Open Access Article
Open Access ArticleRecent advances in the chemistry and biology of azaphilones
Chunmei Chenac,
Huaming Taod,
Weihao Chenac,
Bin Yanga,
Xuefeng Zhou a,
Xiaowei Luo
a,
Xiaowei Luo *b and
Yonghong Liu*abce
*b and
Yonghong Liu*abce
aCAS Key Laboratory of Tropical Marine Bio-resources and Ecology, Guangdong Key Laboratory of Marine Materia Medica, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, P. R. China. E-mail: yonghongliu@scsio.ac.cn; Fax: +86-20-89023174; Tel: +86-20-89023174
bInstitute of Marine Drugs, Guangxi University of Chinese Medicine, Nanning 530200, P. R. China. E-mail: luoxiaowei1991@126.com; Fax: +86-771-4733826; Tel: +86-771-4733826
cUniversity of Chinese Academy of Sciences, Beijing 100049, P. R. China
dSchool of Traditional Chinese Medicine, Southern Medical University, Guangzhou 510515, P. R. China
eWuya College of Innovation, Shenyang Pharmaceutical University, Shenyang 110016, P. R. China
First published on 10th March 2020
Abstract
Azaphilones have continuously aroused considerable attention owing to their structural diversity and significant biological activities recently. This review attempts to give a comprehensive summary of recent progress on the isolation, identification, and biological activity, along with synthetic and biosynthetic studies of azaphilones reported from October 2012 to December 2019. Herein, a total of 252 compounds predominantly originated from 32 genera of fungi, such as Penicillium (20%) and Talaromyces (11%), were included in this research with citations of 105 references. Among these azaphilones, approximately half of them were found with various biological activities, of which over 40% displayed cytotoxic/anti-tumor effects. This review would shed light on the future development and research of azaphilones.
1. Introduction
Natural products have been evidenced as important sources of drug entities and 49% of small molecules functioning as antitumor agents are directly or indirectly derived from natural products.1 Polyketides represent one of the most structurally diverse classes of natural products arising from simple aromatics to highly modified complex ones, meanwhile fungi have been regarded as a major source of bioactive polyketides.2Azaphilones or azaphilonoids, known as fungal pigments, are also fungal polyketides, featuring the isochroman scaffold containing a pyrone–quinone bicyclic core and a quaternary carbon center.3,4 Besides, they have been proved with various activities, such as enzyme inhibitions, antimicrobial, cytotoxic, antioxidative, and anti-inflammatory activities.3,4 Due to their structural diversity and promising bioactivity, in 2013, J. Gao et al. summarized the information on 373 azaphilones of 18 categories covered between the end of 1932 and September 2012.3
Recently, a series of azaphilones with novel structures and remarkable bioactivities were reported. Penicilones A–D (192–195) with different configurations at the quaternary carbon center, were found with anti-MRSA activity.5 Peyronellones A and B (21–22) were identified as a pair of unusual tetracyclic caged adducts of azaphilones and pyruvic acid with hypoxia-protective activity.6 Four unusual dimers of azaphilones and furanone derivatives via Michael addition, citrifurans A–D (24–27), were found with inhibitory activities against LPS-induced NO production.7
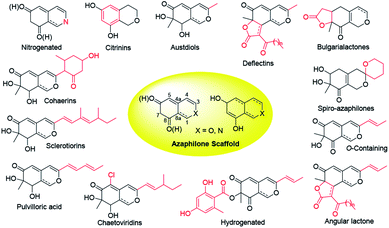
Over last seven years, 252 newly reported naturally-derived azaphilones are classified into 13 types based on structural patterns (Fig. 1), including citrinin-types, austdiols, deflectin-types, bulgarialactone-types, hydrogenated spiro-azaphilones, O-containing Monascus pigments, angular lactone-types, hydrogenated ones, chaetoviridins and chaephilones, pulvilloric acid-types, sclerotiorins, cohaerins, and nitrogenated ones.3 They were majorly obtained from fungi examplified by Penicillium sp. and Talaromyces sp. with a large range of biological activities, such as cytotoxicity,8 antitumor activity,9 antimicrobial activity,10 anti-inflammatory,11 enzyme inhibitions,12 antioxidant activity,6 antiviral activity,13,14 antileishmanial activity,15 brine shrimp toxicity,16 insecticidal activity,17 and hypoxia-protective activity.6 Given the continuing interests of azaphilones arising from chemists and pharmacologists, recent advances in the chemistry and biology of azaphilones covered from October 2012 to December 2019 were concluded in this review, focusing on fungal sources, isolation, structural identification, biological activities, chemical synthesis and biosynthesis.
2. Azaphilones
2.1 Citrinin-type azaphilones
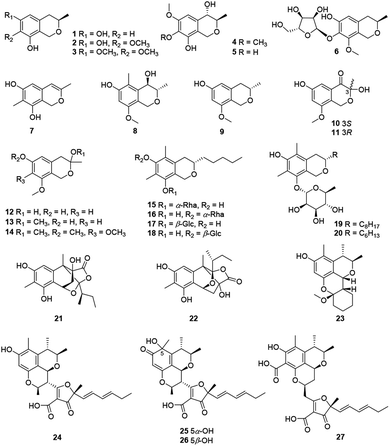
Recently, twenty-seven citrinin-type azaphilones (1–27) were isolated from several fungal strains (Fig. 2 and Table 1). Most of them were monomeric citrinin derivatives like annulohypoxylomans A–C (1–3),11 annulohypoxylomanols A and B (4–5),11 annulohypoxyloside (6),11 and the novel pentaketide (7).18| No. | Species | Activity |
|---|---|---|
| Annulohypoxylomans A–C (1–3) | Annulohypoxylon truncatum JS540![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11 11 |
|
| Annulohypoxylomanols A–B (4–5) | Annulohypoxylon truncatum JS540![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11 11 |
|
| Annulohypoxyloside (6) | Annulohypoxylon truncatum JS540![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11 11 |
|
| 7 | Talaromyces stipitatus ATCC 10500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 18 18 |
|
| (3S,4R)-6-Hydroxy-8-methoxy-3,5-dimethyl isochromanol (8) | Penicillium sp.19 | Cytotoxic (L5178Y: 26.20% at 10 μg mL−1) |
| (3S)-6-Hydroxy-8-methoxy-3-methylisochroman (9) | Penicillium sp.19 | Cytotoxic (L5178Y: 16.10% at 10 μg mL−1) |
| (S)-3,6-Dihydroxy-8-methoxy-3-methylisochroman-4-one (10) | Aspergillus fumigatus20 | Cytotoxic (MV4-11: 38.39 μM) |
| (R)-3,6-Dihydroxy-8-methox-3-methylisochroman-4-one (11) | Aspergillus fumigatus20 | |
| 8-Methoxy-3-methylisochromane-3,6-diol (12) | Aspergillus fumigatus20 | Cytotoxic (MV4-11: 30.00 μM) |
| 3,8-Dimethoxy-3-methylisochroman-6-ol (13) | Aspergillus fumigatus20 | |
| 3,6,7,8-Tetramethoxy-3-methylisochromane (14) | Aspergillus fumigatus20 | |
| Colletobredin A (15) | Colletotrichum aotearoa BCRC 09F0161![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21 21 |
Anti-inflammatory (inhibit NO production: 182.2 μM) |
| Colletobredins B–D (16–18) | Colletotrichum aotearoa BCRC 09F0161![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21 21 |
|
| Monascuspilorin (19) | Monascus pilosus BCRC 38072![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22 22 |
