Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Structure characterization and anticoagulant activity of a novel polysaccharide from Leonurus artemisia (Laur.) S. Y. Hu F
Cheng Hu ,
Hao-Xuan Li,
Meng-Ting Zhang and
Li-Fang Liu*
,
Hao-Xuan Li,
Meng-Ting Zhang and
Li-Fang Liu*
State Key Laboratory of Natural Medicines, Department of Chinese Medicines Analysis, School of Traditional Chinese Pharmacy, China Pharmaceutical University, No. 24 Tongjia Lane, Nanjing 21198, China. E-mail: liulifan69@126.com; Fax: +86 25 8618 5136; Tel: +86 25 8618 5136
First published on 10th January 2020
Abstract
An acidic polysaccharide, named LAP-1, was extracted and isolated from Leonurus artemisia (Laur.), and was further purified with ion exchange chromatography and gel chromatography. The extraction conditions of the crude polysaccharides were optimized by single-factor experiments and response surface methodology. The primary structure of the purified polysaccharide was measured by FT-IR, GC-MS, and NMR. The results showed that LAP-1 was mainly composed of galacturonic acid (GalA), mannose (Man), xylose (Xyl), rhamnose (Rha), arabinose (Ara), glucose (Glc), galactose (Gal), fucose (Fuc), ribose (Rib), and glucuronic acid (GlcA) in the molar ratio of 8.74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.45
3.45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.02
1.02![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.11
2.11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5.60
5.60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.73
4.73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.08
1.08![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.09
1.09![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.47. Primary structure analysis results indicated that LAP-1 contained characteristic glycosyl linkages such as →1)-α-D-Manp, →1)-α-D-Glcp, →1)-α-D-Arap-(2→, →1)-β-D-Galp-(3→, →1)-β-D-Manp-(4→, →1)-β-D-Galp-(4→, →1)-β-D-Glcp-(4→, →1)-β-D-GalAp-(4→, →1)-β-D-GlcAp-(4→, →1)-β-D-Manp-(4,6→, →1)-β-D-Manp-(3,4→. The Mw/Mn (PDI), Mn, Mz and Mw of LAP-1 were determined to be 1.423, 6.979 × 103 g mol−1, 1.409 × 104 g mol−1, and 9.930 × 103 g mol−1 by HPSEC-MALLS-RID and DLS. SEM, TEM and AFM results indicated that LAP-1 was a highly branched structure. LAP-1 showed mild anticoagulant activity, low toxicity, and less spontaneous bleeding compared with heparin sodium. These results demonstrated the effective coagulation activity of Leonurus artemisia polysaccharides. Thus, the purified LAP-1 could be explored as a promising anticoagulant agent for the treatment of coagulation disorders.
1.47. Primary structure analysis results indicated that LAP-1 contained characteristic glycosyl linkages such as →1)-α-D-Manp, →1)-α-D-Glcp, →1)-α-D-Arap-(2→, →1)-β-D-Galp-(3→, →1)-β-D-Manp-(4→, →1)-β-D-Galp-(4→, →1)-β-D-Glcp-(4→, →1)-β-D-GalAp-(4→, →1)-β-D-GlcAp-(4→, →1)-β-D-Manp-(4,6→, →1)-β-D-Manp-(3,4→. The Mw/Mn (PDI), Mn, Mz and Mw of LAP-1 were determined to be 1.423, 6.979 × 103 g mol−1, 1.409 × 104 g mol−1, and 9.930 × 103 g mol−1 by HPSEC-MALLS-RID and DLS. SEM, TEM and AFM results indicated that LAP-1 was a highly branched structure. LAP-1 showed mild anticoagulant activity, low toxicity, and less spontaneous bleeding compared with heparin sodium. These results demonstrated the effective coagulation activity of Leonurus artemisia polysaccharides. Thus, the purified LAP-1 could be explored as a promising anticoagulant agent for the treatment of coagulation disorders.
1. Introduction
Leonuri herba, also called motherwort herb in China, is a famous traditional Chinese medicine (TCM) obtained from the fresh or dried aerial part of Leonurus artemisia (Laur.) S. Y. Hu F. As recorded in the Chinese Pharmacopoeia (2015 edition), it has been a commonly used herb for treating irregular menstruation, dysmenorrhea, amenorrhea, lochia, edema of acute nephritis and blood system diseases.1 Recent studies have also demonstrated its outstanding functions in promoting blood circulation and regulating menstruation, diuresis and detumescence.2Leonurus artemisia contains many chemical constituents with a variety of pharmacological effects, such as alkaloids for antioxygenation, diterpenoids for anti-platelet agglutinating, flavonoids for uterine excitation and volatile oil for bacteriostasis.3 Most of the previous literature has attributed its anticoagulant activity to its small molecule components such as leonurine, genkwanin, and aethylparabenum.
To date, there has been no report about the relationship between its anticoagulant activity and polysaccharides in Leonurus artemisia. Nevertheless, the anticoagulant function of herbal polysaccharides should not be ignored, especially since most of the archaic TCM decoctions were prepared via water boiling. More recently, several herbal polysaccharides have been demonstrated to have significant anticoagulant activity.4 However, as one of the abundant components in Leonurus artemisia, the anticoagulant effect of Leonurus artemisia polysaccharides has seldom been reported. As is well known, the structural properties of polysaccharides such as ratios of constituent monosaccharides,5 molecular size, types and chain conformations, and features of glycosidic linkages are closely correlated to their bioactivities.6,7
Therefore, this work aimed to extract, purify and investigate the fundamental structure information, such as the chemical properties of polysaccharides from Leonurus artemisia, in addition to examining its anticoagulant activity.
