Open Access Article
Open Access ArticleEnteric pH responsive cargo release from PDA and PEG coated mesoporous silica nanoparticles: a comparative study in Drosophila melanogaster†
Nidhi Sapre‡
 a,
Rusha Chakraborty‡
a,
Rusha Chakraborty‡ b,
Poorvi Purohitc,
Suresh Bhatc,
Gaurav Das
b,
Poorvi Purohitc,
Suresh Bhatc,
Gaurav Das *b and
Sneha R. Bajpe
*b and
Sneha R. Bajpe *a
*a
aSymbiosis Centre for Nanoscience and Nanotechnology, Symbiosis International (Deemed University) (SIU), Pune, India. E-mail: sneha.bajpe@scnn.edu.in
bNational Centre for Cell Science, Pune, India. E-mail: gauravdas@nccs.res.in
cCSIR-National Chemical Laboratory, Pune, India
First published on 23rd March 2020
Abstract
Physiological stimulus-specific cargo release from nanoparticle carriers is a holy grail of drug delivery research. While the majority of such work is carried out in vitro with cell lines, widespread use of common mammalian model systems – mice and rats – is difficult due to the associated cost and regulatory restrictions. Here we use the inexpensive, easily reared, excellent genetic model system Drosophila melanogaster to test pH responsive cargo release from widely used mesoporous silica nanoparticles (MSNs) coated with pH sensitive polydopamine (PDA) and polyethylene glycol (PEG) polymers. We synthesized 650 ± 75 nm diameter PDA or PEG coated mesoporous silica nanoparticles loaded with a fluorescent dye and fed to individual adult flies. Subsequently, the passage of the particles were monitored through the fly gut. As in mammals, the fly intestine has multiple pH specific zones that are easily accessible for imaging and also genetic, biochemical or physiological manipulations. We observed that both the species of MSNs ruptured around the acidic (pH < 4.0) middle midgut of the flies. PEG coated particles showed sharper specificity of release in the acidic middle midgut of flies than the PDA coated ones and had less tendency to clump together. Our results clearly show that the Drosophila gut can be used as a model to test pH responsive biocompatible materials in vivo. Our work paves the way for greater use of Drosophila as an in vivo complete systemic model in drug delivery and smart materials research. It also suggests that such specific delivery of chemical/biological cargo can be exploited to study basic biology of the gut cells and their communication with other organs.
1 Introduction
Maintaining tight control over the amount of drug released as-and-when needed by the human body is a long-standing challenge for drug delivery research.1 Functional materials such as zeolites,2,3 metal–organic frameworks,4–7 zeolitic imidazole frameworks8,9 etc., are some of the most popular class of materials used in drug delivery research.10 These materials, due to their biocompatibility and high porosity can be used as effective drug carriers. Among the various nano-sized carrier materials studied for drug delivery, mesoporous silica is perhaps the most popular due to its easy synthesis, high porosity and stability.11–16 Mesoporous silica nanoparticles (MSNs) are porous solids with a large number of honeycomb-like pores that can accommodate a wide variety of cargo molecules. These biocompatible materials have large surface area (>900 m2 g−1), large pore volume (>0.9 cm3 g−1) and tunable pore size (2–10 nm).16 Despite the established biocompatibility,18 non-functionalized MSNs lack the ability to specifically and efficiently react to external physiological stimuli and could disintegrate leading to premature cargo-release.18–20 Temporal or location specific cargo-release can be enhanced by (i) introducing additional molecular gates17,19 into the silica matrix that open on receiving stimuli such as changes in pH,20 redox,21 temperature17 etc., or (ii) by coating the silica nanoparticles with a stimulus responsive polymer.12,20,22–24 As an example, change in pH during cellular endocytosis can cause disintegration of responsive polymers surrounding carrier nanoparticles to release their cargo in a controlled manner.18,20,25,26 Among the various polymers used, polyethylene glycol (PEG)24,27–30 is popular due to its biocompatibility and biodegradability.26,31 One other polymer used frequently in nanomaterial coating is polydopamine (PDA);23,32–37 a polymeric oxidation product of the catecholaminergic neurotransmitter dopamine. It has a molecular structure similar to that of the naturally occurring pigment melanin (eumelanin) and possesses the ability to be deposited on all types of inorganic, organic and superhydrophobic surfaces.32,38 In vivo testing of functional drug-delivering materials in a biological system is critical to ascertain its suitability for biological applications. Usually, such testing of new materials and drugs are carried out in animal or human derived cell lines.39–42 While this approach has been helpful, the effect of these molecules in context of a complete organism with distinct organs and a holistic, systemic milieu cannot be modeled with cell lines.The fruit fly or vinegar fly Drosophila melanogaster, is a versatile genetic model that can be grown easily and inexpensively and offer a well-developed genetic tool kit to manipulate all its genes, cell types, tissues and organs.43–45 Importantly, flies and humans have homologous genes and organs, making it possible to model human diseases and study their underlying genetic, cellular and physiological mechanisms.46–50 For the same reasons flies also lend themselves for rapid testing of materials, including nanomaterials or nanoparticles, for their toxicity, biocompatibility and mode of action.51–59 However, there has almost been no effort to use flies for in vivo screening of smart carrier materials and stimulus responsive drug release.58,60 The fly gut bear remarkable similarity with the human gut.61 Both have similar developmental origin, harbour stem cells in its luminal epithelial lining, have neuronal innervations and secrete digestive enzymes in distinct zones that break down nutrients and facilitate their absorption through the gut epithelium. Importantly, like the mammalian gut, the fly gut has distinct pH zones that range from highly acidic in the middle midgut (pH < 4) to neutral in the foregut, crop & anterior midgut (pH ∼ 7.4) and to alkaline (pH ∼ 9–11) in the posterior midgut & hindgut.62–64 Here, we hypothesized that we can utilize the fly gut pH microenvironments to test zone-specific cargo release from smart carrier materials. We report a comparative study of cargo release from fluorescent dye loaded MSNs coated with two pH responsive polymers, PDA and PEG in the Drosophila gut. Our results indicate that both these particles readily release their cargo in the middle midgut (MMG) acidic zone and are not toxic to flies. Our studies poise the Drosophila melanogaster gut as an accessible model to screen pH stimulus specific cargo release in a well characterised genetic model organism.