|
| Monascupurpurin (20) | Monascus purpureus BCRC 31499![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23 23 |
|
| Peyronellone A (21) | Peyronellaea glomerate6 | Antioxidant (ABTS: 88.8 μM, DPPH: 126.3 μM) |
| Peyronellone B (22) | Peyronellaea glomerate6 | Antioxidant (ABTS: 95.2 μM, DPPH: 83.5 μM); hypoxia-protective activity (improved the survival rate of H/R-treated human umbilical vein endothelial cells from 35% to 70% at 5 μM) |
| Penicitol A (23) | Penicillium chrysogenum HND11-24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24 24 |
Cytotoxic (HeLa: 4.6 μM, BEL-7402: 10.5 μM, HEK-293: 6.7 μM, HCT-116: 5.6 μM, A549: 7.6 μM) |
| Citrifuran A (24) | Aspergillus sp.7 | Anti-inflammatory (inhibit NO production: 18.3 μM) |
| Citrifuran B (25) | Aspergillus sp.7 | Anti-inflammatory (inhibit NO production: 22.6 μM) |
| Citrifuran C (26) | Aspergillus sp.7 | Anti-inflammatory (inhibit NO production: 25.3 μM) |
| Citrifuran D (27) | Aspergillus sp.7 | |
| Fusaraisochromenone (28) | Fusarium sp. PDB51F5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 25 |
|
| Felinone A (29) | Beauveria felina EN-135![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 16 16 |
Brine shrimp toxic (61.4% at 100 μg mL−1) |
| Xylariphilone (30) | Xylariales sp. PSU-ES163,30 Hypoxylon sp. BCRC 12F0687![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 31 31 |
Anti-inflammatory (inhibit NO production: 17.5 μM,31 inhibit IL-6 production: 5.3 μM, inhibit IL-12 p40 production: 19.4 μM); anti-tumor (inhibit TNF-α production: 37.6 μM)11 |
| Aspergillusone C (31) | Aspergillus clavatus32 | Cytotoxic (MCF-7: 2.5 μM, A549: 41.9 μM) |
| 7(S)-7-Hydroxy-3,7-dimethyl-isochromene-6,8-dione (32) | Nigrospora sp. YE3033![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 26 26 |
|
| Chlamyphilone (33) | Pochonia chlamydosporia17 | Insecticidal (Acyrthosiphon pisum: LD50 = 175 μg mL−1, MIC = 150 μg mL−1) |
| Nemanecins A–C (34–36) | Nemania sp. BCC 30850![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 27 27 |
|
| Perangustols A–B (37–38) | Cladosporium perangustum FS62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 28 28 |
|
| Dothideomynone C (40) | Dothideomycete sp. CRI7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33 33 |
Cytotoxic (A549: 46.5 μg mL−1, HuCCA-1: 48.1 μg mL−1, MOLT-3: 17.4 μg mL−1) |
| Dothideomynones B (39), D–F (41–43) | Dothideomycete sp. CRI7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 34 34 |
|
| Mycoleptone A (44) | Mycoleptodiscus indicus15 | Cytotoxic (PC3: 10.0 μM); antileishmanial (Leishmania major: 28.5 μM) |
| Mycoleptone B (45) | Mycoleptodiscus indicus15 | Cytotoxic (PC3: 7.1 μM); antileishmanial (Leishmania major: 21.7 μM) |
| Mycoleptone C (46) | Mycoleptodiscus indicus15 | |
| Deflectin C1 (47) | Aspergillus deflectus NCC0415![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 36 36 |
Inhibit enzyme (SHP2: 29.3 μM, PTP1B: 40.4 μM) |
| Deflectin C2 (48) | Aspergillus deflectus NCC0415![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 36 36 |
Inhibit enzyme (SHP2: 21.1 μM, PTP1B: 19.8 μM) |
| Deflectin C3 (49) | Aspergillus deflectus NCC0415![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 36 36 |
Inhibit enzyme (SHP2: 16.2 μM, PTP1B: 16.5 μM) |
| Deflectin D1 (50) | Aspergillus deflectus NCC0415![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 36 36 |
Inhibit enzyme (SHP2: 19.2 μM, PTP1B: 19.2 μM) |
| Deflectin D2 (51) | Aspergillus deflectus NCC0415![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 36 36 |
Inhibit enzyme (SHP2: 7.0 μM, PTP1B: 6.1 μM) |
| Deflectin E (52) | Aspergillus deflectus NCC0415![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 36 36 |
Inhibit enzyme (SHP2: 16.0 μM, PTP1B: 24.0 μM) |
| 8,11-Didehydrochermesinone B (53) | Nigrospora sp. YE3033![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 26 26 |
|
| Colletotrichone A (54) | Colletotrichum sp. BS4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 37 |
Antimicrobial (Bacillus subtilis: 0.1 μg mL−1, Escherichia coli: 1.0 μg mL−1) |
| Colletotrichone B (55) | Colletotrichum sp. BS4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 37 |
Antimicrobial (Staphylococcus aureus: 5.0 μg mL−1) |
| Colletotrichone C (56) | Colletotrichum sp. BS4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 37 |
Antimicrobial (Escherichia coli: 5.0 μg mL−1) |
| Coniellin A (57) | Coniella fragariae9 | Anti-tumor (MDA-MB-231: 21.5 μM, inhibit NF-κB activation: 4.4 μM) |
| Coniellin B (58) | Coniella fragariae9 | Anti-tumor (MDA-MB-231: 19.6 μM) |
| Coniellin C (59) | Coniella fragariae9 | Anti-tumor (MDA-MB-231: 21.0 μM) |
| Coniellin D (60) | Coniella fragariae9 | Anti-tumor (MDA-MB-231: 18.6 μM, inhibit NF-κB activation: 37.8 μM) |
| Coniellin E (61) | Coniella fragariae9 | Anti-tumor (MDA-MB-231: 79.3 μM, inhibit NF-κB activation: 29.4 μM) |
| Coniellin F (62) | Coniella fragariae9 | Anti-tumor (inhibit NF-κB activation: 70.7 μM) |
| Coniellin G (63) | Coniella fragariae9 | Anti-tumor (MDA-MB-231: 21.6 μM, inhibit NF-κB activation: 11.3 μM) |
| Coniellins H–I (64–65) | Coniella fragariae35 | |
| 5′,6′-Dihydroxyacetosellin (66) | Epicoccum nigrum strain 749![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38 38 |
|
| Monakaocinol (67) | Monascus kaoliang39 | |
| Monascuspirolide A (68) | Monascus purpureus BCRC 38110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 42 42 |
Anti-inflammatory (inhibit NO production: 17.5 μM) |
| Monascuspirolide B (69) | Monascus purpureus BCRC 38110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 42 42 |
Anti-inflammatory (inhibit NO production: 23.5 μM) |
| Thielavialides A–E (70–74) | Thielavia sp. PA0001![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 40 |
|
| 5-O-Acetyl-epi-pestafolide A (75) | Trichocladium sp.41 | |
| 5-epi-Pestafolide A (76) | Trichocladium sp. co-cultured with Bacillus subtilis41 | |
| Peniazaphilin A (77) | Penicillium sp. CPCC 400786![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13 13 |
Antiviral (HIV: 60.4 μM) |
| Monapilosusazaphilone (78) | Monascus pilosus103 | |
| Monascusazaphilone A (79) | Monascus purpureus BCRC 38108![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
Anti-inflammatory (inhibit NO production: 4.6 μg mL−1); cytotoxic (LPS-induced RAW264.7: cell viability 83%) |
| Monascusazaphilone B (80) | Monascus purpureus BCRC 38108![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
Anti-inflammatory (inhibit NO production: 8.88 μg mL−1); cytotoxic (LPS-induced RAW264.7: cell viability 85%) |