2. Materials and methods
2.1 Materials and reagents
Leonurus artemisia samples were obtained from Jiangxi Xinzheng Pharmaceutical Co., Ltd., Pingxiang, China. The sephacryl S-200 and diethylaminoethylcellulose-52 (DEAE-52) were obtained from Whatman International, Ltd. (Kent, UK). Ethanol, sulfuric acid, hydrochloric acid, phenol, ethyl ether and other reagents were purchased from the Nanjing Chemical Reagent Co., Ltd. (Nanjing, China). All of the chemicals reagents and chemicals were of analytical grade.2.2 Ultrasonic-assisted extraction of Leonurus artemisia polysaccharide
The dried aerial part of the Leonurus artemisia samples were ground and filtered through an 80 mesh. Ethanol was used for defatting and decoloring at 60 °C for 3 h in a Soxhlet extractor system and then centrifuged at 8000 rpm for 15 min. The precipitate was dried through vacuum dehydration. Then, the ultrasonic-assisted extraction of the polysaccharides was performed in an ultrasonic cleaner at 40 kHz (KH-250DV, Kunshan Hachuang Ultrasonic Instrument Co., Ltd., Kunshan, China).| Extraction yield (%) = (S × W1/W0) × 100 | (1) |
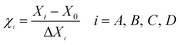 | (2) |
The experimental runs and the values of the four independent variables for BBD are showed in Table 1. The interrelationships and relationships between the independent variables and the response are indicated in the second-order polynomial model (eqn (3)) for predicting the optimized conditions.
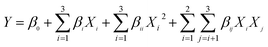 | (3) |
| Run | Coded variable levels | Yield of LAP (%) | |||
|---|---|---|---|---|---|
| Extraction time (min) | Extraction power (W) | Extraction temperature (°C) | Ratio of water to material (mL g−1) | ||
| Factor 1: A | Factor 2: B | Factor 3: C | Factor 4: D | ||
| 1 | −1 | 0 | −1 | 0 | 2.16 ± 0.19 |
| 2 | −1 | 0 | 0 | 1 | 2.30 ± 0.55 |
| 3 | 1 | 0 | 0 | −1 | 2.06 ± 0.22 |
| 4 | 0 | 0 | 1 | −1 | 2.15 ± 0.36 |
| 5 | 0 | 0 | 0 | 0 | 3.35 ± 0.11 |
| 6 | 0 | 1 | −1 | 0 | 2.75 ± 0.24 |
| 7 | 1 | −1 | 0 | 0 | 2.50 ± 0.15 |
| 8 | −1 | 1 | 0 | 0 | 2.79 ± 0.42 |
| 9 | 0 | −1 | −1 | 0 | 2.72 ± 0.07 |
| 10 | 0 | −1 | 0 | −1 | 2.05 ± 0.19 |
| 11 | 0 | 0 | 0 | 0 | 3.29 ± 0.19 |
| 12 | −1 | −1 | 0 | 0 | 2.26 ± 0.38 |
| 13 | 1 | 1 | 0 | 0 | 2.67 ± 0.22 |
| 14 | 0 | 1 | 1 | 0 | 3.12 ± 0.23 |
| 15 | 0 | −1 | 1 | 0 | 2.85 ± 0.11 |
| 16 | −1 | 0 | 0 | −1 | 1.72 ± 0.46 |
| 17 | 0 | 1 | 0 | −1 | 2.29 ± 0.06 |
| 18 | 0 | −1 | 0 | 1 | 2.86 ± 0.37 |
| 19 | 0 | 0 | 1 | 1 | 2.73 ± 0.09 |
| 20 | 0 | 0 | 0 | 0 | 3.23 ± 0.09 |
| 21 | −1 | 0 | 1 | 0 | 2.66 ± 0.22 |
| 22 | 0 | 0 | 0 | 0 | 3.34 ± 0.34 |
| 23 | 1 | 0 | 0 | 1 | 2.27 ± 0.17 |
| 24 | 1 | 0 | 1 | 0 | 2.56 ± 0.09 |
| 25 | 0 | 0 | −1 | −1 | 2.10 ± 0.58 |
| 26 | 1 | 0 | −1 | 0 | 2.49 ± 0.09 |
| 27 | 0 | 1 | 0 | 1 | 2.96 ± 0.21 |
| 28 | 0 | 0 | −1 | 1 | 2.76 ± 0.25 |
| 29 | 0 | 0 | 0 | 0 | 3.30 ± 0.29 |
The related experimental data were calculated and analyzed by Design-Expert software (version 11.0, Stat-Ease Inc., Minneapolis, USA). The validity of the statistical experimental design was verified by confirmation experiments under the optimized conditions. The LAP samples obtained from the optimized conditions were used for further purification steps.
2.3 Purification of polysaccharide from crude Leonurus artemisia polysaccharides
Leonurus artemisia polysaccharides (LAP) was first deproteinated and depigmented by macroporous adsorptive resin D101. Then, further deproteinization was performed with the Sevag method 5 times, followed by dialysis (cut-off Mw 3500 Da) against distilled water for 72 h. The polysaccharides components of the dialysates were precipitated by ethanol. The precipitate was sequentially washed with ethanol, acetone and ether, followed by freeze-drying. Ion-exchange chromatography (DEAE-32) with a gradient of 0–2.0 mol L−1 NaCl and a Sephacryl S-200 column were then used to purify the obtained sample. During the purification process, an acid polysaccharide showed a high concentration and purity compared with other fractions. The resultant fraction named LAP-1 was concentrated, collected, and freeze-dried for further research.2.4 Characterization of LAP-1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 16 was used as the mobile phase. The column temperature was set at 30 °C, and the flow rate was set at 1 mL min−1. The ratio was calculated by each monosaccharides standard by the above methods.
16 was used as the mobile phase. The column temperature was set at 30 °C, and the flow rate was set at 1 mL min−1. The ratio was calculated by each monosaccharides standard by the above methods.Then, formic acid (88%) and trifluoroacetic acid were used to hydrolyze the methylated samples for 6 h and 8 h at 100 °C, separately. After being reduced by NaBH4 and acetylated, the products were determined on a gas chromatography-mass spectrometer (6890N-5975B, Agilent, USA) that was equipped with an HP-5MS quartz capillary column (30 m × 0.25 mm × 0.25 μm). Temperature rising program: 1 °C min−1, 160–180 °C, 3 °C min−1 to 220 °C, held for 10 min. The mass charge ratio range was set at 25–500.
