Open Access Article
Open Access ArticleDual signal amplification for microRNA-21 detection based on duplex-specific nuclease and invertase†
Xitong Huang,
Zhiming Xu,
Ji-Hua Liu,
Bo-Yang Yu * and
Jiangwei Tian
* and
Jiangwei Tian *
*
State Key Laboratory of Natural Medicines, Jiangsu Key Laboratory of TCM Evaluation and Translational Research, Research Center for Traceability and Standardization of TCMs, School of Traditional Chinese Pharmacy, China Pharmaceutical University, Nanjing 211198, P. R. China. E-mail: boyangyu59@163.com; jwtian@cpu.edu.cn
First published on 18th March 2020
Abstract
MicroRNA-21 (miRNA-21) is a significant biomarker which is closely related to some kinds of diseases, such as cancer, cardiovascular disease and kidney disease. Therefore, the detection of miRNA-21 is of great importance and can provide essential information for disease diagnosis. In this study, we report a facile, sensitive assay for miRNA-21 detection using personal glucose meters (PGM). Biotinylated DNA strand linked invertase (Inv) is conjugated on the surface of streptavidin-coated magnetic beads (MBs) to form a MBs–DNA–Inv complex. Target miRNA-21 in the detection system is captured by the MBs–DNA–Inv probe through DNA/RNA hybridization. The duplex-specific nuclease (DSN) enzyme specifically cleaves the DNA to recycle the target miRNA and release invertase, thereby triggering the dual signal amplification and ensuring high sensitivity. Besides, we establish a linear relationship between PGM and different concentrations of miRNA-21 in the range of 10 to 200 pM. The limit of detection is 1.8 pM, which is more sensitive than some of the previous reports. In addition, the biosensor exhibits excellent sequence selectivity and single-base mutation can be discriminated. Moreover, the expression of miRNA-21 is confirmed in urine from mice by our method, which is in good accordance with the qRT-PCR result. Therefore, a dependable, low-cost strategy for the detection of miRNA has been established and it meets the latest analytical demands for miRNA determination that is suitable for the public.
1 Introduction
MicroRNAs (miRNAs) are members of a short noncoding RNA (18–25 nucleotides) pairing to the 3′ untranslated region or messenger RNAs and regulates the diverse gene expression.1 The specificity of cells and tissues is the main feature of miRNA expression, and it is highly stable in plasma and serum.2 These characteristics determine its unique advantages in disease diagnosis and prognosis evaluation. Recent studies indicate that microRNA-21 (miRNA-21) is closely related to the pathophysiological processes of some kinds of diseases, such as cancer, cardiovascular disease and kidney disease.3 Therefore, accurate detection of miRNA-21 is significant for scientific research, diagnosis and the treatment of disease.However, traditional methods for miRNA analysis such as northern blotting analysis, quantitative reverse transcription-polymerase chain reaction (qRT-PCR), oligonucleotide microarrays, and next-generation sequencing (NGS) remain challenging for the sensitivity and selectivity for miRNA quantitation.4 For instance, the detection limits of northern blotting are usually low, qRT-PCR is more calculated for the quantification of longer RNA targets considering the relatively low selectivity during reverse transcription (RT) and PCR process. In the process of qRT-PCR, small contaminants may also be amplified to produce false positives. The application of NGS is still restricted by the relatively high rate of detection errors, high demands for apparatus, computer performance, personnel capability and expensive cost for one test.5 Therefore, developing a strategy with low demands for instrument and operational skills, but high sensitivity and selectivity to miRNA detection is meaningful and desirable.
As a widely used personal diagnosis equipment at home, a personal glucose meter (PGM) benefits from its portable size, simple operation and reliable quantitative results.6 In 2011, Lu et al. creatively linked PGM with functional DNA sensors to achieve portable, low-cost and quantitative detection of targets beyond glucose.7 Later on, many groups have used PGM as the signal collectors. For example, Su et al. used PGM for copper(II) detection based on click chemistry.8a Xu et al. used PGM for DNA detection based on isothermal circular strand-displacement8b and Xie et al. used the PGM for DNA detection based on polyamidoamine dendrimer signal amplification.8c However, few studies have used PGM for miRNA detection.