2 Experimental section
2.1 Chemicals
Tetraethyl orthosilicate (TEOS) (cat. no. 8.00658), hexadecyl trimethyl ammonium bromide (CTAB) (cat. no. 8.14119), PEG400 (cat. no. 8.07485), hydrochloric acid (cat. no. 1.93401.0521) were purchased from Merck, India. Ammonia solution (cat. no. 78719) was purchased from SRL Chemicals, India. Potassium hydroxide (cat. no. Q26705) was purchased from Qualigens, India. Dopamine hydrochloride (cat. no. H8502) and bromothymol blue (cat. no. 114413) was purchased from Sigma Aldrich, India. Rhodamine B (cat. no. GRM980) was purchased from Hi-Media, India. Ethanol (cat. no. 32221) was purchased from Honeywell, India. Amaranth FCF food grade dye was purchased from Vidhi, Pune, India (batch no. PAM13568). Agarose (cat. no. 180720), low melting agarose (cat. no. AGAL0050) and PBS tablets (cat. no. 2810301) were purchased from MP Biomedicals, India. All chemicals were used without further purification. Milli-Q® and RO water were used for all experiments.2.2 Synthesis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water solution containing 10 mL of ammonia (10 mL, 25 wt%). This reaction mixture was heated at 35 °C, and 10 mL TEOS was added rapidly under vigorous stirring. The reaction mixture was stirred for 24 h. The reaction product was collected by vacuum filtration and washed thrice with 35% ethanol
water solution containing 10 mL of ammonia (10 mL, 25 wt%). This reaction mixture was heated at 35 °C, and 10 mL TEOS was added rapidly under vigorous stirring. The reaction mixture was stirred for 24 h. The reaction product was collected by vacuum filtration and washed thrice with 35% ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water and dried at 60 °C under vacuum, overnight. The white powder was then dispersed in acidified ethanol (400 μL of concentrated HCL in 200 mL of ethanol) and stirred at 30 °C for 2 h. This step was repeated thrice for removal of template and then the sample was washed three to four times with water and dried at 60 °C under vacuum, overnight.
water and dried at 60 °C under vacuum, overnight. The white powder was then dispersed in acidified ethanol (400 μL of concentrated HCL in 200 mL of ethanol) and stirred at 30 °C for 2 h. This step was repeated thrice for removal of template and then the sample was washed three to four times with water and dried at 60 °C under vacuum, overnight.2.2.3.1 PEG coated RhB@MSN. 1 g of RhB@MSNs were added to 100 mL of water and sonicated for 5–10 minutes. After sonication, 10% of PEG-400 was added to this solution and the whole mixture was stirred for six hours. The mixture was then washed several times with water, followed by centrifugation at 4000 rpm at 25 °C for 10 minutes. The final product was collected and dried in a desiccator.
2.2.3.2 PDA coated RhB@MSN. 1 g of RhB@MSNs were added to Tris–HCL buffer (10 mM, pH 8.5) containing dopamine hydrochloride (500 mg of dopamine in 500 mL) and sonicated for 5–10 minutes. The mixture was then stirred for six hours. The mixture was then washed several times with water, followed by centrifugation at 4000 rpm at 25 °C for 10 minutes. The final greyish black product was collected and dried at 40 °C under vacuum, overnight.
2.3 Physicochemical characterization
Dynamic light scattering (DLS) & zeta potential measurements were performed on Anton Paar Litesizer 500, Anton Paar, GmbH. The samples were measured for 1 min and auto fitting of the correlation function was performed using the Anton Paar DLS software Kalliope. Zeta potential measurements were performed using an Omega cuvette. Scanning electron microscopy (SEM) was performed on Carl Zeiss EVO 18, Zeiss, USA. The powder samples were gold sputtered before measurement. Transmission Electron Microscopy (TEM) was performed on Philips CM200; operating voltage: 20–200 kV, resolution: 2.4 Å. The samples were suspended in water and loaded on a Cu grid and air dried before measurement. X-ray photoelectron spectroscopy (XPS) measurements were performed using K-Alpha + XPS instrument, Thermo Fisher, UK, with Al Kα (monochromatic) using 6 mA beam current at 12 kV. The instrument was calibrated with Ag 3d 352 eV. The samples were drop casted on a silicon wafer and mounted on a standard SS plate with conductive carbon tape and loaded into the sample chamber with a vacuum pressure of 1.2 × 10−8 mbar. Sample spot size was 400 μm and the reference binding energy was C 1s (284.8 eV). Brunauer–Emmett–Teller (BET) surface area analysis by Nitrogen adsorption and desorption were carried out using Quadrasorb SI, Quantachrome Instruments, Anton Paar, GmbH. Pore size distributions were calculated from adsorption isotherm using the BJH method. Fourier transform infrared (FTIR) measurements were performed on a PerkinElmer Spectrum-GX, PerkinElmer, USA, at resolution 4 (nm Å−1), scan range from 4000–600 cm−1, the total number of scans 16 and samples were scanned directly in ATR mode. All graphs were plotted using Igor Pro software, Wavemetrics, USA. Confocal microscopy images for the synthesised nanoparticles were obtained on a Zeiss LSM 880 confocal microscope, Zeiss, USA. 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilutions of 1 mg mL−1 PEG/PDA coated RhB@MSN samples were prepared in 1% low melting (LM) agarose (MP Biomedicals, cat. no. AGAL0050) and 20 μL of each solution was added on a clean glass slide. The added droplets were immediately covered with a coverslip and allowed to spread. Coverslips were sealed with transparent nail polish and imaged with Airyscan 2.0 scanner using 40× (plan-apochromat/1.4 oil DIC M27) and 100× (α plan-apochromat/1.46 oil DIC M27 Elyra) objectives. pH of various solutions were checked using Hanna (HI-5521-02) pH meter, Hanna Instruments, USA.
10 dilutions of 1 mg mL−1 PEG/PDA coated RhB@MSN samples were prepared in 1% low melting (LM) agarose (MP Biomedicals, cat. no. AGAL0050) and 20 μL of each solution was added on a clean glass slide. The added droplets were immediately covered with a coverslip and allowed to spread. Coverslips were sealed with transparent nail polish and imaged with Airyscan 2.0 scanner using 40× (plan-apochromat/1.4 oil DIC M27) and 100× (α plan-apochromat/1.46 oil DIC M27 Elyra) objectives. pH of various solutions were checked using Hanna (HI-5521-02) pH meter, Hanna Instruments, USA.
2.4 In vitro RhB release studies
Three different pH solutions were prepared: starting with 1 mM HCl (pH 3.0) and then adjusting pH by the addition of 1 mM KOH to obtain solutions of pH – 7.4 and 9.0. 5 mg of samples were weighed separately in different vials to which 2 mL of different pH solutions were added and the vials were rotated using a rotating mixer for different time periods (between 0 to 5 h). All samples were then centrifuged at 4000 rpm for 10 minutes at 25 °C and absorbance measurements were performed on the supernatants at 553 nm wavelength. The blank was subtracted from each measurement. The measurements were repeated thrice per sample and the samples were measured in duplicates.2.5 Fly experiments
2.5.2.1 Feeding. To deprive flies of food and water, 25–30 females were transferred to an empty vial with dry filter paper and kept in an incubator at 25 °C and ∼60% humidity for 12–14 hours. Resultant hungry and thirsty flies were then transferred to a fresh empty vial. The flies were ice anesthetized and 10–15 healthy flies were immobilized on a glass slide using nail polish with their wings stuck on the surface of the glass slide with their abdomens facing up (see ESI Fig. SI-8†). Flies were allowed to recover for 30 minutes at 25 °C. A 1 μL droplet of 0.1 mg mL−1 of PEG coated and 0.02 mg mL−1 of PDA coated RhB@MSN solution in RO water (adjusted to pH 7.0) were fed to individual flies using a 0.1–3 μL micro pipette for 10 seconds. Only flies that fed continuously for ∼10 seconds were used for further experimentation. Subsequent to feeding, flies on the glass slide were placed in a petri dish with a wet filter paper at the bottom to avoid dehydration and kept at 25 °C for 30 min.