2.5 Additional structural analysis of LAP-1
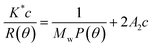 | (4) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
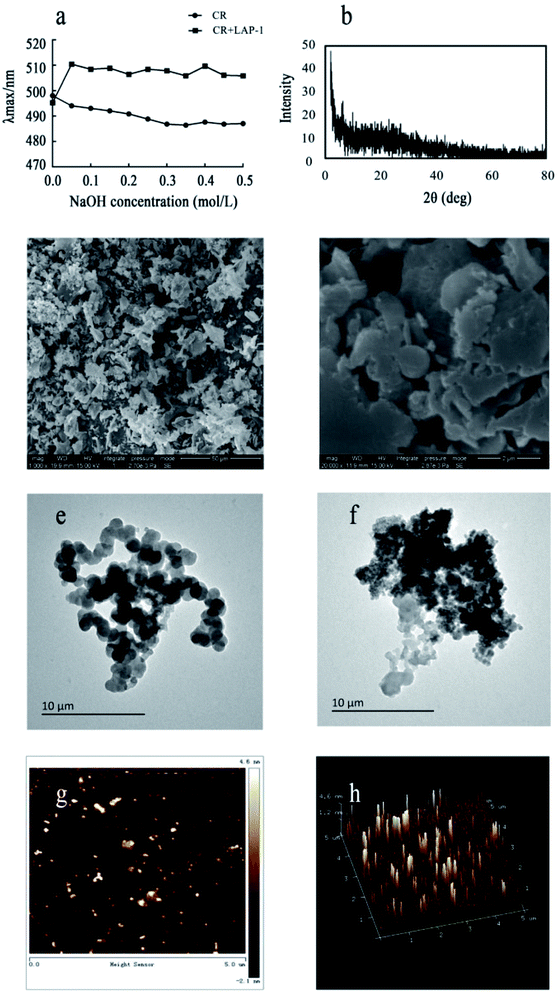
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The control group included the same concentration of Congo red and a sodium hydroxide solution without LAP-1. After reaction for 10 min at room temperature, the absorbance was determined from 400 to 600 nm. A diffractometer Bruker AXS D8 (Bruker, Germany) was used to determine the crystalline characteristics of LAP-1. The scattering angles (2θ) were set at 5–80° with Ni-filtered Cu Kα radiation (λ = 1.5406 Å), and the current and voltage were set at 40 mA and 40 kV, respectively.
1. The control group included the same concentration of Congo red and a sodium hydroxide solution without LAP-1. After reaction for 10 min at room temperature, the absorbance was determined from 400 to 600 nm. A diffractometer Bruker AXS D8 (Bruker, Germany) was used to determine the crystalline characteristics of LAP-1. The scattering angles (2θ) were set at 5–80° with Ni-filtered Cu Kα radiation (λ = 1.5406 Å), and the current and voltage were set at 40 mA and 40 kV, respectively.2.6 Anticoagulant activity in vitro
Prothrombin time (PT), activated partial thromboplastin time (aPTT), and thrombin time (TT) of LAP-1 were evaluated by the method published with some modifications.14 Briefly, 20 μL sample solutions of different concentrations were mixed with 80 μL of citrated healthy human plasma (0.109 mol L−1 sodium citrate, ratio: 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). Then, 100 μL pretreated PT reagent, aPTT reagent and TT reagent were added to the mixture and incubated at 37 °C for 180 s. Clotting time was determined by an automatic coagulation analysis system (CS-5100, Sysmex, Japan). Normal saline (0.90%) and heparin sodium (0.002 mg mL−1) were used as the negative control and positive drug group, respectively.
1, v/v). Then, 100 μL pretreated PT reagent, aPTT reagent and TT reagent were added to the mixture and incubated at 37 °C for 180 s. Clotting time was determined by an automatic coagulation analysis system (CS-5100, Sysmex, Japan). Normal saline (0.90%) and heparin sodium (0.002 mg mL−1) were used as the negative control and positive drug group, respectively.
2.7 Statistical analysis
All data are shown as the mean ± standard deviation (SD) of three determinations. The statistical analysis was calculated by using SPSS 22.0. P < 0.05 is defined as significant and P < 0.01 is defined as highly significant.3. Results and discussion
3.1 Optimization of extraction conditions
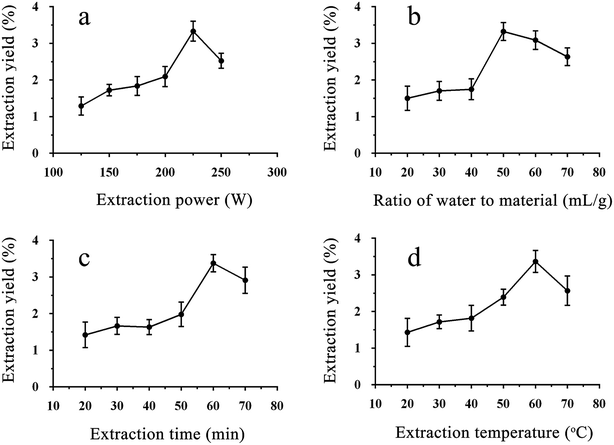
An appropriate temperature and time were beneficial for the yield of crude polysaccharides, while too much time and too high of a temperature had the opposite effect, as shown in Fig. 1c and d and the previous literature.15 The yield reached its peak at 60 °C and 60 min, and lower temperatures and shorter times made it hard to facilitate cell wall fragmentation.15 However, the curve decreased after the peak, and the reason for this could be that a higher temperature and too much time can damage the structure of crude polysaccharides due to the ultrasonic power.16
According to the above results, extraction powers of 200–250 W, extraction times of 50–70 min, extraction temperatures of 50–70 °C, and ratios of water to material from 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 60
1 to 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were selected for the subsequent experiments.
1 were selected for the subsequent experiments.