| Berkchaetoazaphilone C (81) | Pleurostomophora sp.49 | |
| Monascuskaodione (82) | Monascus kaoliang44 | |
| Monascuspurone (83) | Monascus ruber45 | |
| Monasfluol B (84) | Monascus spp.46 | |
| Monascusazaphilone C (85) | Monascus purpureus BCRC 38108![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
Anti-inflammatory (inhibit NO production: 6.77 μg mL−1); cytotoxic (LPS-induced RAW264.7: cell viability 86%) |
| Acetyl-monasfluol B (86) | Monascus ruber47 | |
| MC-2, MC-4 (87–88) | Monascus purpureus (mppC mutant)48 | |
| Berkchaetoazaphilone A (89) | Pleurostomophora sp.49 | Anti-tumor (inhibit TNF-α production: 95% at 100 μM, inhibit IL-1β production: 95% at 100 μM); anti-inflammatory (inhibit IL-6 production: 100% at 100 μM, inhibit IL-33 production: 100% at 100 μM); inhibit enzyme (caspase 1: 150 μM, MMP-3: 130 μM) |
| Berkchaetoazaphilone B (90) | Pleurostomophora sp.49 | Cytotoxic (Y79: 1.1 μM, MOLT-4: 10 μM, RPMI-8226: 10 μM, SR: 10 μM, LOX IMVI: 10 μM, CCRF-CEM: 10 μM); anti-tumor (inhibit TNF-α production: 95% at 100 μM, inhibit IL-1β production: 95% at 100 μM); anti-inflammatory (inhibit IL-6 production: 100% at 100 μM, inhibit IL-33 production: 100% at 100 μM); inhibit enzyme (caspase 1: 25 μM, MMP-3: 15 μM) |
| Lenormandin A (91) | Hypoxylon lenormandii50 | Antimicrobial (Rhodotorula glutinis: 33.3 μg mL−1, Bacillus subtilis: 67.0 μg mL−1, Staphylococcus aureus: 67.0 μg mL−1) |
| Lenormandin B (92) | Hypoxylon lenormandii50 | Cytotoxic (L929: 18.0 μg mL−1); antimicrobial (Mycobacterium sp.: 67.0 μg mL−1, Rhodotorula glutinis 67.0 μg mL−1) |
| Lenormandin C (93) | Hypoxylon lenormandii50 | Cytotoxic (L929: 32.0 μg mL−1); antimicrobial (Rhodotorula glutinis: 67.0 μg mL−1) |
| Lenormandin D (94) | Hypoxylon lenormandii50 | Antimicrobial (Bacillus subtilis: 67.0 μg mL−1, Staphylococcus aureus: 67.0 μg mL−1) |
| Lenormandin E (95) | Hypoxylon lenormandii50 | Cytotoxic (L929: 22.0 μg mL−1); antimicrobial (Rhodotorula glutinis: 67.0 μg mL−1) |
| Peyronellone F (96) | Peyronellaea glomerate6 | |
| Lenormandin F (97) | Hypoxylon lenormandii50 | Antimicrobial (Rhodotorula glutinis: 67.0 μg mL−1) |
| Lenormandin G (98) | Hypoxylon lenormandii50 | Antimicrobial (Rhodotorula glutinis: 67.0 μg mL−1) |
| Phialomustin A (99) | Phialophora mustea52 | Cytotoxic (MIAPaCa2: 35 μM, A549: 98 μM, HCT-116: 8 μM, T47D: 10 μM) |
| Phialomustin C (100) | Phialophora mustea52 | Cytotoxic (MIAPaCa2: 38 μM, HCT-116: 100 μM, T47D: 7 μM); antimicrobial (Candida albicans: 14.3 μM, Aspergillus fumigatus: 60.6 μM, Aspergillus parasiticus: 35.2 μM, Aspergillus flavus: 88.4 μM) |
| Phialomustin D (101) | Phialophora mustea52 | Cytotoxic (MIAPaCa2: 60 μM, HCT-116: 30 μM, T47D: 9.2 μM); antimicrobial (Candida albicans: 73.6 μM) |
| Fragirubrins A–E (102–106) | Hypoxylon fragiforme51 | |
| (+)-6′′-Hydroxymitorubrinol acetate (107) | Hypoxylon rubiginosum56 | Cytotoxic (L929: 21 μg mL−1) |
| (+)-6′′-Hydroxymitorubrinol (108) | Hypoxylon rubiginosum56 | |
| 6′′-Hydroxy-(R)-mitorubrinic acid (109) | Aspergillus sp. 16-5C53 | |
| Purpurquinone D (110) | Aspergillus sp. 16-5C53 | |
| Talarophilones A–B (111–112) | Talaromyces sp. CMB-W045![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 54 54 |
|
| Pinophilins D–F (113–115) | Penicillium pinophilum XS-20090E18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 55 55 |
|
| Rutilins C–D (116–117) | Hypoxylon fragiforme51 | |
| Pinazaphilone A (118) | Penicillium sp. HN29-3B1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 12 |
|
| Montagnuphilone B (120) | Montagnulaceae sp. DM0194![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 57 57 |
Anti-inflammatory (inhibit NO production: 39.58 μM) |
| Montagnuphilones A (119), C–D (121–122), F–G (124–125) | Montagnulaceae sp. DM0194![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 57 57 |
|
| Montagnuphilone E (123) | Montagnulaceae sp. DM0194![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 57 57 |
Anti-inflammatory (inhibit NO production: 25.48 μM) |
| Glaziellin A (126) | The fruiting body of Glaziella splendens14 | Antiviral (H1N1: 230.6 μM, H3N2: 235.8 μM, H5N1: 165.4 μM) |
| Comazaphilone G (127) | Penicillium variabile58 | Anti-inflammatory (inhibit NO production: 4.35 μM) |
| Comazaphilone H (128) | Penicillium variabile58 | Anti-inflammatory (inhibit NO production: 40.52 μM) |
| Pinazaphilone B (129) | Penicillium sp. HN29-3B1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 12 |
Inhibit enzyme (α-glucosidase: 28.0 μM) |
| Pinophilin G (130) | Penicillium pinophilum59 | Antimicrobial (Vibrio parahemolyticus: 25.0 μM) |
| Talaraculone A (131) | Talaromyces aculeatus60 | Inhibit enzyme (α-glucosidase: 78.6 μM) |
| Talaraculone B (132) | Talaromyces aculeatus60 | Antimicrobial (Vibrio anguillarum: 0.26 μg mL−1); inhibit enzyme (α-glucosidase: 22.9 μM) |
| Talaraculones C–F (133–136) | Talaromyces aculeatus60 | |
| Pleosporalone E (137) | Pleosporales sp. CF09-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 61 61 |
Antimicrobial (Vibrio alginolyticus: 25 μg mL−1) |
| Pleosporalone F (138) | Pleosporales sp. CF09-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 61 61 |
Antimicrobial (Vibrio alginolyticus: 25 μg mL−1) |
| Pleosporalone G (139) | Pleosporales sp. CF09-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 61 61 |
Antimicrobial (Vibrio anguillarum: 13 μg mL−1, Vibrio parahemolyticus: 6.3 μg mL−1) |
| Pleosporalone H (140) | Pleosporales sp. CF09-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 61 61 |
Antimicrobial (Vibrio anguillarum: 6.3 μg mL−1, Vibrio parahemolyticus: 25 μg mL−1) |
| epi-Pinophilin B (141) | Aspergillus fumigatus 14–27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 104 104 |
|
| Chaetoviridin J (142) | Chaetomium globosum63 | Anti-tumor (inhibit NF-κB activation: 32.6% at 50 μM); anti-inflammatory (inhibit NO production: 95.4%, at 50 μM) |
| Chaetoviridin K (143) | Chaetomium globosum63 | Anti-tumor (inhibit NF-κB activation: 33.4% at 50 μM); anti-inflammatory (inhibit NO production: 39.4%, at 50 μM) |
| Chaephilones A–B (144–145) | Chaetomium globosum62 | |
| Chaephilone C (146) | Chaetomium sp. NA-S01-R1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 64 64 |
Cytotoxic (A549: 15.7 μM, HeLa: 7.7 μM, Hep G2: 20.2 μM); antimicrobial (Vibrio vulnificus: 32.2 μg mL−1, Vibrio campbellii: 30.1 μg mL−1, MRSA: 7.6 μg mL−1) |
| Nigbeauvin A (147) | Nigrospora oryzae co-cultured with Beauveria bassiana65 | Antimicrobial (Bacillus bassiana: 128 μg mL−1, Nigrospora oryzae: 512 μg mL−1, Bacillus subtilis: 128 μg mL); anti-inflammatory (inhibit NO production: 37%, at 50 μM) |