2.5.2.2 Dissection and mounting. Nanoparticle fed flies were transferred to a vial containing 70% ethanol for 30 seconds and anesthetized. After anesthetization, their guts were dissected and washed 2–3 times with 1× PBS before mounting as described above. No more than 2 guts were mounted between two glass cover-slip (18 mm × 18 mm × 1 mm, Bluestar) spacers were glued with nail polish ∼10 mm apart onto the slide and a third cover-slip was carefully placed over the sample. Mounting media was added gently from one of the open edges and allowed to wick completely to just submerge the sample. Finally, excess mounting media was removed with a tissue and the cover-slip was sealed on the edges using a transparent nail polish and allowed to dry before imaging.
2.5.2.3 Confocal imaging. The samples above were imaged using Zeiss LSM 880 confocal microscope using 10× (plan-apochromat/0.45 M27) and 20× (plan-apochromat/0.8 M27) objectives. Z-stacks of individual samples were acquired with tile-scanning for each optical section. Images were processed with Fiji66 to derive maximum intensity projection of Z-stacks and to adjust contrast and brightness. Figures were put together at the highest possible resolution using Keynote (Apple Inc. Cupertino, California) and level functions were used on the digital images to make them consistent with visual observation. No quantitative image analysis was carried out on such a level adjusted image.
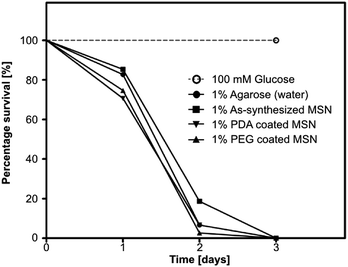
2.5.2.4 Survival analysis. 5 mL of 1% agarose solution was poured in fly vials and allowed to solidify. Vials were wiped thoroughly to remove excess water from the walls and a piece of nonabsorbent cotton was placed inside each vial (ESI Fig. SI-9†) to avoid fly contact with agarose medium. In small feeding caps 250 μL of 1% PDA coated MSN, 1% PEG coated MSN, as-synthesized MSN, 100 mM glucose and only water samples in 1% low melting (LM) agarose were added with 0.8% amaranth FCF dye as an ingestion indicator. One cap each was fixed on the inside wall of a vial with double sided tape. 25 healthy female flies were mildly CO2 anesthetized and added to each vial. All conditions were set in triplicates. The flies were allowed to revive and vials were kept at 25 °C. Number of surviving flies were counted and transferred to fresh vials with freshly prepared nanoparticles in 1 % LM agarose every 24 hours.
2.6 Statistical analysis
Data is represented as mean ± SD (Fig. 3). Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software, USA). Non-parametric Kruskal–Wallis test was performed for in vitro release studies for estimating difference of RhB dye release from a dye loaded nanoparticle at different pH at a given time point. A p-value of <0.05 was assumed to indicate statistically significant dye release at different pH environments. Survival assay data was plotted using the Kaplan–Meier method and curves were compared using log-rank (Mantel–Cox) test (Fig. 6). See ESI Table SI-1† for detailed results of statistical analysis.3 Results and discussion
3.1 Characterization of MSNs
A series of mesoporous silica nanoparticles (MSNs) were synthesized by changing the solvent mixture ratio to obtain particles of various sizes ranging from 200 nm to 900 nm. We chose particles with an average size of 650 ± 75 nm for ease of confocal microscopy imaging in vivo after feeding adult flies. As-synthesized MSN samples were then loaded with 0.005% RhB, and coated with PDA or PEG for 6 hours. All samples were characterized by DLS, SEM, TEM, FTIR and XPS. BET surface area analysis of as-synthesized MSNs show a surface area of ∼482 m2 g−1 and estimated pore diameter of 5.7 nm (see ESI Fig. SI-1A and B).† Since the MSNs were not calcined at higher temperatures after synthesis, a reduction in surface area could be attributed to incomplete evacuation of the template. However, since our primary goal was to observe delivery in Drosophila gut, partial presence of template and reduced loading capacity were not deterrents to our purpose. Surface coating by PDA or PEG was monitored by zeta (ζ) potential measurements which clearly show a shift in the net surface charge from around −26 mV for as-synthesized MSN particles (typical for MSNs)11,67 to around −32 mV and −37 mV for PDA and PEG coated RhB@MSN particles respectively, confirming the alteration in surface properties of the synthesized MSNs by coating. Higher dispersibility of the coated samples also suggest a shift to higher negative charge of the particles. The average particle size of all the samples along with the ζ potential values are listed in Table 1.| Measured parameters | As-synthesized MSNs | PDA coated RhB@MSN | PEG coated RhB@MSN |
|---|---|---|---|
| Particle diameter (nm) | 819.5 | 945.48 | 962 |
| Zeta potential (mV) | −26.63 | −32.14 | −37.02 |
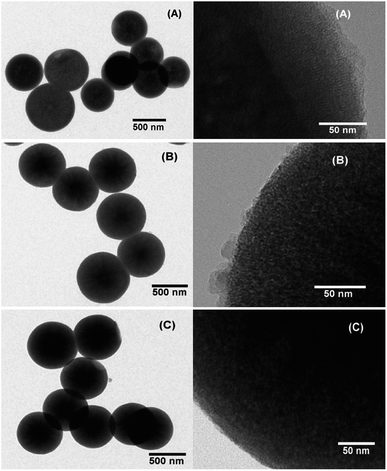
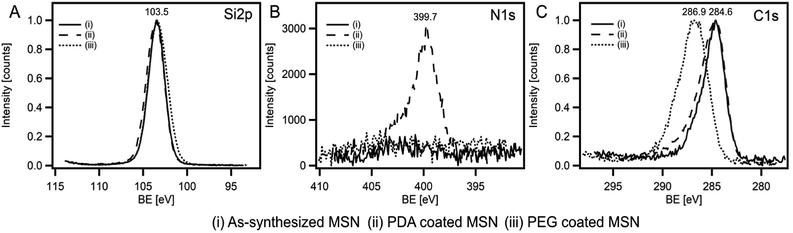