Inspired by pioneering works, we propose a miRNA detection strategy that combined a PGM with a functional nucleic acid sensor for miRNA quantitative detection. To further upgrade the detection sensitivity, an amplification strategy of duplex-specific nuclease (DSN) and invertase (Inv) is applied. Invertase is a perfect mediator for quantification of target using PGM because it converts sucrose to glucose and its highly efficient enzymatic conversion contributes to the sensitivity of the assay.9 On the other hand, DSN subsequently selectively cleaves the DNA in DNA/RNA hybrid to promise for target miRNA recycling, which will trigger the next rounds of reactions to implement signal amplification.10 In the sensing system, which is illustrated in Scheme 1, biotinylated DNA strand linked invertase is conjugated on the surface of streptavidin-coated magnetic beads (MBs) to form MBs–DNA–Inv complex. Target miRNA-21 in the detection system is captured by MBs–DNA–Inv probe through DNA/RNA hybridization. The DSN enzyme specifically cleaves the DNA to recycle the target miRNA and release invertase. As a result, there is more free invertase released from the MBs–DNA–Inv conjugates, target miRNA will recycle to trigger the next rounds of reactions and achieve signal amplification. After magnetic separation of MBs, the supernatant containing the released invertase is transferred into sucrose solution and sucrose was catalysed into glucose. Finally, the concentration of miRNA-21 is transformed to the level of glucose for monitoring of PGM. A reliable and convenient miRNA-21 assay is expected by making use of DSN and the target-triggered release of invertase from a functional MBs–DNA–Inv conjugate. The PGM combined with the DSN holds enormous potential for sensitive and low-cost miRNA quantitative detection.
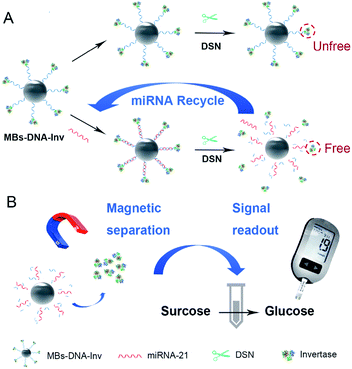 | ||
| Scheme 1 Schematic diagram of the MBs–DNA–Inv and DSN-assisted signal amplification platform for the detection of miRNA-21. | ||
2 Experimental
2.1 Chemicals and materials
Streptavidin-MBs (1 μM in diameter), sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (sulfo-SMCC), Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) were purchased from Aladdin (Shanghai, China). Grade VII invertase obtained from baker's yeast (S. cerevisiae) was brought from Sigma (St. Louis, MO). Duplex-specific nuclease (DSN, Evrogrn, cat. no. EA002) was purchased from Newborn Company (Shenzhen, China). HPLC-purified, synthetic oligonucleotides used in this work were synthesized from Sangon Biotech (Shanghai, China). Detail sequences of the oligonucleotides DNA and microRNA molecules were listed in Table S1.† They were denatured at 95 °C for 5 min and naturally cooled down to room temperature before use. Diethyl pyrocarbonate (DEPC) treated water was used in the preparation of aqueous solutions. Express miRNA extraction kit (cat. no. R4310-02) was purchased from Magen Gene (Guangzhou, China) and miRNA qRT-PCR Starter kit (cat. no. C10211-2) was obtained from RiboBio (Guangzhou, China). Other chemicals were of analytical grade and obtained from standard reagent suppliers and used directly. All solutions were prepared with Milli-Q water (resistivity ¼ 18 MU cm) from a Millipore system.2.2 Instrumentation
