, Q. Han, W. Chen, H. Li and W. Yuan, Nat. Commun., 2018, 9, 1–16 CrossRef CAS.
- S. Shah, P. T. Yin, T. M. Uehara, S. T. D. Chueng, L. Yang and K. B. Lee, Adv. Mater., 2014, 26, 3673–3680 CrossRef CAS.
- Y. Chen, M. B. Taskin, Z. Zhang, Y. Su, X. Han and M. Chen, Biomater. Sci., 2019, 7, 2165–2173 RSC.
- L. Luckenbill-Edds, Brain Res. Rev., 1997, 23, 1–27 CrossRef CAS.
- Y. Zhu, S. Murali, W. Cai, X. Li, J. W. Suk, J. R. Potts and R. S. Ruoff, Adv. Mater., 2010, 22, 3906–3924 CrossRef CAS.
- Y. C. Shin, S. J. Song, S. W. Hong, S. J. Jeong, W. Chrzanowski, J. C. Lee and D. W. Han, Nanomaterials, 2017, 7, 369 CrossRef.
- X. Zhou, B. Liu, Y. Chen, L. Guo and G. Wei, Mater. Adv., 2020, 1, 2163–2181 RSC.
- E. J. Lee, J. H. Lee, Y. C. Shin, D.-G. Hwang, J. S. Kim, O. S. Jin, L. Jin, S. W. Hong and D.-W. Han, Biomater. Res., 2014, 18, 18–24 CrossRef.
- H. H. Yoon, S. H. Bhang, T. Kim, T. Yu, T. Hyeon and B. S. Kim, Adv. Funct. Mater., 2014, 24, 6455–6464 CrossRef CAS.
- S. H. Kang, Y. C. Shin, E. Y. Hwang, J. H. Lee, C. S. Kim, Z. Lin, S. H. Hur, D. W. Han and S. W. Hong, Mater. Horiz., 2019, 6, 1066–1079 RSC.
- M. Mazaheri, O. Akhavan and A. Simchi, Appl. Surf. Sci., 2014, 301, 456–462 CrossRef CAS.
- K. Zhang, H. Zheng, S. Liang and C. Gao, Acta Biomater., 2016, 37, 131–142 CrossRef CAS.
- J. Wang, Y. Cheng, L. Chen, T. Zhu, K. Ye, C. Jia, H. Wang, M. Zhu, C. Fan and X. Mo, Acta Biomater., 2019, 84, 98–113 CrossRef CAS.
- S. Unal, S. Arslan, T. Gokce, B. M. Atasoy, B. Karademir, F. N. Oktar and O. Gunduz, Eur. Polym. J., 2019, 115, 157–165 CrossRef CAS.
- J. Song, H. Gao, G. Zhu, X. Cao, X. Shi and Y. Wang, Carbon, 2015, 95, 1039–1050 CrossRef CAS.
- H. Koh, T. Yong, C. Chan and S. Ramakrishna, Biomaterials, 2008, 29, 3574–3582 CrossRef CAS.
- L. Wang, Y. Wu, T. Hu, P. X. Ma and B. Guo, Acta Biomater., 2019, 96, 175–187 CrossRef CAS.
- B. G. Keselowsky, D. M. Collard and A. J. García, Biomaterials, 2004, 25, 5947–5954 CrossRef CAS.
- M. Lampin, R. Warocquier-Clérout, C. Legris, M. Degrange and M. Sigot-Luizard, J. Biomed. Mater. Res., Part A, 1997, 36, 99–108 CrossRef CAS.
- X. K. Kong, Q. W. Chen and Z. Y. Lun, J. Mater. Chem. A, 2014, 2, 610–613 RSC.
- K. Nalampang, R. Molloy and W. Punyodom, Polym. Adv. Technol., 2007, 18, 240–248 CrossRef CAS.
- K. K. Gupta, N. Pal, P. K. Mishra, P. Srivastava, S. Mohanty and P. Maiti, J. Biomed. Mater. Res., Part A, 2014, 102, 2600–2612 CrossRef.