In order to confirm the particle shape and size, scanning electron microscope (SEM) and transmission electron microscope (TEM) measurements were performed. SEM and TEM images clearly revealed spherical particles with a size of 635 ± 75 nm for the as-synthesized MSNs, 642 ± 60 nm for the PDA coated RhB@MSNs and 655 ± 55 nm for the PEG coated RhB@MSN samples (see Fig. 1, SI-3 and SI-4†). The difference between the coated and uncoated samples can be clearly seen from the TEM images (Fig. 1). The significant difference in particle diameter obtained by DLS when compared to TEM is attributed to agglomeration of the samples in water during DLS measurements. Although TEM images clearly show the difference in surface morphology for PDA or PEG coated samples when compared to the as-synthesized MSN particles, we performed XPS measurements in order to precisely establish the presence of PDA or PEG coating. Fig. 2 shows the XPS spectra for (i) as-synthesized MSN, (ii) PDA coated MSN and (iii) PEG coated MSN samples. As can be seen in Fig. 2A, all samples exhibit distinct peak at 103.5 eV,68 characteristic of Si–O bonds. In case of the PDA coated MSN sample, the N 1s spectrum clearly shows the presence of N–H bond with a distinct peak at 399.7 eV confirming the presence of an amine group35,69,70 on the MSN surface which is clearly absent for both PEG coated and as-synthesized MSN sample (Fig. 2B). Finally, in the case of PEG coated samples, C–O bonds in PEG results in a shift of the characteristic C–C peak from 284.6 eV to 286.8 eV (Fig. 2C) establishing the surface coupling of MSN by PEG.71,72
3.2 In vitro tests of the nanoparticles
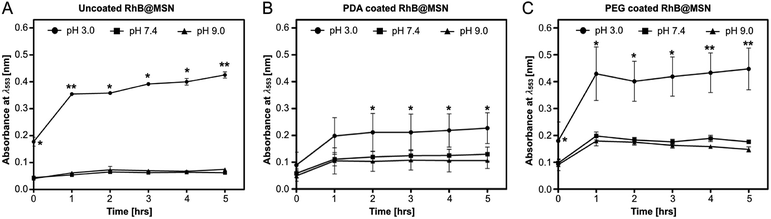
In order to investigate the in vitro release of PDA and PEG coated RhB@MSNs, the synthesis optimized samples were suspended in three different pH (3.0, 7.4 and 9.0) solutions and RhB release was monitored hourly for 5 hours. We performed control measurements on uncoated RhB@MSN samples (Fig. 3A) as well. Clearly, both PDA and PEG coated samples show higher release at pH 3.0 as compared to pH 7.4 or 9.0 (Fig. 3B and C) and most RhB is released within two hours in both cases. To confirm the pH-triggered cargo release of both PDA and PEG in acidic pH, we performed ζ potential measurements of the samples under the same pH conditions. Control measurements on uncoated RhB@MSN and as-synthesized MSN samples were also performed. Table 2 summarizes the ζ potential for both PDA and PEG coated samples along with the uncoated MSN and as-synthesized MSNs at different pH.| Samples | pH – 3.0 | pH – 7.4 | pH – 9.0 |
|---|---|---|---|
| As-synthesized MSN | 33.73 | −25.2 | −34.86 |
| Uncoated RhB@MSN | 34.45 | −20.02 | −36.19 |
| PDA coated RhB@MSN | 31.15 | −26.31 | −33.12 |
| PEG coated RhB@MSN | 29.42 | −29.83 | −39.33 |
As expected, all samples show positive potential at pH 3.0, suggesting protonation in acidic medium. Also, the particles are in an electrostatically stable configuration at pH 3.0 (≥+30 mV) and pH 9.0 (≤−30 mV) suggesting particle stability and less agglomeration in solution. The release profile in combination with ζ potential values for the uncoated RhB@MSN sample in pH 3.0 (Fig. 3A) reveals that there is decreased electrostatic interaction between the positively charged RhB and negatively charged silica surface in acidic medium11 resulting in burst release of the dye within an hour. At neutral pH, the surface of uncoated RhB@MSN has a negative surface charge density resulting in further entrapment of RhB molecules in the MSN matrix and hence inhibiting its release. In the case of PDA coated RhB@MSN sample at pH 3.0 (Fig. 3B), the electrostatic interaction between the positively charged dye molecules and the protons in the acidic medium is reduced due to the presence of a polymeric barrier. As previously reported by DeMuth et al.,11 and Casasús et al.,73 the presence of a multilayer polyamine group on the surface of MSN would certainly inhibit the release of RhB molecules due to higher charge density. The polydopamine layer will expand due to the coulombic repulsion between the protonated amine groups causing it to expand in such a way that it reduces the effective pore diameter of the MSN and hence hinder the release of RhB from the pores.11,73 Also, the PDA coated RhB@MSN sample has higher negative potential at neutral pH resulting in higher release of the dye when compared to uncoated RhB@MSN. However, when compared to the ζ potential value of the PEG coated RhB@MSN at pH 7.4, lower negative potential (−26 mV) suggests agglomeration of the particles in solution. In the case of PEG coated RhB@MSN at pH 3.0 (Fig. 3C) the total release of RhB is similar to that of the uncoated RhB@MSN (Fig. 3A). The increased total release of the dye molecules when compared to that of PDA coated RhB@MSN sample could be due to the hydrophilic nature of PEG. It is also possible that the use of low molecular weight PEG (PEG-400) would result in the formation of a thin layer of coating when compared to the PDA coated sample, resulting in more RhB release.74 Finally at pH 9.0, all samples show negative potential ≤−30 mV suggesting that the particles are electrostatically stable in solution and hence less or no RhB release is observed. A schematic representation of the proposed mechanism of RhB release from the uncoated and coated RhB@MSN samples is shown in Fig. 4.