The signal of glucose concentration was detected by a Roche ACCU-CHEK personal glucose meter. Dynamic light scattering (DLS) characterization was performed using a Zetasizer Nano ZS90 (Malvern Instruments, UK) by means of a 90 Plus/BI-MAS equipment (Brookhaven, USA). Zeta potential measurement was performed on a Malvern Zetasizer-Nano Z instrument. UV-vis absorption spectra were collected on an UV-2550 UV-vis spectrophotometer (Shimadzu Company, Japan). All pH measurements were carried out with a Sartorius basic pH-meter. Gel separation were achieved by Model VE180 micro-vertical electrophoresis tank from Shanghai Tianneng Technology Co, Ltd. Gel Imaging System was from BIO-RAD Molecular Imager (USA). The HE staining images were acquired on a digital pathology slice scanner using NanoZoomer 2.0 RS (Hamamatsu, China). The concentration of nucleic acid was detected by NanoDrop 2000 (Thermo Fisher Scientific, USA). The reverse transcription was performed on Mastercycler nexus (Eppendorf, Germany) and qRT-PCR were carried out with QuantStudio TM 3 Real-Time PCR Instrument (Thermo Scientific, USA).2.3 Synthesis of the DNA–Inv conjugate
Firstly, 30 μL of DNA strand dissolved in diethylpyrocarbonate (DEPC) treated water (1 mM), 1 mL of sodium phosphate buffer (1 M, pH 5.5) and 1 mL of TCEP solution (30 mM) were mixed and incubated for 1 h at room temperature. Then it was purified by ultrafiltration using an Amicon-10K ultrafiltration tube 8 times. Briefly, 8 mg invertase and 1 mg sulfo-SMCC were dissolved in Buffer A (0.1 M NaCl, 0.1 M sodium phosphate buffer, pH 7.3). Then vortexed for 5 minutes and rolled for 2 h at room temperature. Excess sulfo-SMCC was removed by centrifugation; the product of sulfo-SMCC-Inv was purified by Amicon-100K 8 times. The purified sulfo-SMCC-Inv solution was mixed with the above SH-DNA solution, and allowed to stand at room temperature for 48 hours; then being purified by Amicon-100K 8 times using Buffer A to remove unreacted DNA strand.2.4 Preparation of the MBs–DNA–Inv complex
A solution of streptavidin-coated magnetic beads (MBs) in a microtube (1 mL 1 mg mL−1) was placed close to a magnetic rack for 1 minute. The clear solution was discarded and replaced by 1 mL of Buffer B (0.1 M NaCl, 0.1 M sodium phosphate buffer, pH 7.3, 0.05% Tween-20). This buffer exchange procedure was repeated twice. Then, 12 μL of 0.5 mM DNA–Inv (about 20 mg mL−1) conjugate in Buffer B in water was added to the 1 mL MBs solution and mixed on a roller for 60 minutes at room temperature. Excess DNA–Inv conjugate was washed off by Buffer B five times and was recycled for further use by condensing the washing solutions using an Amicon-100 K. The MBs residue from 100 μL MBs solution after removal of solvent was used for each test.2.5 Detection of miRNA-21 with PGM
20 μL MBs–DNA–Inv was added 10 μL of varying concentrations of miRNA-21, 2 μL of DSN buffer (50 mM Tris–HCl, pH 8.0, 5 mM MgCl2, and 1 mM DTT), 7 μL of DEPC-treated water, and 1 μL of DSN (1 U μL−1). The mixed solution incubating at 45 °C for 50 min allowing for the target miRNA-21 recycling assisted by DSN. Then the solution was placed next to the magnetic rack. The clear supernatant was dropped into a tube with 100 μL of sucrose in Buffer A (1 M). Before PGM measurements, the mixture was heated to 55 °C for 40 min. Finally, the obtained mixed solution was detected by a PGM.2.6 Extraction and detection of miRNA from urine
Male Institute for Cancer Research (ICR) mice were purchased from the Laboratory Animal Center and Institute of Comparative Medicine at Yangzhou University. All animal study protocols were designed in according to guidelines set by the National Institute of Health Guide for the Care and Use of Laboratory Animals and approved by the Animal Ethical Experimentation Committee of China Pharmaceutical University. The total RNA was extracted from mice urine by the expression of the miRNA extraction kit (cat. no. R4310-02) and the quality of the miRNA was assessed using NanoDrop. To quantify the expression of mature miRNA-21, the following procedure was carried out. The extracted miRNA was separately detected by our proposed method and qRT-PCR. Total RNA was reverse-transcribed to cDNA and then quantified by qRT-PCR using miRNA qRT-PCR Starter kit (cat. no. C10211-2). The qRT-PCR reaction conditions was described as follows: 95 °C for 10 min, followed by 40 cycles with a 2 s interval at 95 °C, 20 s interval at 60 °C and 10 s interval at 70 °C.3 Results and discussion