| Nigbeauvin B (148) | Nigrospora oryzae co-cultured with Beauveria bassiana65 | Anti-inflammatory (inhibit NO production: 39%, at 50 μM) |
| Nigbeauvins C–E (149–151) | Nigrospora oryzae co-cultured with Beauveria bassiana65 | |
| Nigirpexins A–C (152–154) | Nigrospora oryzae co-cultured with Irpex lacteus66 | |
| Nigirpexin D (155) | Nigrospora oryzae co-cultured with Irpex lacteus66 | Antimicrobial (Irpex lacteus: 256 μg mL−1, Nigrospora oryzae: 512 μg mL−1, Bacillus subtilis: 512 μg mL−1) |
| Isonigirpexin C (156) | Nigrospora oryzae cocultured with Irpex lacteus67 | |
| Dechloroisochromophilone II (157) | Penicillium multicolor CM01![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 70 |
|
| epi-Isochromophilone III (158) | Penicillium multicolor CM01![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 70 |
Cytotoxic (NCl-H187: 6.2 μg mL−1, KB: 6.9 μg mL−1, MCF-7: 10.6 μg mL−1) |
| 159 | Penicillium 303![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 71 71 |
Cytotoxic (MDA-MB-435: 24.62 μg mL−1, HepG2: 17.92 μg mL−1, HCT-116: 11.09 μg mL−1, A549: 16.63 μg mL−1) |
| Hypocrellone A (160) | Hypocrella sp. isolate WYTY-21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 72 72 |
|
| Eupenicilazaphilone A (161) | Eupenicillium sp. 6A-9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 73 73 |
Cytotoxic (MCF-7: 49.95 μM) |
| Eupenicilazaphilone B (162) | Eupenicillium sp. 6A-9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 73 73 |
Cytotoxic (MCF-7: 40.71 μM, A549: 63.32 μM) |
| Eupenicilazaphilone C (163) | Eupenicillium sp. 6A-9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 73 73 |
Cytotoxic (MCF-7: 36.88 μM, A549: 43.96 μM) |
| Geumsanols A–B (164–165), D–E (167–168) | Penicillium sp. KCB11A109![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 74 74 |
|
| Geumsanol C (166) | Penicillium sp. KCB11A109![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 74 74 |
Cytotoxic (HL-60: 88.9 μM) |
| Penidioxolanes A–B (169–170) | Penicillium sp. KCB12C078![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 68 68 |
|
| Penicilazaphilone C (171) | Penicillium sclerotiorum M-22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75 75 |
Cytotoxic (B-16: 0.065 mM, SGC7901: 0.720 mM); antimicrobial (Staphylococcus aureus: 31.25 μg mL−1, Pseudomonas aeruginosa: 62.5 μg mL−1, Klebsiella pneumonia: 16.53 μg mL−1, Escherichia coli: 16.53 μg mL−1) |
| Penicilazaphilones D–E (172–173) | Penicillium sclerotiorum76 | |
| Sclerketide B (174) | Penicillium sclerotiorum CHNSCLM-0013![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 77 77 |
Anti-inflammatory (inhibit NO production: 3.4 μM) |
| Helicusin E (175) | Bartalinia robillardoides strain LF550![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 78 78 |
|
| Isochromophilone XI (176) | Bartalinia robillardoides strain LF550![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 78 78 |
Antimicrobial (Bacillus subtilis: 55.6 μM, Staphylococcus lentus: 78.4 μM, Trichophyton rubrum: 41.5 μM); inhibit enzyme (PDE4: 8.30 μM) |
| Bromophilone A (177) | Penicillium canescens79 | Cytotoxic (L5178Y: 13.9 μM, A2780: 37 μM) |
| Bromophilone B (178) | Penicillium canescens79 | Cytotoxic (L5178Y: 8.9 μM, A2780: 2.7 μM) |
| Isochromophilonol (179) | Chaetomium cupreum RY202![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 105 105 |
Cytotoxic (KB: 9.63 μg mL−1, NCI-H187: 27.18 μg mL−1) |
| Ochrephilonol (180) | Chaetomium cupreum RY202![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 105 105 |
Cytotoxic (KB: 30.2 μg mL−1) |
| Isochromophilone A (181) | Diaporthe sp. SCSIO 41011![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8 |
Cytotoxic (ACHN: 27 μM, 786-O: 34 μM, OS-RC-2: 45 μM) |
| Isochromophilones B–C (182–183) | Diaporthe sp. SCSIO 41011![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8 |
|
| Isochromophilone D (184) | Diaporthe sp. SCSIO 41011![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8 |
Cytotoxic (786-O: 38 μM, OS-RC-2: 44 μM) |
| Isochromophilone E (185) | Diaporthe sp. SCSIO 41011![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8 |
Cytotoxic (ACHN: 14 μM, 786-O: 8.9 μM, OS-RC-2: 13 μM) |
| Isochromophilone F (186) | Diaporthe sp. SCSIO 41011![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8 |
Cytotoxic (ACHN: 13 μM, 786-O: 10 μM, OS-RC-2: 38 μM) |
| Sclerotiorins A–C (187–189) | Penicillium sclerotiorum OUCMDZ-3839![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 69 69 |
|
| Cohaerin G (190) | Annulohypoxylon cohaerens81 | Antimicrobial (Nocardia sp.: 33.3 μg mL−1) |
| Cohaerin H (191) | Annulohypoxylon cohaerens81 | Cytotoxic (L929: 4.9 μg mL−1) |
| Cohaerin I (192) | Annulohypoxylon cohaerens81 | Cytotoxic (L929: 0.4 μg mL−1); antimicrobial (Nocardia sp.: 33.3 μg mL−1) |
| Cohaerin K (193) | Annulohypoxylon cohaerens81 | Antimicrobial (Nocardia sp.: 16.6 μg mL−1, Staphylococcus aureus: 16.6 μg mL−1) |
| Minutellin A (194) | Annulohypoxylon minutellum82 | Cytotoxic (L929: 5.1 μg mL−1, KB3.1: 5.3 μg mL−1); antimicrobial (Micrococcus luteus: 16.7 μg mL−1, Bacillus subtilis: 66.7 μg mL−1) |
| Minutellin B (195) | Annulohypoxylon minutellum82 | Antimicrobial (Bacillus subtilis: 66.7 μg mL−1) |
| Minutellin C (196) | Annulohypoxylon minutellum82 | Cytotoxic (L929: 10 μg mL−1, KB3.1: 25 μg mL−1); antimicrobial (Micrococcus luteus: 66.7 μg mL−1, Bacillus subtilis: 66.7 μg mL−1, Rhodoturula glutinis: 33.3 μg mL−1, Mucor hiemalis: 66.7 μg mL−1) |
| Minutellin D (197) | Annulohypoxylon minutellum82 | Antimicrobial (Micrococcus luteus: 16.7 μg mL−1, Bacillus subtilis: 33.3 μg mL−1, Staphylococcus aureus: 33.3 μg mL−1) |
| Penicilone A (198) | Penicillium janthinellum HK1-6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
|
| Penicilone B (199) | Penicillium janthinellum HK1-6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
Cytotoxic (SMMC-7721: 32 μM); antimicrobial (Staphylococcus aureus: 3.13 μg mL−1, Enterococcus faecalis: 3.13 μg mL−1, Enterococcus faecium: 3.13 μg mL−1, Escherichia coli: 3.13 μg mL−1)83 |
| Penicilone C (200) | Penicillium janthinellum HK1-6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
Cytotoxic (SMMC-7721: 21 μM); antimicrobial (Staphylococcus aureus: 6.25–12.5 μg mL−1, Enterococcus faecalis: 12.5 μg mL−1, Enterococcus faecium: 12.5 μg mL−1)83 |
| Penicilone D (201) | Penicillium janthinellum HK1-6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
Cytotoxic (SMMC-7721: 27 μM); antimicrobial (Staphylococcus aureus: 3.13–12.5 μg mL−1, Enterococcus faecalis: 6.25 μg mL−1, Enterococcus faecium: 12.5 μg mL−1)83 |