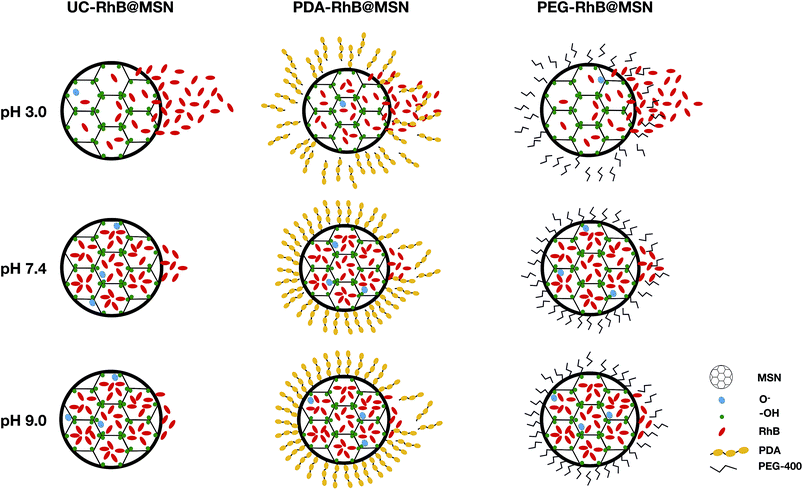 | ||
| Fig. 4 Schematic representation of proposed in vitro RhB release mechanism for the uncoated and coated MSNs. Note: UC = uncoated. | ||
3.3 In vivo tests of the nanoparticles
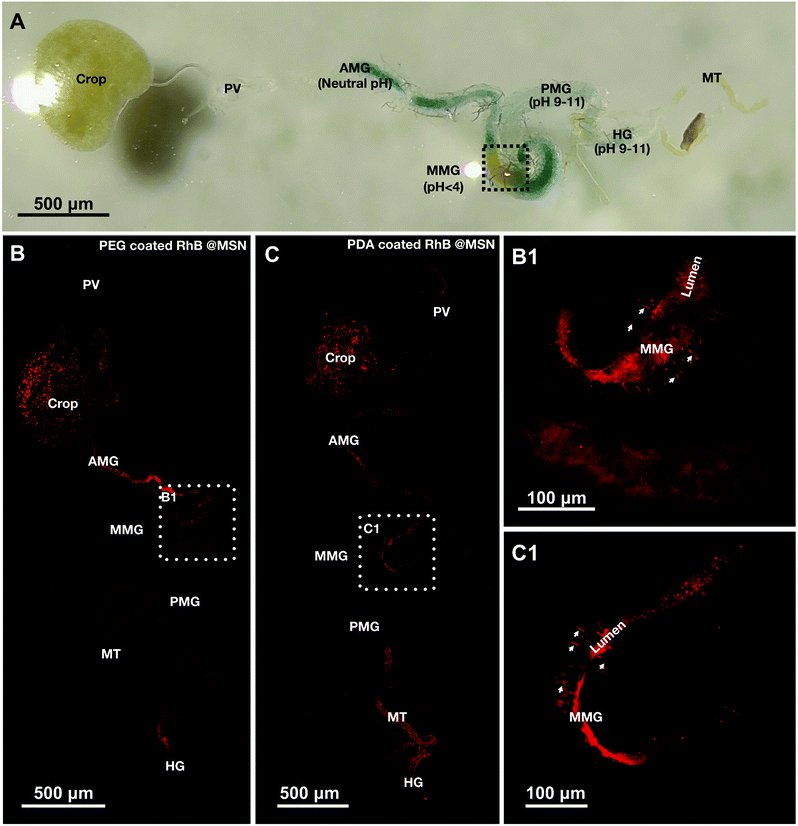
To determine the pH responsive release and safety of synthesized MSNs coated with PDA and PEG in a whole organism model we chose adult Drosophila melanogaster. Almost all organ systems and cell types in the fly have been extensively studied in the context of development, cellular function and physiology.75–77 Recently, flies are being increasingly used to study toxic impact of different classes of nanoparticles on cells, organs, physiology and ultimately survival and reproductive capacity of flies.51,52,54–56,78–84 However, not many studies have focused on testing smart, stimulus responsive materials in flies to understand conserved mechanisms of nanocarrier based cargo/drug delivery. Only two recent studies have utilized the larval or adult fly gut for testing drug carrier materials.58,60 The Drosophila gut is divided into three main regions: the foregut, midgut and the hindgut (Fig. 5A) with distinct acidic, neutral and alkaline zones like in mammals.62,64 Ferguson et al.,58 demonstrated recently in adult Drosophila suzukii, a pest species, that acidic pH-responsive poly(2-vinylpyridine) microcapsules release their cargo in the fly midgut. To further test the feasibility of the fly gut as a model system for testing smart cargo release from biocompatible nanoparticles, we tested the pH-triggered release of our PDA and PEG coated MSNs and performed in vivo comparative study of the release, in the better characterized and genetically tractable Drosophila melanogaster species. As mentioned in the experimental section, 0.02 mg mL−1 of PDA and 0.1 mg mL−1 of PEG coated RhB@MSNs were fed to female flies and then they were kept for 30 minutes in a humid petri dish before dissection. The passage of these nanoparticles in the fly gut were visualized by confocal imaging of dissected guts. We observed that intact and discrete PEG coated RhB@MSN particles were present in the insect crop, where recently ingested food is stored before passing down through the midgut. Very low background fluorescence indicated that hardly any RhB@MSN particle rupture and RhB release in the neutral pH of the crop (Fig. 5B). Discrete fluorescent signals from PEG coated RhB@MSN particles were also observed in the neutral pH of anterior midgut (AMG), in spite of saturated fluorescent signals due to particle accumulation, indicating that they were still intact at that zone (Fig. 5B). Remarkably, in the acidic middle midgut (MMG), with a pH < 4.0, discrete red fluorescent signal was lost and replaced by diffused red signal (area marked by white dotted box and inset in Fig. 5B and B1). This strongly suggests that, as in vitro, RhB release in vivo was in the MMG due to the highly acidic pH environment. Interestingly, the MMG also shows distinct red filamentous staining pattern, perpendicular to the gut lumen and spanning the gut walls. Based on previous studies of gut morphology,64,85 we think these filamentous staining patterns are caused by paracellular (i.e., in between the cells) transport of water soluble RhB dye, across the MMG epithelial cell layer (Fig. 5B1 and C1). Midguts of mammals and insects including flies, exhibit both transcellular transport (through the cells) and paracellular (in between cells) transport of small molecules, nutrients, drugs and insecticides.61,86,87 Stressors like injury and cold temperature shock has been observed to induce such paracellular leakage of dyes in flies.88,89 In our case, it is possible that the stress of food and water deprivation before nanoparticle feeding may result in the observed paracellular dye seepage from the MMG lumen. The preferred mode of transport across gut epithelium is likely influenced by the size, charge and orientation of the cargo molecule. A study in the lepidopteran larvae midgut epithelium with two insecticidal peptides reported preferential transcellular transport for a fluorescein isothiocyanate conjugated Aedes aegypti peptide trypsin modulating oostatic factor (FITC-Aea-TMOF) and paracellular transport of a Rhodamine labeled neuropeptide proctolin (Rh-proctolin).90 We intend to conduct future studies to elucidate the role of nutrient deprivation states and the physicochemical properties of specific cargoes on transport across the fly gut epithelium.The release of PDA coated RhB@MSN along the gut is similar to the PEG coated RhB@MSN in many respects and follows the same general pattern from crop to MMG (Fig. 5C and C1). The notable differences are (i) PDA coated RhB@MSN showed clumping all the way upto the MMG and (ii) higher background fluorescence in the crop and AMG. The more discrete nature of the PEG coated RhB@MSN compared to PDA coated RhB@MSN might be attributed to the higher electrostatic repulsion between individual particles due to higher negative surface charge at neutral pH of the former than the latter (Table 2). The higher background fluorescence of the PDA coated RhB@MSNs could be due to relatively higher levels (than the PEG coated RhB@MSNs preparation) of leftover un-encapsulated RhB in our preparation.