| Y = 3.30 + 0.054A + 0.11B + 0.090C + 0.29D − 0.092AB − 0.11AC − 0.092AD + 0.060BC − 0.034BD − 0.020CD − 0.59A2 − 0.16B2 − 0.26C2 − 0.61D2 | (5) |
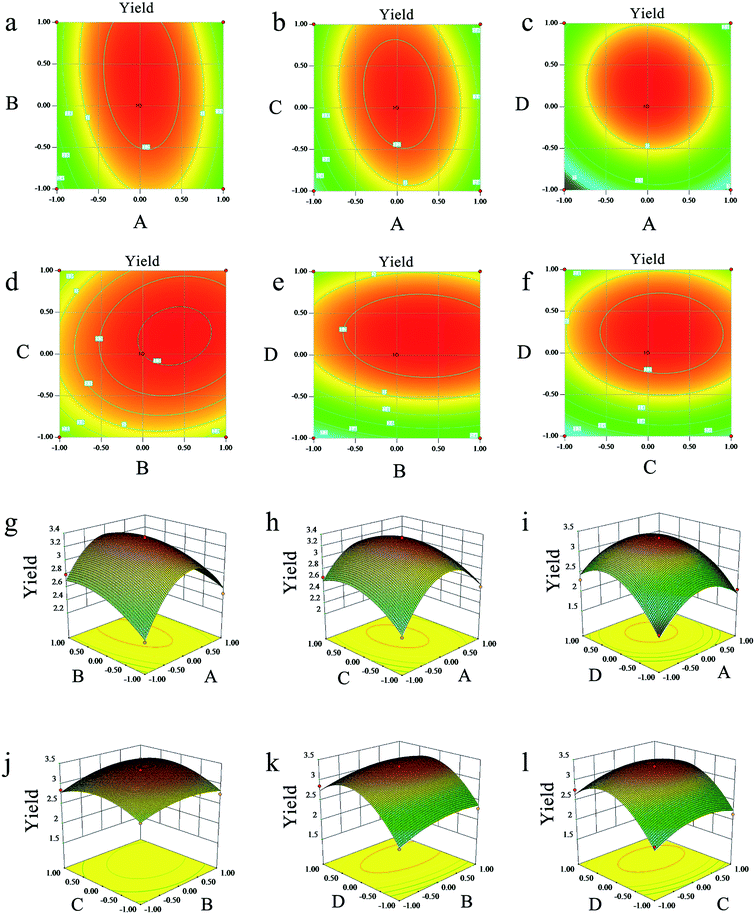
As shown in Table 2, the p-value, determinant coefficient, and coefficient of variation were <0.01, 0.9734 and 5.95. These results showed that the polynomial model equation has good reliability and precision, that it can explain the relationship between the response value and the parameters exactly and that the quadratic terms have significant effects. The interaction of the dependent variable and other factors were illustrated by 2-D contours and 3-D response surfaces as shown in Fig. 2. While the ultrasonic power and the ratio stayed at a zero level, the response value increased with higher temperatures and longer times at the beginning and then declined. Other variables and response values showed similar situations.
| Source | Coefficient estimate | Sum of squares | Degree of freedom | Standard error | Mean square | F-value | P value |
|---|---|---|---|---|---|---|---|
| a Means significance (significance level 0.01).b Means significance (not significant). | |||||||
| Model | 3.3 | 5.400 | 14 | 0.046 | 0.390 | 36.590 | <0.0001a |
| A | 0.054 | 0.035 | 1 | 0.030 | 0.035 | 3.280 | 0.092 |
| B | 0.11 | 0.150 | 1 | 0.030 | 0.150 | 14.270 | 0.002a |
| C | 0.09 | 0.096 | 1 | 0.030 | 0.096 | 9.130 | 0.009a |
| D | 0.29 | 1.020 | 1 | 0.030 | 1.020 | 97.060 | <0.0001a |
| AB | −0.092 | 0.034 | 1 | 0.051 | 0.034 | 3.230 | 0.094 |
| AC | −0.11 | 0.047 | 1 | 0.051 | 0.047 | 4.490 | 0.053 |
| AD | −0.092 | 0.034 | 1 | 0.051 | 0.034 | 3.240 | 0.093 |
| BC | 0.06 | 0.014 | 1 | 0.051 | 0.014 | 1.370 | 0.262 |
| BD | −0.034 | 0.005 | 1 | 0.051 | 0.005 | 0.430 | 0.522 |
| CD | −0.02 | 0.002 | 1 | 0.051 | 0.002 | 0.150 | 0.703 |
| A2 | −0.59 | 2.240 | 1 | 0.040 | 2.240 | 212.130 | <0.0001a |
| B2 | −0.16 | 0.170 | 1 | 0.040 | 0.170 | 16.450 | 0.001a |
| C2 | −0.26 | 0.440 | 1 | 0.040 | 0.440 | 41.550 | <0.0001a |
| D2 | −0.61 | 2.420 | 1 | 0.040 | 2.420 | 229.730 | <0.0001a |
| Residual | 0.150 | 14 | 0.011 | ||||
| Lack of fit | 0.140 | 10 | 0.014 | 5.950 | 0.052b | ||
| Pure error | 0.009 | 4 | 0.002 | ||||
| Cor total | 5.550 | 28 | |||||
| R2 | 0.9734 | ||||||
| Adj R2 | 0.9486 | ||||||
| Pred R2 | 0.8538 | ||||||
| Adeq precision | 22.160 | ||||||
| CV% | 3.91 | ||||||
The validation of this model was also proven by the predicted optimal experiments. The extraction conditions experiments showed that reaction temperature, reaction time, ultrasonic power and ratio of water to materials after response surface optimization were 62 °C, 60 min, 230 W, and 3.15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. The extraction yield of LAP was 3.45 ± 0.27% under the predicted optimal conditions, which is consistent with the predicted optimal yield based on this model.
1, respectively. The extraction yield of LAP was 3.45 ± 0.27% under the predicted optimal conditions, which is consistent with the predicted optimal yield based on this model.
3.2 Characteristics of LAP-1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.45
3.45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.02
1.02![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.11
2.11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5.60
5.60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.73
4.73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.08
1.08![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.09
1.09![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
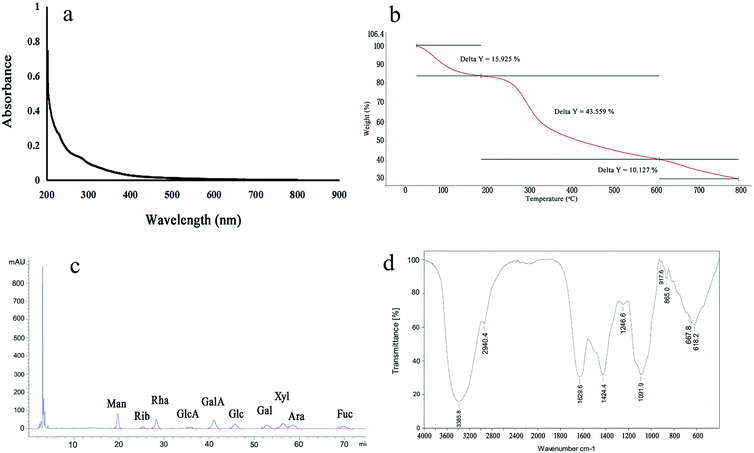
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.47. According to the results, LAP-1 showed a varied monosaccharide composition. The galacturonic acid, glucose, galactose were the major monosaccharides in LAP-1 which revealed that there were a large number of glycosidic linkages of galacturonic acid, glucose, galactose exist in LAP-1. Accurate glycosidic linkages identification was carried on follow-up experiments.