| Penicilone G (202) | Penicillium janthinellum HK1-6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 83 83 |
Cytotoxic (SMMC-7721: 21 μM); antimicrobial (Staphylococcus aureus: 12.5–50 μg mL−1, Enterococcus faecalis: 25 μg mL−1, Enterococcus faecium: 25 μg mL−1) |
| Penicilone H (203) | Penicillium janthinellum HK1-6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 83 83 |
Cytotoxic (SMMC-7721: 24 μM); antimicrobial (Staphylococcus aureus: 3.13–12.5 μg mL−1, Enterococcus faecalis: 3.13 μg mL−1, Enterococcus faecium: 12.5 μg mL−1) |
| Meliasendanin A (204) | The fruits of Melia toosendan84 | Antioxidant (ABTS: 62.8 μM) |
| Pleosporalone A (205) | Pleosporales sp. CF09-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 85 85 |
Antimicrobial (Botrytis cinereal: 0.39 μM, Rhizopus oryzae: 0.78 μM, Phytophthora capsici: 0.78 μM) |
| Pleosporalone B (206) | Pleosporales sp. CF09-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 61 61 |
Antimicrobial (Alternaria brassicicola: 1.6 μg mL−1, Botryosphaeria dothidea: 1.3 μg mL−1, Fusarium oxysporum: 1.6 μg mL−1) |
| Pleosporalone C (207) | Pleosporales sp. CF09-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 61 61 |
Antimicrobial (Alternaria brassicicola: 6.3 μg mL−1, Botryosphaeria dothidea: 3.1 μg mL−1, Fusarium oxysporum: 25 μg mL−1) |
| Chaetomugilide A (208) | Chaetomium globosum DAOM 240359,89 Chaetomium globosum TY1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 88 88 |
Cytotoxic (HePG2: 1.7 μM);88 antimicrobial (Pseudomonas putida: < 20 μM, Bacillus subtilis: <20 μM Saccharomyces cerevisiae: 20–200 μM)89 |
| Chaetomugilide B (209) | Chaetomium globosum TY1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 88 88 |
Cytotoxic (HePG2: 19.8 μM) |
| Chaetomugilide C (210) | Chaetomium globosum DAOM 240359,89 Chaetomium globosum TY1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 88 88 |
Cytotoxic (HePG2: 53.4 μM)88 |
| Isochromophilone XIII (211) | Chaetomium globosum DAOM 240359![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 89 89 |
Antimicrobial (Pseudomonas putida: 20–200 μM, Bacillus subtilis: 20–200 μM) |
| Chaetoviridide A (212) | Chaetomium sp. NA-S01-R1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 64 64 |
Cytotoxic (A549: 15.2 μM, HeLa: 12.3 μM, Hep G2: 3.9 μM); antimicrobial (Vibrio vulnificus: 30.5 μg mL−1, Vibrio rotiferianus: 7.3 μg mL−1, Vibrio campbellii: 32.7 μg mL−1, MRSA: 15.5 μg mL−1) |
| Chaetoviridide B (213) | Chaetomium sp. NA-S01-R1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 64 64 |
Cytotoxic (A549: 16.3 μM, HeLa: 5.6 μM, Hep G2: 18.2 μM); antimicrobial (Vibrio vulnificus: 7.4 μg mL−1, Vibrio rotiferianus: 31.3 μg mL−1, Vibrio campbellii: 32.3 μg mL−1, MRSA: 7.3 μg mL−1) |
| Chaetoviridide C (214) | Chaetomium sp. NA-S01-R1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 64 64 |
Cytotoxic (A549: 23.1 μM, HeLa: 17.7 μM, Hep G2: 22.2 μM); antimicrobial (Vibrio vulnificus: 15.7 μg mL−1, Vibrio rotiferianus: 15.3 μg mL−1, MRSA: 7.6 μg mL−1) |
| N-Glutarylchaetoviridin A (215) | Chaetomium globosum HDN151398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 90 |
Cytotoxic (HL-60: 10.3 μM, K562: 20.3 μM, BEL-7402: 23.9 μM) |
| N-Glutarylchaetoviridin B (216) | Chaetomium globosum HDN151398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 90 |
|
| N-Glutarylchaetoviridin C (217) | Chaetomium globosum HDN151398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 90 |
Cytotoxic (HL-60: 11.1 μM, K562: 11.7 μM, BEL-7402: 10.9 μM, HCT-116: 11.3 μM, HeLa: 22.1 μM, L-02: 18.2 μM, MGC-803: 6.6 μM, HO8910: 9.7 μM, SH-SY5Y: 26.5 μM, NCl-H1975: 11.2 μM, U87: 18.3 μM, MDA-MB-231: 13.2 μM) |
| Peniazaphilones A–D (218–221) | Penicillium sp. ZJ-27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 86 86 |
|
| Sclerotiorin D (222) | Penicillium sclerotiorum OUCMDZ-3839,69 Penicillium sclerotiorum CHNSCLM-0013![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 77 77 |
Anti-inflammatory (inhibit NO production: 2.7 μM)77 |
| Isochromophilone X (223) | Bartalinia robillardoides strain LF550![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 78 78 |
Inhibit enzyme (PDE4: 11.7 μM) |
| Penazaphilone A (224) | Penicillium sclerotiorum cib-411![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 91 91 |
Anti-inflammatory (inhibit NO production: 15.29 μM) |
| Penazaphilones B–D, (225–227), G (230), I (232) | Penicillium sclerotiorum cib-411![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 91 91 |
|
| Penazaphilone E (228) | Penicillium sclerotiorum cib-411![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 91 91 |
Anti-inflammatory (inhibit NO production: 9.34 μM) |
| Penazaphilone F (229) | Penicillium sclerotiorum cib-411![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 91 91 |
Anti-inflammatory (inhibit NO production: 9.50 μM) |
| Penazaphilone H (231) | Penicillium sclerotiorum cib-411![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 91 91 |
Anti-inflammatory (inhibit NO production: 7.05 μM) |
| Berkchaetorubramine (233) | Pleurostomophora sp.49 | Inhibit enzyme (caspase 1: 50 μM, MMP-3: 45 μM) |
| (6-[(Z)-2-Carboxyvinyl]-N-GABA-PP-V) (234) | Talaromyces albobiverticillius 30548![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 87 87 |
|
| Atrorosins A, C–I, K–N, Q–R, T, V–W, Y (235–252) | Talaromyces atroroseus92 |
The fungus Penicillium sp., collected in the sediment of the hyper saline lake Wadi El-Natrun in Egypt, produced two novel cytotoxic citrinin analogues, (3S,4R)-6-hydroxy-8-methoxy-3,5-dimethyl isochromanol (8) and (3S)-6-hydroxy-8-methoxy-3-methylisochroman (9).19 An endophytic fungus from Cordyceps sinensis, Aspergillus fumigatus, gave five new isochromanes (10–14), among which 10 and 11 were determined as a pair of enantiomers purified by chiral HPLC methods.20 Meanwhile, 10 and 12 exhibited moderate activities against MV4-11 cell line with IC50 values of 38.4 and 30.0 μM, respectively.20
As the first report on isochroman glycoside metabolites from the genus Colletotrichum, colletobredins A–D (15–18) were extracted from Colletotrichum aotearoa BCRC 09F0161, an endophytic fungus found in the leaves of an endemic Formosan plant Bredia oldhamii Hook. f. (Melastomataceae).21 Colletobredin A (15) exhibited weak NO inhibitory activity in LPS activated murine macrophage RAW264.7 cells.21 Besides, Monascus pilosus BCRC 38072 and Monascus purpureus BCRC 31499 also produced the isochroman glycoside metabolites, monascuspilorin (19)22 and monascupurpurin (20),23 respectively.