We also fed flies uncoated RhB@MSNs and looked at the release profile of RhB. In accordance with the in vitro release data (Fig. 3A), some RhB release and paracellular dye leakage in the epithelium was also observed in the acidic MMG (ESI Fig. SI-7B†). As explained in the previous section, this could be due to the fact that RhB is positively charged and could fit in an electrostatically stable configuration inside the negatively charged interior of the MSNs at neutral and alkaline pH. At acidic pH the decreased electrostatic interaction between the positively charged RhB and the silica surface results in a burst release of the dye molecule. However, a lot of discrete and also aggregated dye signals (very likely the particles themselves) are observed in the MMG and in the crop (ESI Fig. SI-7B†). The aggregation could due to the lower negative potential of the uncoated RhB@MSN (Table 2). The overall brighter signal seen in the image is in line with our observation that more dye is retained in uncoated RhB@MSN and that some RhB cargo is lost during the coating process of RhB@MSNs with PEG/PDA. Finally, feeding flies with free RhB as a control convincingly shows that RhB dye as possibly other cargo (the various dye loaded & coated/uncoated MSNs), is retained preferentially in the crop and then in the MMG (ESI Fig. SI-7A, A1, SI-7B and B1†) and also shows paracellular leak in between the MMG epithelial cells. Overall, our results suggest that coating with polymers could improve desirable properties of the cargo loaded MSNs. It reduces aggregation and potentially regulates cargo release from the MSNs. In other such applications, polymer coating increases longevity of systemically circulating nanoparticles and functionalize particles to be stimuli responsive.24,91
3.4 Survival analysis upon nanoparticle feeding
The quantification of survival was performed by placing the flies in vials with 1% agarose bed and small plastic feeding caps (see ESI Fig. SI-9†) loaded with different MSN species along with control samples. Flies were exposed to PDA/PEG coated and as-synthesized MSN particles in low melting (LM) agarose. Only 1% LM agarose (water) and 100 mM glucose in 1% LM agarose served as controls. A food grade indicator dye, Amaranth, was included in all samples to check for ingestion. Number of surviving flies was recorded every 24 hours. All flies on the agarose (water) control failed to survive beyond 3 days due to lack of nutrition (Fig. 6). The nanoparticle fed flies followed the same trend for survival as that of the water control, suggesting that as-synthesized MSNs as well as PDA and PEG coated MSNs do not provide nutrition but were also likely non-toxic at 1% (10 mg mL−1) concentration as they did not kill flies earlier than the water control.4 Conclusions
In this work, we have compared pH-responsive release of RhB from mesoporous silica nanoparticles coated with two polymers: polydopamine (PDA) and polyethylene glycol (PEG) in the Drosophila melanogaster gut. We observe that both these types of carrier particles show specific release of RhB cargo in the highly acidic (pH < 4.0) middle midgut of flies. PEG coated particles show better release profile and clump less compared to the PDA coated species. Interestingly, uncoated RhB@MSNs also exhibit acid responsive cargo release like the coated MSNs. Our survival tests show that the various nanoparticles (coated and uncoated) are nontoxic at the conditions tested and are not a source of nutrition to flies. The use of Drosophila melanogaster to verify enteric pH responsive release provides an inexpensive and importantly, a genetic whole organism model to monitor drug release from a large range of functionalized materials. Most work on testing the long-term tissue or organ specific effects of the drug delivering nanoparticles are carried out on common laboratory rodents like mice and rats. Such studies require extensive animal experimentation clearance. In contrast we believe flies offer a rapid mode of carrying out similar studies, at a far lower cost and the ability to further explore the effect of nanoparticle exposure on specific molecular pathways, gene expression, physiology and behaviour. In addition to assessing the effect of the carrier material, it is not a long stretch to see that flies can easily be co-opted to study the biology of the cargo molecule interacting with their target cells/organs and also to monitor the systemic non-specific effect on cells and organs away from their delivery site as they circulate inside the organism over time.57 The expeditious nature of such studies in flies will also allow faster fine tuning of the already-existing materials to function optimally. We also anticipate that cleverly designed functional materials can be used to uncover fundamental physiological, cellular, molecular mechanisms underlying proper gut function and its communication with other organs in flies.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge financial support from SIU and NCCS. We thank Dr B. L. V. Prasad, CSIR-National Chemical Laboratory, Pune, for fruitful discussions and feedback. We also thank Dr C. P. Vinod and the CMC facility at CSIR-National Chemical Laboratory, Pune, for help with XPS measurements. We thank Dr Amit Yadav for help with SEM facility use at NCMR, Pune. We thank the Sophisticated Analytical Instrument Facility (SAIF), IIT Bombay, for help with TEM measurements. We are grateful to NCCS imaging facility for assistance during long hours of imaging. Finally, we thank Dr Das and Dr Bajpe's research group members for feedback and support.Notes and references
- M. Bruneau, S. Bennici, J. Brendle, P. Dutournie, L. Limousy and S. Pluchon, J. Controlled Release, 2019, 294, 355–371 CrossRef CAS PubMed.
- R. Amorim, N. Vilaça, O. Martinho, R. M. Reis, M. Sardo, J. Rocha, A. M. Fonseca, F. Baltazar and I. C. Neves, J. Phys. Chem. C, 2012, 116, 25642–25650 CrossRef CAS.
- M. G. Rimoli, M. R. Rabaioli, D. Melisi, A. Curcio, S. Mondello, R. Mirabelli and E. Abignente, J. Biomed. Mater. Res., Part A, 2008, 87, 156–164 CrossRef PubMed.
- C. Orellana-Tavra, R. J. Marshall, E. F. Baxter, I. A. Lázaro, A. Tao, A. K. Cheetham, R. S. Forgan and D. Fairen-Jimenez, J. Mater. Chem. B, 2016, 4, 7697–7707 RSC.
- W. Cai, J. Wang, C. Chu, W. Chen, C. Wu and G. Liu, Adv. Sci., 2019, 6, 1801526 CrossRef PubMed.
- P. Horcajada, C. Serre, G. Maurin, N. A. Ramsahye, F. Balas, M. Vallet-Regí, M. Sebban, F. Taulelle and G. Férey, J. Am. Chem. Soc., 2008, 130, 6774–6780 CrossRef CAS PubMed.
- P. Horcajada, C. Serre, M. Vallet-Regí, M. Sebban, F. Taulelle and G. Férey, Angew. Chem., Int. Ed. Engl., 2006, 118, 6120–6124 CrossRef.
- Y. Guo, J. Wang, D. Zhang, T. Qi and G. L. Li, Colloids Surf., A, 2019, 561, 1–8 CrossRef CAS.
- C.-Y. Sun, C. Qin, X.-L. Wang, G.-S. Yang, K.-Z. Shao, Y.-Q. Lan, Z.-M. Su, P. Huang, C.-G. Wang and E.-B. Wang, Dalton Trans., 2012, 41, 6906–6909 RSC.
- G. Férey, Chem. Soc. Rev., 2008, 37, 191–214 RSC.
- P. DeMuth, M. Hurley, C. Wu, S. Galanie, M. R. Zachariah and P. DeShong, Microporous Mesoporous Mater., 2011, 141, 128–134 CrossRef CAS.
- M. Sönmez, D. Ficai, A. Ficai, L. Alexandrescu, M. Georgescu, R. Trusca, D. Gurau, M. A. Titu and E. Andronescu, Int. J. Pharm., 2018, 549, 179–200 CrossRef PubMed.
- X.-Y. Lou, Y.-P. Li and Y.-W. Yang, Biotechnol. J., 2019, 14, e1800354 CrossRef PubMed.
- Y. Lu, Y. Yang, Z. Gu, J. Zhang, H. Song, G. Xiang and C. Yu, Biomaterials, 2018, 175, 82–92 CrossRef CAS PubMed.
- B. G. Trewyn, I. I. Slowing, S. Giri, H.-T. Chen and V. S.-Y. Lin, Acc. Chem. Res., 2007, 40, 846–853 CrossRef CAS PubMed.