3.1 Characterization of DNA–Inv conjugate and MBs–DNA–Inv
The synthetic approach to prepare the DNA–Inv conjugate used the maleimide-thiol reaction by the heterobifunctional linker (sulfo-SMCC).7a First of all, the purified DNA–Inv conjugate was characterized by UV-vis absorption spectra and PAGE images. As shown in Fig. S1,† the UV-vis absorption spectrum of the purified DNA–Inv conjugate overlaid well with the sum of the spectra of its two components, DNA and invertase, suggesting successful conjugation. The successful conjugation had also been confirmed by SDS-PAGE, while the migration of the DNA–Inv conjugate band was less than that of invertase because the conjugation increased the molecular weight of the enzyme (Fig. S2†). The hydration diameter of the conjugate was detected by DLS. The increased average hydration diameter from 1.09 ± 0.027 μM to 1.90 ± 0.12 μM (Fig. 1A) suggested the successful conjunction of MBs–DNA–Inv and a uniform distribution. Besides, the zeta-potential of was reduced from −8.54 ± 0.23 mV to −17.7 ± 0.51 mV after conjunction (Fig. 1B), which indicated that the probe was highly dispersible in aqueous solution.11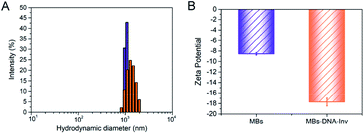 | ||
| Fig. 1 (A) Size distribution of MBs (blue) and MBs–DNA–Inv (orange) determined by DLS. (B) Zeta potentials for MBs and MBs–DNA–Inv. Data are means ± SD (n = 3). | ||
3.2 Optimization of experimental parameters
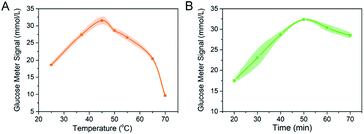
The performance of our miRNA detection method was determined by the efficiency of DSN process. In order to obtain the high performance of detection platform, length of DNA strand, the reaction temperature and incubation time were further optimized. To ensure the proper spacer length for target miRNA-21 to interact with the invertase, the length of the DNA strand in designed platform was optimized. DNA strand 1–5 with different lengths of adenine domain (5, 10, 15, 20, 25) were modified to the 5′ end of the DNA strand respectively (detail sequences were listed in Table S1†). After the reaction, the supernatant was transferred to 1 M sucrose solution and reacting for 40 min. The signal reading was measured by PGM, and the cleaving efficiency under various ratio synthesis conditions was compared by the amount of glucose finally produced. The results showed that the cleavage efficiency was high for the DNA strand with 20 adenine domains. With longer spacer (strand 5), PGM showed no significant change in signal (Fig. S3†). Thereby, we decided to choose DNA strand 4 to synthesis the MBs–DNA–Inv platform in the next experiments.The detection sensitivity of our platform for the miRNA-21 was improved by DSN-oriented signal amplification. Incubation time and temperature are the two crucial parameters for the DSN performance because these factors will influence the enzyme activity, hybridization efficiency and stability of DNA/RNA hybrids. A higher or lower hybridization temperature to Tm value would lead to an ineffective or nonspecific hybridization. Therefore, a series of reaction temperature (25 °C, 37 °C, 45 °C, 50 °C, 55 °C, 65 °C, and 70 °C) was studied. As showed in Fig. 2A, the PGM reached a maximal value at 45 °C because higher temperature might decrease invertase activity, and this temperature was selected for subsequent detection.