Among citrinin dimers, peyronellones A–B (21–22), a pair of rare tetracyclic caged adducts of azaphilone with pyruvic acid, were isolated from Peyronellaea glomerate with antioxidative abilities, stereochemistry of which were deduced by Rh2(OCOCF3)4-induced ECD experiments and calculations.6 Significantly, 22 (5 μM) could inhibit hypoxia/reoxygenation (H/R)-induced late-stage apoptosis of human umbilical vein endothelial cells and displayed the same hypoxia-protective activity as the positive control verapamil.6
Chemical investigation of a mangrove-derived fungus Penicillium chrysogenum HND11-24, led to the characterization of penicitol A (23), a citrinin dimer with a novel tetracyclic skeleton, which was found with cytotoxicity against HeLa, BEL-7402, HEK-293, HCT-116, and A549 cell lines (IC50 = 4.6–10.5 μM).24 Citrifurans A–D (24–27), four uncommon heterodimers of azaphilone and furanone derivatives, were discovered from Aspergillus sp., meanwhile structure of 24 was confirmed by single-crystal X-ray diffraction, among which 24–26 displayed moderate inhibitory on NO production in RAW 264.7 macrophages with IC50 values of 18.3, 22.6, and 25.3 μM, respectively.7
2.2 Austdiol-type azaphilones
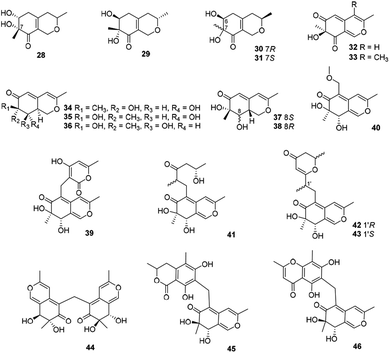
This subgroup of azaphilones is characterized by an austdiol core with nineteen members (28–46) (Fig. 3 and Table 1), including fusaraisochromenone (28),25 7(S)-7-hydroxy-3,7-dimethyl-isochromene-6,8-dione (32),26 nemanecins A–C (34–36),27 and perangustols A and B (37–38).28Felinone A (29) was obtained from a marine bryozoan derived fungus Beauveria felina EN-135 with weak activity against brine shrimp with the lethal rate of 61.4% (100 μg mL−1),16 while absolute configuration of which was revised by chemical synthesis.29 Moreover, xylariphilone (30), the diastereoisomer of 29, was metabolized by the seagrass-derived fungus Xylariales sp. PSU-ES163 and Hypoxylon sp. BCRC12F 0687,30,31 which exhibited significant inhibitory effects on the productions of NO (IC50 = 17.5 μM)31 and also demonstrated inhibitory activities towards IL-6 (IC50 = 5.3 μM), IL-12 p40 (IC50 = 19.4 μM), and TNF-α (IC50 = 37.6 μM).11 Another diastereoisomer of 29, aspergillusone C (31) was produced by Aspergillus clavatus with cytotoxicity against MCF-7 and A549 cell lines with IC50 values of 2.5 and 41.9 μM, respectively.32
Recently, Cavallo's group described the isolation and determination of an insecticidal azaphilone, chlamyphilone (33), yielded by Pochonia chlamydosporia.17 Chemical analysis of the fungus Dothideomycete sp. CRI7 afforded five austdiol-like azaphilones, dothideomynones B–F (39–43), while compounds 41–43 were obtained from the fermentation broths under deionized water supplemented with potassium bromide.33,34 Of particular note, 40 exhibited cytotoxicity against HuCCA-1, A549, and MOLT-3 with IC50 values of 48.1, 46.5, and 17.4 μg mL−1, respectively.33
Mycoleptodiscus indicus, an endophytic fungus of the officinal plant Borreria verticillate from the South American, was found to produce three azaphilones with an unusual methylene bridge, mycoleptones A–C (44–46).15 Amongst, 44 and 45 showed cytotoxicity against PC3 cell line with IC50 values of 10.0 and 7.1 μM, respectively.15
2.3 Deflectin-type azaphilones
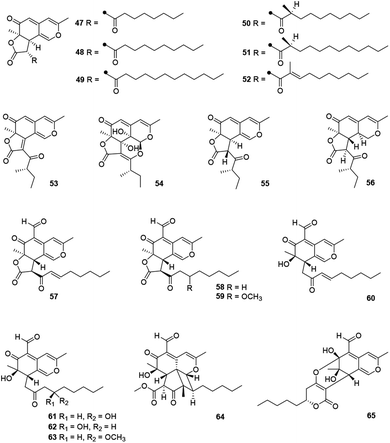
This angular deflectin-type azaphilones included fourteen compounds (47–65) (Fig. 4 and Table 1) were characterized with a methyl at C-3 and an angular γ-lactone ring, along with a ketone aliphatic chain at C-2′ like 8,11-didehydrochermesinone B (53),26 or at C-8 such as coniellins H–I (64–65).35Chromatographic separation of crude extracts from Aspergillus deflectus NCC0415 gave six deflectins with inhibitory activity against protein tyrosine phosphatases, SHP2 and PTP1B, deflectins C1–C3, D1–D2, and E (47–52), absolute configurations of which were assigned by ECD analyses and the GIAO 13C NMR calculation.36 And it was proposed that the ketone aliphatic side chain in 47–52 had an impact on the enzyme inhibition activities.36
Chemical investigations of the endophytic fungus Colletotrichum sp. BS4 resulted in the achievements of colletotrichones A–C (54–56), while 54 exhibited notable antibacterial potencies against Escherichia coli and Bacillus subtilis with MIC values of 1.0 and 0.1 μg mL−1, respectively.37 Additionally, antibacterial activity of 55 was comparable to the standard antibiotics against Staphylococcus aureus and 56 displayed inhibitory activity against Escherichia coli with the MIC value of 5.0 μg mL−1.37
Furthermore, coniellins A–I (57–65) were obtained from the goose dung derived fungus Coniella fragariae.9,35 And coniellins H and I (64–65) shared a unique tetracyclic core and an aldehyde group at C-5.35 Remarkably, 57 demonstrated inhibition of NF-κB activation in MDA-MB-231 with an IC50 value of 4.4 μM and reduced the migration of tumor cells with 60% inhibition at 5 μM and 98% inhibition at 10 μM after 24 h.9
2.4 Bulgarialactone-type azaphilones
5′,6′-Dihydroxyacetosellin (66) and monakaocinol (67) were classified into this subgroup harboring an extensively conjugated and a linear γ-lactone ring, which were isolated from the marine-derived fungi Epicoccum nigrum (strain 749)38 and Monascus kaoliang,39 respectively.2.5 Hydrogenated spiro-azaphilones
Ten compounds belonged to hydrogenated spiro-azaphilones with a five- or six-membered ring spiroketal system on the azaphilone skeleton (Fig. 5 and Table 1), such as thielavialides A–E (70–74),40 5-O-acetyl-epi-pestafolide A (75),41 and 5-epi-pestafolide A (76).41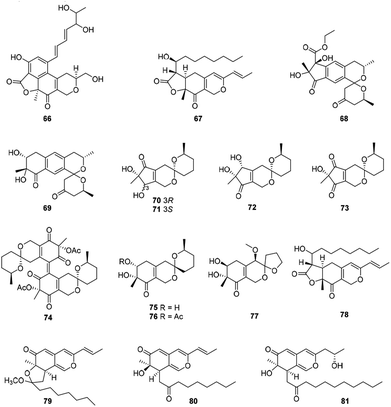 | ||
| Fig. 5 Chemical structures of bulgarialactone-type, hydrogenated spiro-azaphilones, and O-containing Monascus pigments (66–81). | ||
Herein, monascuspirolides A and B (68–69) bearing with an unique 5′,6′-dihydrospiro[isochromane-1,2′-pyran]-4′(3′H)-one pattern, were isolated from Monascus purpureus BCRC 38110. Of note, compounds 68 and 69 indicated stronger NO inhibitory activity than that of the positive control quercetin with the IC50 values of 17.5 and 23.5 μM, respectively.42 Moreover, the fungus Penicillium sp. CPCC 400786 yielded peniazaphilin A (77) with anti-HIV activity (IC50 = 60.4 μM).13
2.6 O-Containing Monascus pigments