1.47. According to the results, LAP-1 showed a varied monosaccharide composition. The galacturonic acid, glucose, galactose were the major monosaccharides in LAP-1 which revealed that there were a large number of glycosidic linkages of galacturonic acid, glucose, galactose exist in LAP-1. Accurate glycosidic linkages identification was carried on follow-up experiments.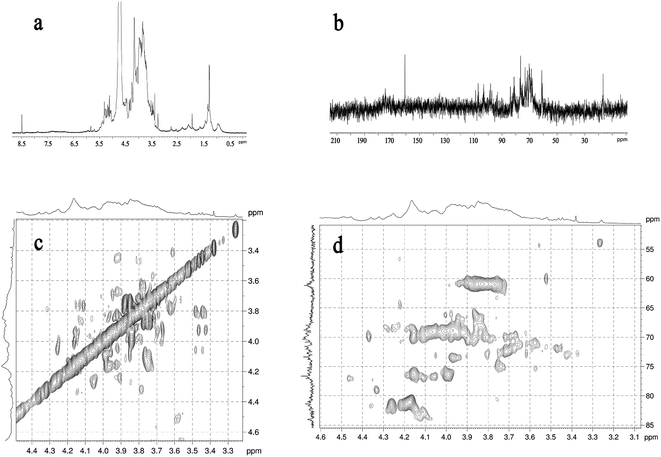 | ||
| Fig. 4 NMR spectrum of LAP-1: (a) 1H NMR spectrum; (b) 13C NMR spectrum; (c) 1H–1H COSY spectrum; (d) HSQC spectrum. | ||
| Glycosyl residues | δ13C/1H (ppm) | |||||
|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | C5 | C6 | |
| H1 | H2 | H3 | H4 | H5 | H6 | |
| →1)-α-D-Manp | 93.93 | 71.59 | 70.84 | 67.99 | 72.79 | 61.04 |
| 5.32 | 3.94 | 3.85 | 3.68 | 3.81 | 3.74/3.86 | |
| →1)-α-D-Glcp | 93.47 | 72.04 | 73.78 | 70.40 | 72.02 | 61.46 |
| 5.29 | 3.53 | 3.71 | 3.42 | 3.83 | 3.76/3.80 | |
| →1)-α-D-Arap-(2→ | 98.48 | 75.23 | 76.71 | 70.07 | 65.99 | |
| 5.22 | 3.26 | 3.43 | 3.63 | 3.33 | ||
| →1)-β-D-Galp-(3→ | 103.26 | 70.37 | 84.01 | 70.00 | 75.69 | 61.35 |
| 4.49 | 3.75 | 3.77 | 4.10 | 3.69 | 3.68/3.74 | |
| →1)-β-D-Manp-(4→ | 103.52 | 73.39 | 75.23 | 80.33 | 75.69 | 61.04 |
| 4.58 | 3.42 | 3.66 | 3.63 | 3.59 | 3.83/3.96 | |
| →1)-β-D-Galp-(4→ | 107.54 | 70.37 | 73.80 | 81.02 | 75.69 | 61.35 |
| 4.49 | 3.75 | 3.77 | 4.16 | 3.69 | 3.68/3.74 | |
| →1)-β-D-Glcp-(4→ | 109.29 | 72.04 | 73.78 | 81.36 | 72.02 | 61.46 |
| 5.09 | 3.44 | 3.67 | 3.60 | 3.59 | 3.83/3.96 | |
| →1)-β-D-GalAp-(4→ | 107.54 | 70.37 | 73.80 | 81.02 | 75.69 | 160.77 |
| 4.49 | 3.75 | 3.77 | 4.16 | 3.69 | 8.48 | |
| →1)-β-D-GlcAp-(4→ | 109.29 | 72.04 | 73.78 | 81.36 | 72.02 | 160.77 |
| 5.06 | 3.44 | 3.67 | 3.60 | 3.59 | 8.48 | |
| →1)-β-D-Manp-(4,6→ | 109.36 | 71.59 | 76.85 | 84.11 | 72.79 | 68.81 |
| 5.15 | 3.46 | 3.88 | 4.45 | 3.52 | 4.06/3.65 | |
| →1)-β-D-Manp-(3,4→ | 109.37 | 70.37 | 81.45 | 81.78 | 72.79 | 60.88 |
| 5.12 | 3.48 | 4.05 | 4.35 | 3.50 | 3.58/3.45 | |
3.3 Additional structural analysis of LAP-1
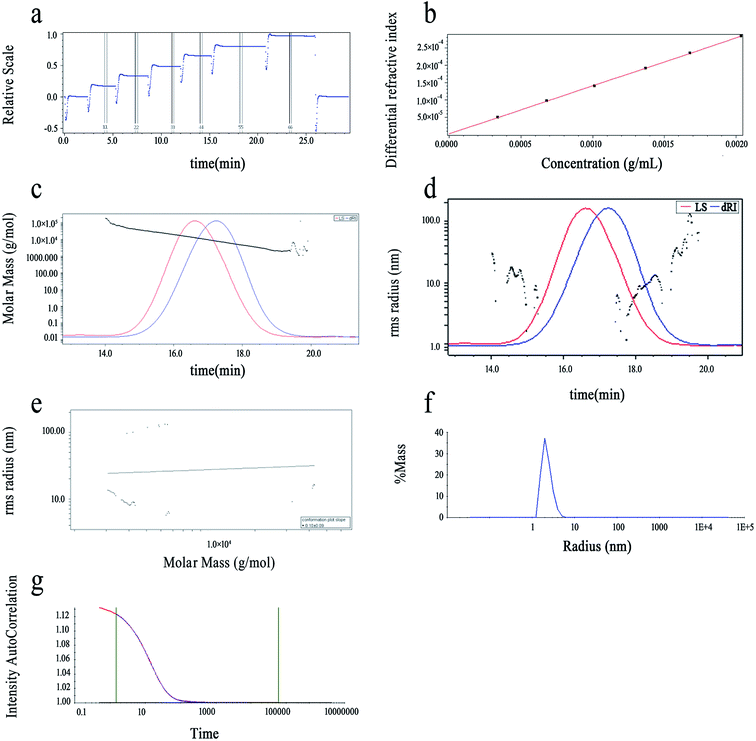
The purity of LAP-1 was verified by a relatively sharp symmetric peak as shown in Fig. 5c. The Mw/Mn (PDI), Mn, Mz and Mw of LAP-1 were measured to be 1.423, 6.979 × 103 g mol−1, 1.409 × 104 g mol−1, and 9.930 × 103 g mol−1. As Fig. 5d indicates, the 〈s2〉 z1/2 (Rg) of LAP-1 is 7.1 (±1.2%) nm. The instability of the RMS radius curves are caused by the compact branched molecules of LAP-1 and chain entanglement between large molecules and small molecules affecting the co-eluting time.