- B. G. Trewyn, S. Giri, I. I. Slowing and V. S.-Y. Lin, Chem. Commun., 2007, 31, 3236–3245 RSC.
- J. Wen, K. Yang, F. Liu, H. Li, Y. Xu and S. Sun, Chem. Soc. Rev., 2017, 46, 6024–6045 RSC.
- L. Yuan, Q. Tang, D. Yang, J. Z. Zhang, F. Zhang and J. Hu, J. Phys. Chem. C, 2011, 115, 9926–9932 CrossRef CAS.
- S. Alberti, G. J. A. A. Soler-Illia and O. Azzaroni, Chem. Commun., 2015, 51, 6050–6075 RSC.
- L. Tan, M.-Y. Yang, H.-X. Wu, Z.-W. Tang, J.-Y. Xiao, C.-J. Liu and R.-X. Zhuo, ACS Appl. Mater. Interfaces, 2015, 7, 6310–6316 CrossRef CAS PubMed.
- L. Maggini, I. Cabrera, A. Ruiz-Carretero, E. A. Prasetyanto, E. Robinet and L. De Cola, Nanoscale, 2016, 8, 7240–7247 RSC.
- X. Li, C. Xie, H. Xia and Z. Wang, Langmuir, 2018, 34, 9974–9981 CrossRef CAS PubMed.
- D. Chang, Y. Gao, L. Wang, G. Liu, Y. Chen, T. Wang, W. Tao, L. Mei, L. Huang and X. Zeng, J. Colloid Interface Sci., 2016, 463, 279–287 CrossRef CAS PubMed.
- J. S. Suk, Q. Xu, N. Kim, J. Hanes and L. M. Ensign, Adv. Drug Delivery Rev., 2016, 99, 28–51 CrossRef CAS PubMed.
- K. Yang, H. Luo, M. Zeng, Y. Jiang, J. Li and X. Fu, ACS Appl. Mater. Interfaces, 2015, 7, 17399–17407 CrossRef CAS PubMed.
- G.-F. Luo, W.-H. Chen, Y. Liu, Q. Lei, R.-X. Zhuo and X.-Z. Zhang, Sci. Rep., 2014, 4, 6064 CrossRef CAS PubMed.
- Y. Hu, Z. Zhao, T. Harmon, P. R. Pentel, M. Ehrich and C. Zhang, Biomaterials, 2018, 182, 72–81 CrossRef CAS PubMed.
- P. Laskar, J. Dey and S. K. Ghosh, Colloids Surf., B, 2016, 139, 107–116 CrossRef CAS PubMed.
- S. Niedermayer, V. Weiss, A. Herrmann, A. Schmidt, S. Datz, K. Müller, E. Wagner, T. Bein and C. Bräuchle, Nanoscale, 2015, 7, 7953–7964 RSC.
- X. Zeng, G. Liu, W. Tao, Y. Ma, X. Zhang, F. He, J. Pan, L. Mei and G. Pan, Adv. Funct. Mater., 2017, 27, 1605985 CrossRef.
- C. Liang, H. Wang, M. Zhang, W. Cheng, Z. Li, J. Nie, G. Liu, D. Lian, Z. Xie, L. Huang and X. Zeng, J. Colloid Interface Sci., 2018, 525, 1–10 CrossRef CAS PubMed.
- M. E. Lynge, R. van der Westen, A. Postma and B. Städler, Nanoscale, 2011, 3, 4916–4928 RSC.
- W. Cheng, J. Nie, N. Gao, G. Liu, W. Tao, X. Xiao, L. Jiang, Z. Liu, X. Zeng and L. Mei, Adv. Funct. Mater., 2017, 27, 1704135 CrossRef.
- W. Cheng, C. Liang, L. Xu, G. Liu, N. Gao, W. Tao, L. Luo, Y. Zuo, X. Wang, X. Zhang, X. Zeng and L. Mei, Small, 2017, 13, 1700623 CrossRef PubMed.
- W. Cheng, J. Nie, L. Xu, C. Liang, Y. Peng, G. Liu, T. Wang, L. Mei, L. Huang and X. Zeng, ACS Appl. Mater. Interfaces, 2017, 9, 18462–18473 CrossRef CAS PubMed.
- Y. Duo, Y. Li, C. Chen, B. Liu, X. Wang, X. Zeng and H. Chen, RSC Adv., 2017, 7, 39641–39650 RSC.
- W. Cheng, X. Zeng, H. Chen, Z. Li, W. Zeng, L. Mei and Y. Zhao, ACS Nano, 2019, 13, 8537–8565 CrossRef CAS PubMed.
- A.-K. Pada, D. Desai, K. Sun, N. Prakirth Govardhanam, K. Törnquist, J. Zhang and J. M. Rosenholm, Int. J. Mol. Sci., 2019, 20, 3408 CrossRef CAS PubMed.
- A. U. Kura, S. Fakurazi, M. Z. Hussein and P. Arulselvan, Chem. Cent. J., 2014, 8, 46 CrossRef PubMed.
- V. Kumar, N. Sharma and S. S. Maitra, Int. Nano Lett., 2017, 7, 243–256 CrossRef CAS.
- A. H.-H. Wong, H. Li, Y. Jia, P.-I. Mak, R. P. d. S. Martins, Y. Liu, C. M. Vong, H. C. Wong, P. K. Wong, H. Wang, H. Sun and C.-X. Deng, Sci. Rep., 2017, 7, 9109 CrossRef PubMed.
- J. L. Wilding and W. F. Bodmer, Cancer Res., 2014, 74, 2377–2384 CrossRef CAS PubMed.
- U. Schäfer and H. Jäckle, Drosophila Melanogaster: A genetic tool, Comparative Genomics, Springer, 2000, pp. 23–41 Search PubMed.
- B. H. Jennings, Mater. Today, 2011, 14, 190–195 CrossRef.
- S. E. Mohr, First in Fly: Drosophila Research and Biological Discovery, Harvard University Press, 2018 Search PubMed.
- U. B. Pandey and C. D. Nichols, Pharmacol. Rev., 2011, 63, 411–436 CrossRef CAS PubMed.
- L. T. Reiter, L. Potocki, S. Chien, M. Gribskov and E. Bier, Genome Res., 2001, 11, 1114–1125 CrossRef CAS PubMed.
- M. Allocca, S. Zola and P. Bellosta, Drosophila melanogaster - Model for Recent Advances in Genetics and Therapeutics, InTech, 2018 Search PubMed.
- B. Ugur, K. Chen and H. J. Bellen, Dis. Models Mech., 2016, 9, 235–244 CrossRef CAS PubMed.
- L. I. Held Jr, Deep homology? Uncanny similarities humans and flies uncovered by Evo-Devo, Cambridge University Press, 2019 Search PubMed.
- X. Liu, D. Vinson, D. Abt, R. H. Hurt and D. M. Rand, Environ. Sci. Technol., 2009, 43, 6357–6363 CrossRef CAS PubMed.