This category consisted of four azaphilones (78–81) were obtained from Monascus sp., characterized with a 1H-isochromene skeleton linked with a propenyl side chain and an acyl chain from the acetyl to the decanoyl unit (Fig. 5 and Table 1).The cultures of the pink mutant of Monascus purpureus BCRC 38108 afforded monascusazaphilones A and B (79–80), which inhibited NO production by macrophages and the inhibition of 79 was stronger than that of the positive control quercetin.43
2.7 Angular lactone-type azaphilones
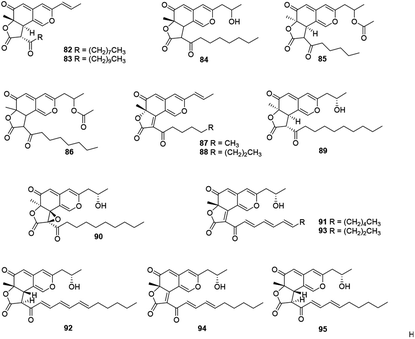
This subgroup including thirteen compounds were characterized by an angular lactone on the azaphilone nucleus with a propenyl side chain and an acyl chain (Fig. 6 and Table 1), for example, monascuskaodione (82),44 monascuspurone (83),45 monasfluol B (84),46 acetyl-monasfluol B (86),47 MC-2 (87), and MC-4 (88).48Monascusazaphilone C (85), carrying with an acetoxy group attached at the side chain, was obtained from Monascus purpureus BCRC 38108, which showed moderate NO inhibitory activity (IC50 = 6.8 μg mL−1).43 The culture broth of an acid mine extremophile strain Pleurostomophora sp. yielded two novel compounds, berkchaetoazaphilones A–B (89–90). Notably, 90 displayed significant anti-inflammatory activity by inhibiting IL-1β, TNFα, and IL-6 production and also exhibited strong cytotoxicity against several human tumor cell lines.49
A series of angular lactone-type azaphilones, named lenormandins A–E (91–95), were obtained from stromata of the xylariaceous fungus, Hypoxylon lenormandii. They were demonstrated weak antimicrobial activities and weak cytotoxicity against mouse fibroblast cell line L929.50
2.8 Hydrogenated azaphilones
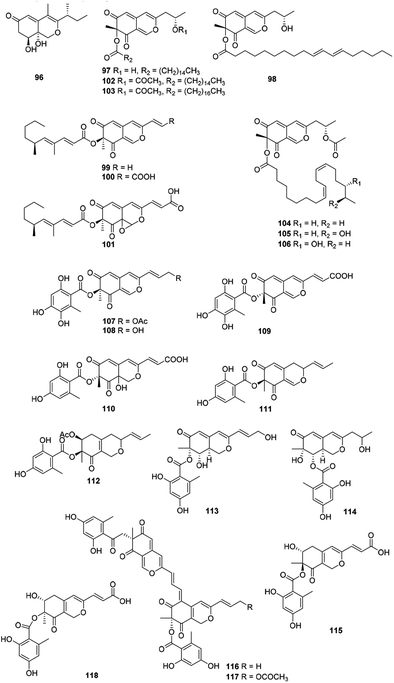
Recently, forty-six hydrogenated azaphilones (96–141) could be divided into three subgroups according to different connections of benzoyl moiety (Fig. 7, 8 and Table 1).3Phialomustins A, C, and D (99–101), three azaphilones sharing with an unprecedented skeleton, were discovered from an endophytic fungus Phialophora mustea. Interestingly, they showed moderate activities toward several human cancer cells, meanwhile 94 and 95 also displayed antifungal activities with IC50 values of 14.3 and 73.6 μM against Candida albicans, respectively.52
Apart from these, (+)-6′′-hydroxymitorubrinol acetate (107) and (+)-6′′-hydroxymitorubrinol (108), were encountered in Hypoxylon rubiginosum, a novel species from northern Thailand. Remarkably, 107 exhibited activity against the mouse fibroblast cell line L929 (IC50 = 21 μg mL−1), whereas 108 was inactive.56
Montagnulaceae sp. DM0194, a fungal endophyte from roots of Persicaria amphibia, produced seven mitorubrins with a benzoyl moiety at C-6, montagnuphilones A–G (119–125). Among them, 123 and 120 showed inhibitions on NO production in LPS-activated RAW264.7 macrophage cells with IC50 values of 25.5 and 39.6 μM, respectively, without cytotoxicity in RAW264.7 cells.57
Glaziellin A (126) was isolated from the fruiting body of Glaziella splendens with weak inhibitory activity against three types of neuraminidases, H1N1, H3N2, and H5N1, with the IC50 values of 230.6, 235.8, and 165.4 μM respectively.14 Chemical study of an endophytic fungus Penicillium variabile, resulted in two novel azaphilones, comazaphilones G and H (127–128), which displayed NO inhibitory activities with IC50 values of 4.35 and 40.52 μM, respectively.58 Another endophytic fungus Penicillium sp. HN29-3B1, harboring in a fresh branch of the mangrove plant Cerbera manghas in the South China Sea, metabolized pinazaphilones A and B (128–129), while 129 exhibited stronger α-glucosidase inhibitory effect (IC50 = 28.0 μM) than that of acarbose (IC50 = 446.7 μM).12
Pinophilin G (130) was isolated from Penicillium pinophilum with antibacterial activity against Vibrio parahemolyticus (MIC = 25.0 μM).59 Fermentation of the saline soil-derived fungus Talaromyces aculeatus was proved to produce talaraculones A–F (131–136), amongst 131 and 135 were the first reported azaphilones with a C4 aliphatic side chain and a methylal group at C-3, respectively. Curiously, 131 and 132 showed stronger inhibitory activity against α-glucosidase with IC50 values of 78.6 and 22.9 μM, respectively, compared to the positive control acarbose (IC50 = 101.5 μM).60
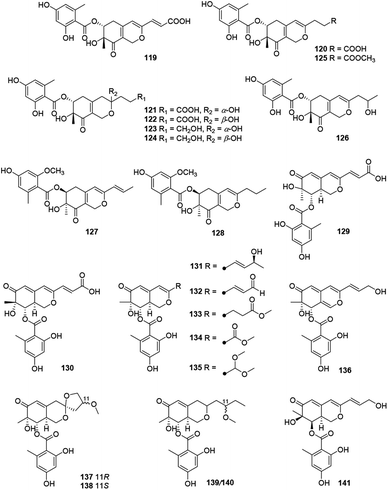
Pleosporalones E–H (137–140) were encountered in an isolate of Pleosporales sp. CF09-1. And the absolute configurations of C-11 in both 137 and 138 were assigned via the GIAO 13C NMR calculations, while the configurations of C-11 in 139 and 140 remained unassigned due to the impracticable calculations.61 They displayed anti-Vibrio anguillarum activities with MIC values from 6.3 to 25 μg mL−1.61
2.9 Chaetoviridins and chaephilones
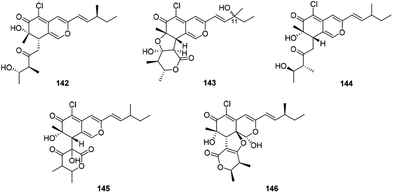
There were five compounds (142–146) obtained from Chaetomium sp. with a chlorine atom at C-5 and a methyl group at C-7, as well as a branched pentenyl side chain at C-3 within this family (Fig. 9 and Table 1), like chaephilones A and B (144–145).62The endophytic fungus Chaetomium globosum, isolated from leaves of Wikstroemia uva-ursi, yielded chaetoviridins J and K (142–143), whilst 143 was obtained as a mixture of unresolvable diastereoisomer failing to be purified by chiral columns. Of note, compound 142 showed obvious activity against NO production while 143 exhibited a weak response.63
In 2018, Wang described the isolation and determination of the chlorinated azaphilone pigment, chaephilone C (146), which was strongly cytotoxic against the HeLa cell line, and also showed anti-MRSA activity compared to the positive control chloramphenicol.64
2.10 Pulvilloric acid azaphilones
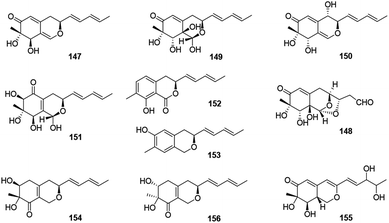
This group consisted of nine novel azaphilones (147–156) were characteristic of an n-pentyl side chain at C-3 (Fig. 10 and Table 1).The co-culture of Nigrospora oryzae and Beauveria bassiana afforded nigbeauvins A–E (147–151), while 148 possessed a novel skeleton with a bicyclic oxygen bridge. Bioactive assays revealed compounds 147 and 148 showed comparable activities on NO production (IC50 = 50 μM).65 However, when Nigrospora oryzae co-cultured with Irpex lacteus, nigirpexins A–D (152–155) and isonigirpexin C (156), the stereoisomer of 142, were obtained.66,67 Among them, 155 showed weak activities in antibacterial and antifungal assays.66
2.11 Sclerotiorin-like azaphilones