The molecular conformation and molecular size of polymers can be determined by index ν and the root-mean-square (RMS) radius 〈s2〉 z1/2 (Rg).26 Branching, sphere, flexible random coil conformation, and rigid rod correspond to the ν values of 0.1–0.4, 0.3, 0.5–0.6, 0.6–1.0, respectively.27 As calculated from Fig. 5e, the ν of LAP-1 was 0.10, κ was 2.83, and the df was 10, indicating that a high branch structure existed in LAP-1.
The chain conformation and structure of the LAP-1 molecule could be measured by its ρ value.28 The ρ value was calculated through the equation: ρ = Rg/Rh and Rh, Rg determined by DLS and SLS, respectively. The ρ values of ≥2, 1.5–1.8 and 0.775 represent extended chains and flexible random coils and spheres, respectively.25 As shown in Fig. 5f and g, the ρ value, Rh and Rg of LAP-1 were 3.09, 2.3 nm, and 7.1 nm. Thus, LAP-1 contains a high branch structure according to the ρ value and the results were consistent with the LLS.
X-ray diffraction (XRD) is a rapid and highly effective method to obtain information about the composition of materials and the structure or morphology of atoms or molecules in materials. The crystal structure causes the incident X-ray beam to diffract in many specific directions. By measuring the angle and intensity of these diffracted beams, crystallographers can generate three-dimensional images of the electron density in crystals. Based on this electron density, the average position of atoms in the crystal, their chemical bonds and various other information can be determined. As Fig. 6b shows, there were no diffraction peaks in the spectra, indicating that LAP-1 is not amorphous in nature. Multifarious spatial structures of pectins with various biological activities exist and a crystalline structure or a triple helical structure are the only two types.
AFM can detect the physical properties of various materials and samples in the nanoregion, including morphology, in the atmosphere and liquid environment, or directly manipulate the nanoregion. It has been widely used for semiconductors, nanofunctional materials, biology, the chemical industry, food, pharmaceutical research and research into various nano-related disciplines and has become a basic tool for nanoscientific research.32 Herein, as shown in Fig. 6g and h, the majority of LAP-1 has a nebula-like structure with a particles aggregation form and scattered on a plane. The height, width and length of the surface typology of LAP-1 were 0.2–4.6 nm, 100–300 nm, and 0.1–2 μm, respectively while the normal size of polysaccharide chains is approximately 1.0 nm. The chain size of LAP-1 indicated that a particular aggregation occurred in LAP-1.33,34 Aggregation of polysaccharides occurs from their side branches entangling each other, and the substitution of the hydroxyl group of polysaccharides can could twisting and converting of the sugar ring conformation, which leads to a decrease of the size of the resulting polysaccharides. The aggregation of polysaccharides could related to some biological activities.
3.4 Anticoagulant activity in vitro
Different stages of the coagulation process can be demonstrated by APTT, TT and PT assays in vitro.35 The coagulation process can be divided into an endogenous coagulation pathway, an exogenous coagulation pathway and a common coagulation pathway.36 APTT assays use brain lipids and activators instead of platelets to detect VIII, IX, XI and excitatory releasing enzymes in the endogenous coagulation pathway to reflect the effects of endogenous factors on coagulation time.37 PT assays are conducted by adding thromboplastin to plasma to reflect the effect of exogenous factors on coagulation time. TT is a simple screening test for fibrin polymerization. It measures the formation time of fibrin from fibrinogen after adding a certain amount of thrombin to plasma.38 The results of the tests of anticoagulant activity of LAP and LAP-1 are summarized in Table 4. Both LAP and LAP-1 showed concentration-dependent anticoagulant activity under the range of experimental concentrations. LAP-1 showed the highest anticoagulant activity at 4 mg mL−1, which had a significant difference from the control groups (P < 0.01 or P < 0.05), while LAP at 4 mg mL−1 demonstrated a smaller difference than saline. A possible explanation for this could be related to the content of sugar and the purity of the polysaccharides. Furthermore, compared with the control group, the APTT, PT and TT increased 1.24-fold, 1.04-fold and 1.14-fold with the addition of LAP-1 at 4 mg mL−1, and the blood clotting time was prolonged to 33.85 s. Based on the above results, the anticoagulant activity of LAP-1 was mediated through inhibiting thrombin activity, preventing the conversion of fibrinogen to fibrin, and inhibiting the intrinsic/common pathways.39,40 Compared with the traditional anticoagulant drug heparin sodium, LAP-1 could inhibit blood coagulation through the exogenous and endogenous coagulation pathways with mild efficacy, low toxicity, less spontaneous bleeding, and no other adverse reactions.| Sample | Concentration (mg mL−1) | APTT(s) | PT(s) | TT(s) |
|---|---|---|---|---|
| a Data in the table were expressed as mean ± SD (n = 3 in each group). *Significantly inhibited compared with the saline group (P < 0.05). **Significantly inhibited compared with the saline group (P < 0.01). | ||||
| Saline | 0.90% | 27.21 ± 1.52 | 14.26 ± 0.28 | 11.20 ± 0.20 |
| Heparin | 0.002 | 33.32 ± 1.21** | 14.87 ± 0.24* | 12.77 ± 0.19** |
| LAP-1 | 0.125 | 27.51 ± 1.44 | 14.46 ± 0.47 | 11.74 ± 0.51 |
| 0.25 | 28.36 ± 1.47 | 14.48 ± 0.50 | 11.90 ± 0.41* | |
| 0.5 | 29.94 ± 0.63* | 14.55 ± 0.37 | 12.01 ± 0.22** | |
| 1 | 31.06 ± 1.00* | 14.61 ± 0.52 | 12.16 ± 0.21** | |
| 2 | 32.18 ± 1.87* | 14.65 ± 0.33 | 12.35 ± 0.46** | |
| 4 | 33.85 ± 1.48** | 14.84 ± 0.28* | 12.73 ± 0.55** | |
| LAP | 0.125 | 27.32 ± 1.50 | 14.01 ± 0.96 | 11.51 ± 0.29 |
| 0.25 | 27.74 ± 0.69 | 14.00 ± 0.02 | 11.52 ± 0.29 | |
| 0.5 | 27.82 ± 0.67 | 14.03 ± 0.73 | 11.56 ± 0.12 | |
| 1 | 28.10 ± 1.65 | 14.08 ± 0.76 | 11.56 ± 0.25 | |
| 2 | 28.83 ± 0.99 | 14.23 ± 0.40 | 11.53 ± 0.61 | |
| 4 | 29.38 ± 0.80* | 14.60 ± 0.45 | 11.80 ± 0.31* | |
In previous studies, the anticoagulant activity of polysaccharides was related to their acid sugar, sulfur structures and aggregation form.35,37,39 LAP-1 contained a higher ratio of galacturonic acid and a large number of →1)-β-D-GalAp-(4→ and →1)-β-D-GlcAp-(4→ compared with other polysaccharides and the spatial stereochemical structure of LAP-1 showed an aggregation form with branches caused by the van der Waals' forces and the hydrogen bonds which different with other plant carbohydrate polymers. The coagulation activity of LAP-1 could attributes to its low molecular weight, glycosidic linkages of galacturonic acid and aggregation form according to the results.