- R. Posgai, M. Ahamed, S. M. Hussain, J. J. Rowe and M. G. Nielsen, Sci. Total Environ., 2009, 408, 439–443 CrossRef CAS PubMed.
- M. Ahamed, R. Posgai, T. J. Gorey, M. Nielsen, S. M. Hussain and J. J. Rowe, Toxicol. Appl. Pharmacol., 2010, 242, 263–269 CrossRef CAS PubMed.
- M. Alaraby, B. Annangi, R. Marcos and A. Hernández, J. Toxicol. Environ. Health, Part B, 2016, 19, 65–104 CAS.
- B. Jovanović, V. J. Cvetković and T. L. Mitrović, Chemosphere, 2016, 144, 43–49 CrossRef PubMed.
- S. A. Pappus and M. Mishra, Adv. Exp. Med. Biol., 2018, 1048, 311–322 CrossRef CAS PubMed.
- F. Barandeh, P.-L. Nguyen, R. Kumar, G. J. Iacobucci, M. L. Kuznicki, A. Kosterman, E. J. Bergey, P. N. Prasad and S. Gunawardena, PLoS One, 2012, 7, e29424 CrossRef CAS PubMed.
- C. T. J. Ferguson, A. A. Al-Khalaf, R. E. Isaac and O. J. Cayre, PLoS One, 2018, 13, e0201294 CrossRef PubMed.
- T. T. Su, Wiley Interdiscip. Rev.: Dev. Biol., 2019, 8, e346 Search PubMed.
- S. Jiang, C. P. Teng, W. C. Puah, M. Wasser, K. Y. Win and M.-Y. Han, ACS Biomater. Sci. Eng., 2015, 1, 1077–1084 CrossRef CAS.
- I. Miguel-Aliaga, H. Jasper and B. Lemaitre, Genetics, 2018, 210, 357–396 CrossRef CAS PubMed.
- G. Overend, Y. Luo, L. Henderson, A. E. Douglas, S. A. Davies and J. A. T. Dow, Sci. Rep., 2016, 6, 27242 CrossRef CAS PubMed.
- M. Strand and C. A. Micchelli, PLoS One, 2013, 8, e80608 CrossRef PubMed.
- S. Shanbhag and S. Tripathi, J. Exp. Biol., 2009, 212, 1731–1744 CrossRef CAS PubMed.
- G. Chen, Z. Teng, X. Su, Y. Liu and G. Lu, J. Biomed. Nanotechnol., 2015, 11, 722–729 CrossRef CAS PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- T. Ribeiro, E. Coutinho, A. S. Rodrigues, C. Baleizão and J. P. S. Farinha, Nanoscale, 2017, 9, 13485–13494 RSC.
- T. Gross, M. Ramm, H. Sonntag, W. Unger, H. M. Weijers and E. H. Adem, Surf. Interface Anal., 1992, 18, 59–64 CrossRef CAS.
- R. A. Zangmeister, T. A. Morris and M. J. Tarlov, Langmuir, 2013, 29, 8619–8628 CrossRef CAS PubMed.
- W. Tamakloe, D. A. Agyeman, M. Park, J. Yang and Y.-M. Kang, J. Mater. Chem. A, 2019, 7, 7396–7405 RSC.
- V. B. Damodaran, C. J. Fee, T. Ruckh and K. C. Popat, Langmuir, 2010, 26, 7299–7306 CrossRef CAS PubMed.
- K. C. Popat, S. Sharma and T. A. Desai, J. Phys. Chem. B, 2004, 108, 5185–5188 CrossRef CAS.
- R. Casasús, E. Climent, M. D. Marcos, R. Martínez-Máñez, F. Sancenón, J. Soto, P. Amorós, J. Cano and E. Ruiz, J. Am. Chem. Soc., 2008, 130, 1903–1917 CrossRef PubMed.
- S.-A. Yang, S. Choi, S. M. Jeon and J. Yu, Sci. Rep., 2018, 8, 185 CrossRef PubMed.
- H. J. Bellen, C. Tong and H. Tsuda, Nat. Rev. Neurosci., 2010, 11, 514–522 CrossRef CAS PubMed.
- A. Singh and K. D. Irvine, Dev. Dyn., 2012, 241, 1–2 CrossRef PubMed.
- Q. Liu and L. H. Jin, Frontiers in Cell and Developmental Biology, 2017, 5, 29 Search PubMed.
- B.-H. Mao, Z.-Y. Chen, Y.-J. Wang and S.-J. Yan, Sci. Rep., 2018, 8, 2445 CrossRef PubMed.
- V. Sundararajan, P. Dan, A. Kumar, G. D. Venkatasubbu, S. Ichihara, G. Ichihara and S. S. Mohideen, Appl. Surf. Sci., 2019, 490, 70–80 CrossRef CAS.
- D. J. Gorth, D. M. Rand and T. J. Webster, Int. J. Nanomed., 2011, 6, 343–350 CAS.
- P. Dan, V. Sundararajan, H. Ganeshkumar, B. Gnanabarathi, A. K. Subramanian, G. D. Venkatasubu, S. Ichihara, G. Ichihara and S. Sheik Mohideen, Appl. Surf. Sci., 2019, 484, 568–577 CrossRef CAS.
- M. C. Chifiriuc, A. C. Ratiu, M. Popa and A. A. Ecovoiu, Int. J. Mol. Sci., 2016, 17, 36 CrossRef PubMed.
- A. Raj, P. Shah and N. Agrawal, Sci. Rep., 2017, 7, 15617 CrossRef PubMed.
- A. Raj, P. Shah and N. Agrawal, PLoS One, 2017, 12, e0178051 CrossRef PubMed.
- V. K. Dimitriadis, J. Insect Physiol., 1991, 37, 167–177 CrossRef.
- S. Denecke, L. Swevers, V. Douris and J. Vontas, Insect Biochem. Mol. Biol., 2018, 103, 22–35 CrossRef CAS PubMed.
- W. H. Karasov, J. Exp. Biol., 2017, 220, 2495–2501 CrossRef PubMed.
- H. A. MacMillan, G. Y. Yerushalmi, S. Jonusaite, S. P. Kelly and A. Donini, Sci. Rep., 2017, 7, 8807 CrossRef PubMed.
- R. J. Katzenberger, S. Chtarbanova, S. A. Rimkus, J. A. Fischer, G. Kaur, J. M. Seppala, L. C. Swanson, J. E. Zajac, B. Ganetzky and D. A. Wassarman, Elife, 2015, 4, e04790 CrossRef PubMed.
- L. Fiandra, M. Casartelli, G. Cermenati, N. Burlini and B. Giordana, J. Insect Physiol., 2009, 55, 10–18 CrossRef CAS PubMed.
- M. A. C. Stuart, W. T. S. Huck, J. Genzer, M. Müller, C. Ober, M. Stamm, G. B. Sukhorukov, I. Szleifer, V. V. Tsukruk, M. Urban, F. Winnik, S. Zauscher, I. Luzinov and S. Minko, Nat. Mater., 2010, 9, 101–113 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra11019d |
| ‡ Equal contribution first author. |
| This journal is © The Royal Society of Chemistry 2020 |