Then we evaluated the optimized incubation time for the DSN. As proved in Fig. 2B, with the extension of reaction time, the PGM signal increased and the highest PGM signal was obtained at 50 min. With the recycle miRNA-21 reaction triggered by DSN, more invertase would release into the supernatant. However, a longer incubation time might reduce the activity of invertase because the DTT contained in reaction buffer might influence the disulfide bond of invertase.12 Hence, in the following studies, the detection of miRNA-21 was carried out at 45 °C for 50 min. Considering that the recommended optimal concentration of DSN kit and reported research was 1 U, we did not optimize the DSN concentration but chose a reaction concentration of 1 U for all experiments.13
3.3 Sensitivity and selectivity of PGM for miRNA-21 detection
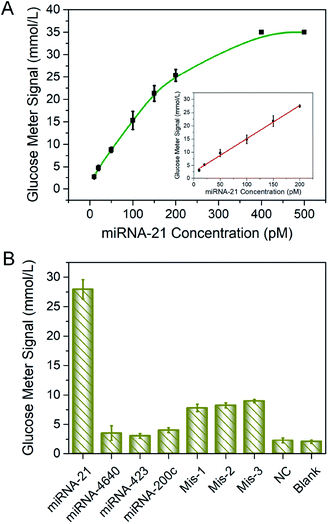
Under optimized conditions, the sensitivity and selectivity of the detection of miRNA-21 by this method was studied. The linear relationship between miRNA-21 concentration and PGM signal was established and was shown in Fig. 3A. As expected, higher levels of the target led to larger PGM signal increases. A good linear range from 10 to 200 pM was obtained with the linear regression equations S = 2.98 + 0.12c, in which S was the value of PGM readings and c was the miRNA-21 concentration, the coefficient of determination (R2) was 0.997. The detection limit for miRNA-21 was calculated to be 1.8 pM (3σ/k, in which was the standard deviation of blank measurements, n = 6, and k was the slope of the linear equation). A performance comparison of the biosensor developed with the other biosensors described in the literature was given in Table S2.†Specificity was also the crucial factor to evaluate the effectiveness and practicability of the proposed strategy. In our method, specific detection of miRNA-21 was achieved based on the design of DNA sequence. Thus, miRNA-21 and several kinds of sequences including miRNA-200c, miRNA-4640, miRNA-423, single-base mismatch-1, single-base mismatch-2, single-base mismatch-3, and negative control (NC) sequences were performed to POCT assay. Detail sequences were shown in Table S1.† As shown in Fig. 3B, only in the presence of the target miRNA-21, significant PGM signal was detected, the responses of other miRNAs were closed to the blank signal, indicating that the interference was almost negligible. The MBs–DNA–Inv platform could effectively differentiate the fully matched target from variants of single-base mismatch. Thus, the designed method obtained high specificity for miRNA detection.
3.4 Detection of miRNA-21 in urine samples
After the determination of critical parameters, we studied the detection of miRNA-21 in urine sample, which was reported to be important in the disease diagnosis.3 Firstly, different amounts of miRNA-21 were mixed with the urine samples and obtained a serious of concentration target (10 pM, 20 pM, 50 pM, 100 pM, 150 pM, 200 pM). A positive linear response between the miRNA-21 concentration (10–200 pM) and PGM signals (S = 3.05 + 0.12c; R2 = 0.995, in which S is the value of PGM readings and c is the miRNA-21 concentration) was shown in Fig. S4,† which indicated that this method under current conditions was suitable for miRNA-21 detection.Next, the detection of miRNA-21 in real diluted mice urine sample was researched using standard addition according the previous reports.14 Different concentrations of miRNA-21 were added into the urine samples (10 pM, 100 pM, 200 pM) from three different healthy mice. The results revealed that the theory of the mass of miRNA-21 was with good agreement between values and observations. As shown in Table 1, the recovery rate varied from 94.7 to 107.0%, providing the strategy to be a possible vista for the detection of miRNAs in real biological samples.
| Sample | Added/pM | Detected/pM | Recovery/% | RSD/% |
|---|---|---|---|---|
| 1 | 10.0 | 10.7 | 107.0 | 4.4 |
| 2 | 100.0 | 106.6 | 106.6 | 2.6 |
| 3 | 200.0 | 189.4 | 94.7 | 3.1 |
Finally, we tested miRNA-21 expression level in 5 urine samples from mice. We compared the results with current benchmark for miRNA detection in the laboratory, that is, qRT-PCR (Fig. S5†). Total miRNA was extracted from each urine sample. Then by using MBs–DNA–Inv and PGM, we acquired the concentration of miRNA-21 under optimal conditions. As demonstrated in Fig. 4, the value obtained by our designed strategy was in consonance with those by qRT-PCR, which clearly demonstrated the potential of our proposed assay for application in real bioanalysis of kidney injury samples.