Anticoagulants can be used to prevent endovascular embolism or thrombosis, stroke or other thrombotic diseases by affecting certain coagulation factors during the process of coagulation.41 Healthy bodies have effective blood coagulation, anticoagulation and fibrinolysis systems, so blood does not coagulate or bleed from the blood vessels, and it always flows freely to complete its function.42 However, when the body is in a high coagulation state or when anticoagulation and fibrinolysis are weakened, thromboembolic diseases occur.43
Thrombosis often causes local blood coagulation in the vascular system, leading to health-related diseases including strokes and heart attacks.44 Risk factors for thrombosis include abnormal hyperlipidemia, hyperglycemia elevated blood pressure and cancer.45 These thrombotic diseases have developed into the main causes of death, so effective anticoagulant drugs are urgently needed. Based on the above data analysis, it can be concluded that LAP-1 has a certain anticoagulant effect in vitro and could be developed as an anticoagulant drug and applied to the treatment of coagulation-related diseases.
4. Conclusions
In this study, a novel pure bioactive polysaccharide was isolated and purified from Leonurus artemisia (LAP-1). The structural characteristics and bioactivities of LAP-1 was under a systematic exploration. Results from this study suggested that LAP-1 (Mw: 9.930 × 103 g mol−1) was mainly composed of galacturonic acid (GalA), mannose (Man), xylose (Xyl), rhamnose (Rha), arabinose (Ara), glucose (Glc), galactose (Gal), fucose (Fuc), ribose (Rib), and glucuronic acid (GlcA). According to IR spectrum, methylation analysis and NMR spectra, LAP-1 was composed of →1)-α-D-Manp, →1)-α-D-Glcp, →1)-α-D-Arap-(2→, →1)-β-D-Galp-(3→, →1)-β-D-Manp-(4→, →1)-β-D-Galp-(4→, →1)-β-D-Glcp-(4→, →1)-β-D-GalAp-(4→, →1)-β-D-GlcAp-(4→, →1)-β-D-Manp-(4,6→, →1)-β-D-Manp-(3,4→. Besides, LAP-1 had a high branch structure measured by MALLS, SEM, TEM, and AFM. In addition, LAP-1 showed an obvious anticoagulant activity with low toxicity compared with the traditional anticoagulant drug heparin sodium. These experimental results provide chemical and bioactive fundamental for the development of LAP-1 as a potential anticoagulant drug and are worth further research.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81673568). We also greatly appreciate the financial support from the priority academic program development of the Jiangsu Higher Education Institutions (PAPD).References
- X. Huang, S. Wang, L. Wang, H. Wang, X. Li and D. Cui, Theriogenology, 2018, 121, 67–71 CrossRef PubMed.
- R. H. Zhang, Z. K. Liu, D. S. Yang, X. J. Zhang, H. D. Sun and W. L. Xiao, Phytochemistry, 2018, 147, 167–183 CrossRef CAS PubMed.
- A. Pitschmann, C. Waschulin, C. Sykora, S. Purevsuren and S. Glasl, Planta Med., 2017, 83, 1233–1241 CrossRef CAS PubMed.
- W. Cai, L. Xie, Y. Chen and H. Zhang, J. Polym. Sci., Part B: Polym. Lett., 1970, 8, 281–288 CrossRef.
- Y. Gao, B. Peng, Y. Xu, J. N. Yang, L. Y. Song, S. X. Bi, Y. Chen, J. H. Zhu, Y. Wen and R. M. Yu, RSC Adv., 2019, 9, 6603–6612 RSC.
- S. S. Ferreira, C. P. Passos, P. Madureira, M. Vilanova and M. A. Coimbra, Carbohydr. Polym., 2015, 132, 378–396 CrossRef CAS PubMed.
- X. Liu, Z. Miao, L. Han, A. Zhou, C. Yong and L. Xin, RSC Adv., 2018, 8, 9243–9252 RSC.
- L. Wang, L. Cheng, F. Liu, T. Li, Z. Yu, Y. Xu and Y. Yang, Molecules, 2018, 23, 1207–1208 CrossRef PubMed.
- M. Dubois, K. A. Gilles, J. K. Hamilton, P. A. Rebers and F. Smith, Anal. Chem., 1956, 28, 350–356 CrossRef CAS.
- N. Blumenkrantz and G. Asboehansen, Anal. Biochem., 1973, 54, 484–489 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- X. Yang, X. Gao, F. Han, B. Xu, Y. Song and R. Tan, Biochimie, 2005, 87, 747–754 CrossRef CAS PubMed.
- S. Hakomori, J. Biochem., 1964, 55, 205–208 CAS.
- L. Liang, L. Ao, T. Ma, Y. Ni, X. Liao, X. Hu and Y. Song, Int. J. Biol. Macromol., 2018, 106, 447–455 CrossRef CAS.