- S. I. Jeong, I. D. Jun, M. J. Choi, Y. C. Nho, Y. M. Lee and H. Shin, Macromol. Biosci., 2008, 8, 627–637 CrossRef CAS.
- S. H. Huang, T. T. Hsu, T. H. Huang, C. Y. Lin and M. Y. Shie, J. Res. Dent. Sci., 2017, 12, 33–43 Search PubMed.
- S.-D. Wang, Q. Ma, K. Wang and H.-W. Chen, ACS Omega, 2018, 3, 406–413 CrossRef CAS.
- J. Wang, Y. Cheng, L. Chen, T. Zhu, K. Ye, C. Jia, H. Wang, M. Zhu, C. Fan and X. Mo, Acta Biomater., 2019, 84, 98–113 CrossRef CAS.
- M. Gonzales, K. Haan, S. E. Baker, M. Fitchmun, I. Todorov, S. Weitzman and J. C. Jones, Mol. Biol. Cell, 1999, 10, 259–270 CrossRef CAS.
- X. Shi, H. Chang, S. Chen, C. Lai, A. Khademhosseini and H. Wu, Adv. Funct. Mater., 2012, 22, 751–759 CrossRef CAS.
- H. Li, K. Fierens, Z. Zhang, N. Vanparijs, M. J. Schuijs, K. Van Steendam, N. L. Feiner Gracia, R. De Rycke, T. De Beer and A. De Beuckelaer, ACS Appl. Mater. Interfaces, 2016, 8, 1147–1155 CrossRef CAS.
- Y. Shuai, C. Mao and M. Yang, Appl. Mater. Interfaces, 2018, 10, 31988–31997 CrossRef CAS.
- G. A. Silva, C. Czeisler, K. L. Niece, E. Beniash, D. A. Harrington, J. A. Kessler and S. I. Stupp, Science, 2004, 303, 1352–1355 CrossRef CAS.
- J. L. Bourke, H. A. Coleman, V. Pham, J. S. Forsythe and H. C. Parkington, Tissue Eng., Part A, 2014, 20, 1089–1095 CrossRef CAS.
- G.-Q. Chen and J. Zhang, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 1–18 CrossRef CAS.
- P. Lu, A. Zehtab Yazdi, X. X. Han, K. Al Husaini, J. Haime, N. Waye and P. Chen, Chem. Res. Toxicol., 2020, 33, 2247–2260 Search PubMed.
- S. J. Song, Y. C. Shin, H. U. Lee, B. Kim, D. W. Han and D. Lim, Nanomaterials, 2018, 8, 408–417 Search PubMed.
- C. Wu, C. Wang, T. Han, X. Zhou, S. Guo and J. Zhang, Adv. Healthcare Mater., 2013, 2, 1613–1619 Search PubMed.
- D. Lander, D. K. Fujii and L. F. Reichardt, Proc. Natl. Acad. Sci. U. S. A., 1985, 82, 2183–2187 CrossRef.
- D. Edgar, R. Timpl and H. Thoenen, EMBO J., 1984, 3, 1463–1468 CrossRef CAS.
- H. G. Sundararaghavan, S. N. Masand and D. I. Shreiber, J. Neurotrauma, 2011, 28, 2377–2387 CrossRef.
- M. Tessier-Lavigne and C. S. Goodman, Science, 1996, 274, 1123–1133 CrossRef CAS.
- S. H. Hsu, C. Y. Chen, P. S. Lu, C. S. Lai and C. J. Chen, Biotechnol. Bioeng., 2005, 92, 579–588 CrossRef CAS.
- W. T. Su, Y. F. Liao, T. W. Wu, B. J. Wang and Y. Y. Shih, J. Biomed. Mater. Res., Part A, 2013, 101, 185–194 CrossRef.
- S. E. Stabenfeldt, A. J. García and M. C. LaPlaca, J. Biomed. Mater. Res., Part A, 2006, 77, 718–725 CrossRef.
- B. Mammadov, R. Mammadov, M. O. Guler and A. B. Tekinay, Acta Biomater., 2012, 8, 2077–2086 CrossRef CAS.
- R. A. Neal, S. G. McClugage III, M. C. Link, L. S. Sefcik, R. C. Ogle and E. A. Botchwey, Tissue Eng., Part C, 2009, 15, 11–21 CrossRef CAS.