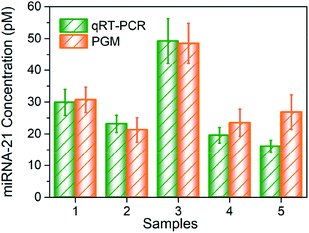 | ||
| Fig. 4 Quantification of the expression of miRNA-21 in five urine samples by our method and qRT-PCR. Data are means ± SD (n = 3). | ||
4 Conclusions
In summary, we designed a dual signal amplification strategy for miRNA-21 detection. This method provided a low detection limit of 1.8 pM for miRNA-21 owning to the amplification of DSN and invertase, the enrichment effect of streptavidin-MBs and the million turnovers of sucrose hydrolysis into glucose. Besides, because the strand was designed for a specific target, the method exhibited excellent sequence selectivity, the single-base mutation was easily discriminated. This approach was universal for miRNA quantitative detection because the designed strategy could be applied for the detection of other free nucleic acids (for example, circulating DNA, mRNA and other microRNAs). The assay method was convenient and practicable for detection at home because PGM could be steered by the demand for intricate apparatus and sophisticated operations. It is of great significance that the application of this method will provide a convenient and dependable, low-cost strategy for the detection of miRNA-21, and it meets the latest analytical demands for miRNA determination that is suitable for the public.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This research was supported by National Natural Science Foundation of China (21775166), Natural Science Foundation for Distinguished Young Scholars of Jiangsu Province (BK20180026), and “Double First-Class” University Project (CPU2018GF06, CPU2018GY32).Notes and references
- (a) V. Ambros, Nature, 2004, 431, 350–355 CrossRef CAS PubMed; (b) D. P. Bartel, Cell, 2004, 116, 281–297 CrossRef CAS PubMed.
- (a) J. Du, X. Q. Cao, L. Zou, Y. Chen, J. Guo, Z. J. Chen, S. S. Hu and Z. Zheng, PLoS One, 2013, 8, e63390 CrossRef CAS PubMed; (b) H. D. Yuan, D. Mischoulon, M. Fava and M. W. Otto, J. Affective Disord., 2018, 233, 68–78 CrossRef CAS PubMed.
- (a) J. Hayes, P. P. Peruzzi and S. Lawler, Trends Mol. Med., 2014, 20, 460–469 CrossRef CAS PubMed; (b) E. Tsitsiou and M. A. Lindsay, Curr. Opin. Pharmacol., 2009, 29, 343–351 Search PubMed; (c) A. Zarjou, S. Yang, E. Abraham, A. Agarwal and G. Liu, Am. J. Physiol. Ren. Physiol., 2011, 301, F793–F801 CrossRef CAS PubMed; (d) J. M. Lorenzen, H. Haller and T. Thum, Nat. Rev. Nephrol., 2017, 7, 286–294 CrossRef PubMed; (e) Y. F. Li, Y. Jing, J. L. Hao, N. C. Frankfort, X. S. Zhou, B. Shen, X. Y. Liu, L. H. Wang and R. S. Li, Protein Cell, 2013, 4, 813–819 CrossRef CAS PubMed; (f) J. Hayes, P. P. Peruzzi and S. Lawler, Trends Mol. Med., 2014, 20, 460–469 CrossRef CAS PubMed.
- (a) E. Koscianska, J. Staregaroslan, L. J. Sznajder, M. Olejniczak, P. Galkamarciniak and W. J. Krzyzosiak, BMC Mol. Biol., 2011, 2, 14 CrossRef PubMed; (b) S. Honda and Y. Kirino, Nucleic Acids Res., 2015, 43, e77 CrossRef PubMed; (c) N. Rosenfeld, R. Aharonov, E. Meiri, S. Rosenwald, Y. Spector, M. Zepeniuk, H. Benjamin, N. Shabes, S. Tabak, A. Levy, D. Lebanony, Y. Goren, E. Silberschein, N. Targan, A. Benari, S. Gilad, N. Sionvardy, A. Tobar, M. Feinmesser and O. Kharenko, Nat. Biotechnol., 2008, 26, 462–469 CrossRef CAS PubMed; (d) A. M. Kietrys, W. A. A. Velema and E. T. Kool, J. Am. Chem. Soc., 2017, 139, 17074–17081 CrossRef CAS PubMed.