- Z. Mzoughi, A. Abdelhamid, C. Rihouey, D. L. Cerf, A. Bouraoui and H. Majdoub, Carbohydr. Polym., 2018, 185, 127–137 CrossRef CAS PubMed.
- G. Chen, K. Chen, R. Zhang, X. Chen, P. Hu and J. Kan, Food Chem., 2018, 245, 1113–1123 CrossRef CAS PubMed.
- P. Seedevi, M. Moovendhan, S. Sudharsan, P. Sivasankar, S. Loganathan, V. Shanmugam and A. Shanmugam, Carbohydr. Polym., 2018, 195, 486–494 CrossRef CAS PubMed.
- S. Cárdenaspérez, J. J. Chanonapérez, N. Güemesvera, J. Cybulska, M. Szymanskachargot, M. Chylinska, A. Kozioł, D. Gawkowska, P. M. Pieczywek and A. Zdunek, Carbohydr. Polym., 2018, 196, 313–321 CrossRef PubMed.
- R. R. Klosterhoff, J. M. Bark, N. M. Glänzel, M. Iacomini, G. R. Martinez, S. M. B. Winnischofer and L. M. C. Cordeiro, Int. J. Biol. Macromol., 2017, 106, 473–480 CrossRef PubMed.
- H. Li, K. Cao, P. Cong, Y. Liu, H. Cui and C. Xue, Carbohydr. Polym., 2018, 190, 87–94 CrossRef CAS PubMed.
- F. Wu, C. Zhou, D. Zhou, S. Ou, X. Zhang and H. Huang, Food Funct., 2018, 9, 294–306 RSC.
- C. W. Cho, C. J. Han, Y. K. Rhee, Y. C. Lee, K. S. Shin and H. D. Hong, Molecules, 2014, 19, 5266–5277 CrossRef PubMed.
- K. Ghai, A. K. Gupta and P. K. Gupta, J. Biol. Act. Prod. Nat., 2012, 2, 250–255 Search PubMed.
- U. Adolphi and W. M. Kulicke, Polymer, 1997, 38, 1513–1519 CrossRef CAS.
- L. Shao, Z. Wu, F. Tian, H. Zhang, Z. Liu, W. Chen and B. Guo, Int. J. Biol. Macromol., 2015, 72, 1429–1434 CrossRef CAS PubMed.
- W. Liu, Y. Liu, R. Zhu, J. Yu, W. Lu, C. Pan, W. Yao and X. Gao, Carbohydr. Polym., 2016, 147, 114–124 CrossRef CAS PubMed.
- Y. Chang, Y. Hu, L. Yu, D. J. Mcclements, X. Xu, G. Liu and C. Xue, Carbohydr. Polym., 2016, 136, 1091–1097 CrossRef CAS PubMed.
- L. Lopez-Torrez, M. Nigen, P. Williams, T. Doco and C. Sanchez, Food Hydrocolloids, 2015, 51, 41–53 CrossRef CAS.
- N. Wang, Y. Chu, F. a. Wu, Z. Zhao and X. Xu, Int. Biodeterior. Biodegrad., 2017, 117, 236–244 CrossRef CAS.
- J. Xu, X. Xu, Y. Liu, H. Li and H. Liu, Environ. Prog. Sustainable Energy, 2016, 35, 345–351 CrossRef CAS.
- J. Chen, C. Lei, S. Lin, C. Liu and P. C. K. Cheung, Food Hydrocolloids, 2015, 46, 1–9 CrossRef.
- T. Mohan, R. Kargl, K. E. Tradt, M. R. Kulterer, M. Braćić, S. Hribernik, K. Stana-Kleinschek and V. Ribitsch, Carbohydr. Polym., 2015, 116, 149–158 CrossRef CAS PubMed.
- D. Salarbashi, S. A. Mortazavi, M. S. Noghabi, B. S. F. Bazzaz, N. Sedaghat, M. Ramezani and I. Shahabi-Ghahfarrokhi, Carbohydr. Polym., 2016, 140, 220–227 CrossRef CAS PubMed.
- Y. Wang, Y. Li, Y. Liu, X. Chen and X. Wei, Int. J. Biol. Macromol., 2015, 77, 76–84 CrossRef CAS PubMed.
- L. Wang, X. Zhang, Y. Niu, A. F. Ahmed, J. Wang and W. Kang, Int. J. Biol. Macromol., 2019, 124, 1230–1237 CrossRef CAS PubMed.
- Z. Ghlissi, F. Krichen, R. Kallel, I. B. Amor, T. Boudawara, J. Gargouri, K. Zeghal, A. Hakim, A. Bougatef and Z. Sahnoun, Int. J. Biol. Macromol., 2019, 123, 335–342 CrossRef CAS PubMed.
- S. Song, L. Wang, L. Wang, Q. Yu, C. Ai, Y. Fu, C. Yan, C. Wen and Z. Zhu, Int. J. Biol. Macromol., 2019, 126, 579–585 CrossRef PubMed.
- Y. E. Libin, L. U. Xu and L. I. Jianrong, Carbohydr. Polym., 2012, 87, 2052–2057 CrossRef.
- X. Qi, W. Mao, Y. Gao, Y. Chen, Y. Chen, C. Zhao, N. Li, C. Wang, M. Yan and C. Lin, Carbohydr. Polym., 2012, 90, 1804–1810 CrossRef CAS PubMed.
- S. J. Yoon, M. A. Yu, Y. R. Pyun, J. K. Hwang, D. C. Chu, L. R. Juneja and P. A. Mourão, Thromb. Res., 2003, 112, 151–158 CrossRef CAS PubMed.
- M. L. Quan, D. Pinto, J. M. Smallheer, W. R. Ewing, K. A. Rossi, J. M. Luettgen, D. A. Seiffert and R. R. Wexler, J. Med. Chem., 2018, 13, 7425–7447 CrossRef PubMed.
- I. Pawlaczyk-Graja, Int. J. Biol. Macromol., 2019, 116, 869–879 CrossRef PubMed.
- J. Weatherald and P. Laveneziana, Eur. Respir. Monogr., 2018, 80, 160–174 CAS.
- Y. Sun, X. Chen, S. Liu, H. Yu, R. Li, X. Wang, Y. Qin and P. Li, Chin. J. Oceanol. Limnol., 2018, 36, 882–891 CrossRef CAS.
- E. Bernardi and G. Camporese, Thromb. Res., 2017, 163, 201–206 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2020 |