- S. Y. Park, J. Park, S. H. Sim, M. G. Sung, K. S. Kim, B. H. Hong and S. Hong, Adv. Mater., 2011, 23, H263–H267 CrossRef CAS.
- J. S. Lee, A. Lipatov, L. Ha, M. Shekhirev, M. N. Andalib, A. Sinitskii and J. Y. Lim, Biochem. Biophys. Res. Commun., 2015, 460, 267–273 CrossRef CAS.
- Z. He, S. Zhang, Q. Song, W. Li, D. Liu, H. Li, M. Tang and R. Chai, Colloids Surf., B, 2016, 146, 442–451 CrossRef CAS.
- F. R. Boroojeni, S. Mashayekhan, H.-A. Abbaszadeh, M. Ansarizadeh, M.-S. Khoramgah and V. R. Movaghar, Int. J. Nanomed., 2020, 15, 3903–3920 CrossRef CAS.
- D. Yang, T. Li, M. Xu, F. Gao, J. Yang, Z. Yang and W. Le, Nanomed. J., 2014, 9, 2445–2455 CrossRef CAS.
- K. Yang, S. J. Yu, J. S. Lee, H. R. Lee, G. E. Chang, J. Seo, T. Lee, E. Cheong, S. G. Im and S. W. Cho, Nanoscale, 2017, 9, 18737–18752 RSC.
- P. N. Hoffman, S. C. Pollock and G. G. Striph, Exp. Neurol., 1993, 119, 32–36 CrossRef CAS.
- L. Dehmelt, P. Nalbant, W. Steffen and S. Halpain, Brain Cell Biol., 2006, 35, 39–56 CrossRef CAS.
- R. E. Leube, M. Moch and R. Windoffer, Curr. Opin., 2015, 32, 13–20 CrossRef CAS.
- D. Lam, H. A. Enright, J. Cadena, S. K. Peters, A. P. Sales, J. J. Osburn, D. A. Soscia, K. S. Kulp, E. K. Wheeler and N. O. Fischer, Sci. Rep., 2019, 9, 1–15 CrossRef CAS.
- W. C. Lee, C. H. Y. Lim, H. Shi, L. A. Tang, Y. Wang, C. T. Lim and K. P. Loh, ACS Nano, 2011, 5, 7334–7341 CrossRef CAS.
- S. Shah, P. T. Yin, T. M. Uehara, S. T. D. Chueng, L. Yang and K. B. Lee, J. Adv. Mater., 2014, 26, 3673–3680 CrossRef CAS.
- T. R. Nayak, H. Andersen, V. S. Makam, C. Khaw, S. Bae, X. Xu, P. L. R. Ee, J. H. Ahn, B. H. Hong and G. Pastorin, ACS Nano, 2011, 5, 4670–4678 CrossRef CAS.
- G. Kumar, C. K. Tison, K. Chatterjee, P. S. Pine, J. H. McDaniel, M. L. Salit, M. F. Young and C. G. Simon Jr., Biomaterials, 2011, 32, 9188–9196 CrossRef CAS.
- A. Farrukh, F. Ortega, W. Fan, N. Marichal, J. I. Paez, B. Berninger, A. Del Campo and M. J. Salierno, Stem Cell Rep., 2017, 9, 1432–1440 CrossRef CAS.
- A. Caceres, J. Mautino and K. S. Kosik, Neuron, 1992, 9, 607–618 CrossRef CAS.
- M. Götz and W. B. Huttner, Nat. Rev. Mol. Cell Biol., 2005, 6, 777–788 CrossRef.
- C. A. Moores, M. Perderiset, C. Kappeler, S. Kain, D. Drummond, S. J. Perkins, J. Chelly, R. Cross, A. Houdusse and F. Francis, EMBO J., 2006, 25, 4448–4457 CrossRef CAS.
- W. L. Lei, S. G. Xing, C. Y. Deng, X. C. Ju, X. Y. Jiang and Z. G. Luo, Cell Res., 2012, 22, 954–972 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ma00733a |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2020 |