- (a) E. Várallyay, J. Burgyán and Z. Havelda, Nat. Protoc., 2008, 3, 190–196 CrossRef PubMed; (b) K. M. Koo, L. G. Carrascosa, M. J. A. Shiddiky and M. Trau, Anal. Chem., 2016, 88, 2000–2005 CrossRef CAS PubMed; (c) H. H. Wang, H. Wang, X. Duan, Y. Sun, X. Wang and Z. Li, Chem. Sci., 2017, 8, 3635–3640 RSC.
- A. Heller and B. Feldman, Chem. Rev., 2008, 108, 2482–2505 CrossRef CAS PubMed.
- (a) Y. Xiang and Y. Lu, Nat. Chem., 2011, 3, 697–703 CrossRef CAS PubMed; (b) Y. Xiang and Y. Lu, Anal. Chem., 2012, 84, 4174–4178 CrossRef CAS PubMed; (c) L. Yan, Z. Zhu, Y. Zou, Y. S. Huang, D. W. Liu, S. S. Jia, D. M. Xu, M. Wu, Y. Zhou and S. Zhou, J. Am. Chem. Soc., 2013, 135, 3748–3751 CrossRef CAS PubMed.
- (a) J. Su, J. Xu, Y. Chen, Y. Xiang, R. Yuan and Y. Q. Chai, Biosens. Bioelectron., 2013, 45, 219–222 CrossRef CAS PubMed; (b) X. T. Xu, K. Y. Liang and J. Y. Zeng, Angew. Chem., 2016, 128, 742–746 CrossRef; (c) J. Fang, G. Guo, Y. J. Yang, W. Yu, Y. Y. Tao, T. Dai, C. J. Yuan and G. M. Xie, Sens. Actuators, B, 2018, 272, 118–126 CrossRef CAS.
- Y. Gu, T. T. Zhang, Z. F. Huang, S. W. Hu, W. Zhao, J. J. Xu and H. Y. Chen, Chem. Sci., 2018, 9, 3517–3522 RSC.
- (a) X. P. Qiu, H. Zhang, H. L. Yu, T. L. Jiang and L. Yang, Trends Biotechnol., 2015, 33, 180–188 CrossRef CAS PubMed; (b) V. E. Anisimova, D. V. Rebrikov, D. A. Shagin, D. V. Rebrikov, A. Dmitry, D. A. Shagin, V. B. Kozhemyako, N. I. Menzorova, D. B. Staroverov, R. Ziganshin, L. L. Vagner, V. A. Rasskazov and S. A. Lukyanov, BMC Biochem., 2008, 9, 1–12 CrossRef PubMed.
- R. L. Xu, Particuology, 2008, 2, 112–115 CrossRef.
- W. N. Kuo, R. N. Kanadia and V. P. Shanbhag, Biochem. Mol. Biol. Int., 1999, 47, 1061–1067 CAS.
- (a) W. P. Peng, Q. Zhao, J. F. Piao, M. Zhao, Y. W. Huang, B. Zhang, W. C. Gao, D. M. Zhou, G. M. Shu, X. Q. Gong and J. Chang, Sens. Actuators, B, 2018, 263, 289–297 CrossRef CAS; (b) B. Tian, J. Ma, Q. Zhen, T. Zardán Gómez de la Torre, M. Donolato, M. F. Hansen, P. Svedlindh and M. Strömberg, ACS Nano, 2017, 11, 1798–1806 CrossRef CAS PubMed; (c) Q. Zhao, J. F. Piao, W. P. Peng, Y. Wang, B. Zhang, X. Q. Gong and J. Chang, ACS Appl. Mater. Interfaces, 2018, 10, 3324–3332 CrossRef CAS PubMed.
- J. Lei, L. Shi, B. Li, C. J. Yang and Y. Jin, Biosens. Bioelectron., 2018, 122, 32–36 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra10657j |
| This journal is © The Royal Society of Chemistry 2020 |