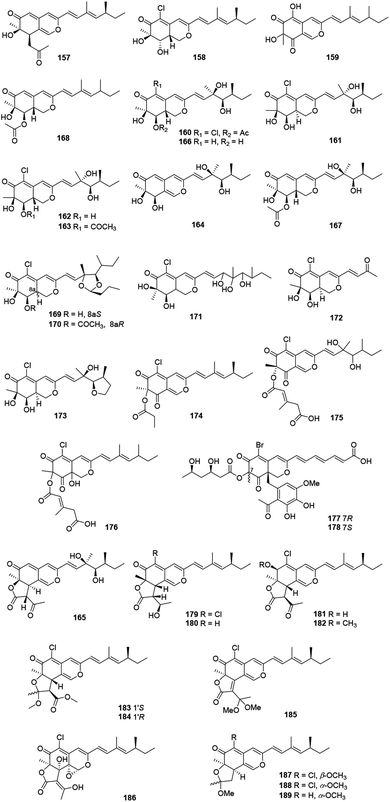
These recently reported sclerotiorin-like azaphilones including thirty-three ones were mainly isolated from Penicillium sp., Emericella sp., and Diaporthe sp. (Fig. 11 and Table 1), like penidioxolanes A–B (169–170)68 as well as sclerotiorins A–C (187–189).69 Dechloroisochromophilone II (157) and epi-isochromophilone III (158) were metabolized by Penicillium multicolor CM01. Significantly, 157 showed AChE inhibitory activity with a minimum inhibition requirement of 0.03 nM, while 158 exhibited notable antimalarial activity and cytotoxicity.70The mangrove endophytic fungus Penicillium sp. 303, produced a novel azaphilone 159, which exhibited weak cytotoxicity toward a series of human cancer lines.71 Isolated from the entomopathogenic fungus Hypocrella sp., hypocrellone A (160) presented moderate cytotoxicity against hepatoma cells BEL-7404 with an IC50 value of 17.4 μM.72 The sponge-derived fungus Eupenicillium sp. 6A-9 metabolized eupenicilazaphilones A–C (161–163), and they were weakly cytotoxic against human cancer lines MCF-7 and A549.73
Derived from a ginseng field, Penicillium sp. KCB11A109 yielded five highly oxygenated azaphilones, geumsanols A–E (164–168). Among them, 168 displayed cytotoxic activities and toxic effects on zebrafish embryos.74 As a novel antineoplastic and antibacterial azaphilone, penicilazaphilone C (171) was found from the marine derived fungus Penicillium sclerotiorum M-22, which showed cytotoxicity against human tumor cells B-16 (IC50 = 0.065 mM) and SGC7901 (IC50 = 0.720 mM), and also indicated significant antibacterial activity against four bacteria.75 Moreover, penicilazaphilones D and E (172–173) were obtained from the sponge-derived fungus Penicillium sclerotiorum and the structure of 172 was further confirmed by single-crystal X-ray diffraction.76
Fermentation of the gorgonian-derived fungus Penicillium sclerotiorum CHNSCLM-0013 produced sclerketide B (174), which denoted significant inhibitory activities against the NO production in the LPS-induced macrophage cell RAW 264.7 and suppressed the expression of iNOS and COX-2 in mRNA level.77 The fungus Bartalinia robillardoides strain LF550, obtained from the Mediterranean sponge Tethya aurantium, metabolized two novel chloroazaphilones, helicusin E (175) and isochromophilone XI (176).78
Inducing the Mediterranian sponge Agelas oroides derived fungus Penicillium canescens by using 5% NaBr, resulted in two new brominated azaphilones, bromophilones A and B (177–178). Notably, 178 showed moderate cytotoxicity against the mouse lymphoma cell line L5178Y and the human ovarian cancer cell line A2780 with IC50 values of 8.9 and 2.7 μM, respectively, while 177 was less active.79 Two new angular types of azaphilones, isochromophilonol (179) and ochrephilonol (180), were identified from Chaetomium cupreum RY202, and 179 exhibited moderated cytotoxicity against KB and NCI-H187 cell lines, while 180 showed weak cytotoxic activity against KB.80 Six new highly oxygenated chloroazaphilone derivatives, namely isochromophilones A–F (181–186), were isolated from the mangrove-derived fungus Diaporthe sp. SCSIO 41011. Among them, 184 showed cytotoxicity against 786-O cells (IC50 = 8.9 μM) and induced apoptosis in 786-O cells in a dose- and time-dependent manner.8
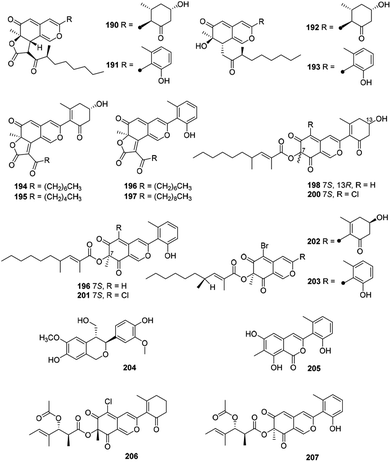
2.12 Cohaerins and related azaphilones
Eighteen novel compounds (190–207) constituted this unique group of azaphilones (Fig. 12 and Table 1). Cohaerins G–I and K (190–193) were produced by Annulohypoxylon cohaerens, along with cohaerins C–F, whose absolute configurations were revised.81 190–193 demonstrated cytotoxicity towards mouse fibroblast L-929 cancer cells, meanwhile, 190, 192, and 193 showed weak activity against the Gram-positive bacteria.81Chemical and biological study of the stromata of Annulohypoxylon minutellum resulted in four novel azaphilones, minutellins A–D (194–197). Meanwhile, they displayed cytotoxicity towards mouse fibroblast L-929 cancer cells and also showed weak activity against the Gram-positive bacteria.82
Four anti-MRSA azaphilones (MIC = 3.13–6.25 μg mL−1), named penicilones A–D (198–201), were obtained from the mangrove marine-derived fungus Penicillium janthinellum HK1-6.5 Interestingly, cultivation of this strain with NaBr led to the isolation of two new brominated azaphilones, penicilones G and H (202–203). Moreover, penicilone H (203) showed antibacterial activity against three bacteria with MIC values ranging from 3.13 to 12.5 μg mL−1.83
With significant antioxidant activity (IC50 = 62.8 μM), meliasendanin A (204) was obtained from the fruits of Melia toosendan.84 The marine-derived fungus Pleosporales sp. CF09-1 yielded three new azaphilone derivatives carrying with aromatic A-ring, pleosporalones A–C (205–207).61,85 Among them, 205 displayed strong antifungal activity against three plant pathogenic fungi with the MIC values ranging from 0.39 to 0.78 μM.85 Additionally, 206 exhibited stronger antifungal activities against Alternaria brassicicola and Fusarium oxysporum with the same MIC value of 1.6 μg mL−1 than that of the positive control ketoconazole, whilst 206 displayed significant activity against Botryosphaeria dothidea (MIC = 3.1 μg mL−1).61
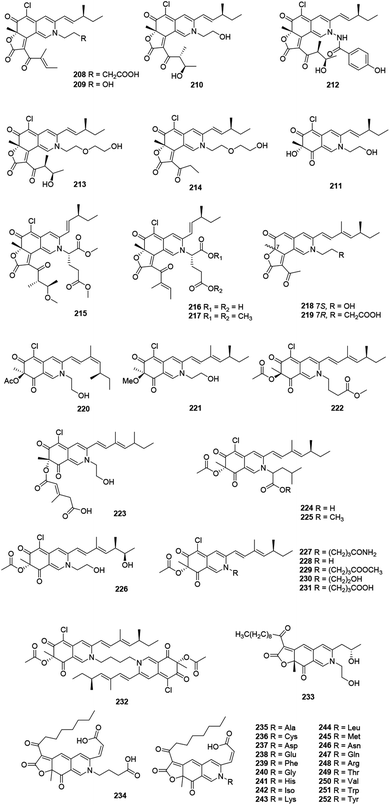
2.13 Nitrogenated azaphilones
This family included forty-five N-containing azaphilones (208–252), which were mainly obtained from Penicillium sp. and Chaetomium sp., (Fig. 13 and Table 1), such as peniazaphilones A–D (218–221),86 berkchaetorubramine (233),49 and (6-[(Z)-2-carboxyvinyl]-N-GABA-PP-V) (234).87The fermentation of an endophytic fungus Chaetomium globosum TY1, isolated from medicinal plant Ginkgo biloba, produced three metabolites, chaetomugilides A–C (208–210), which were cytotoxic against HePG2 with the IC50 values of 1.7, 19.8, and 53.4 μM, respectively.88 Meanwhile, 208 and 210 were also found in Chaetomium globosum (DAOM 240359) derived from an indoor air sample, together with isochromophilone XIII (211). Besides, 208 and 210 showed anti-microbial activities, while 208 (20 μM) reduced the growth of bacteria comparable to the effect of chloramphenicol at the same concentration.89
The culture of Chaetomium sp. NA-S01-R1, derived from the deep sea, yielded chaetoviridides A–C (212–214). Notably, 212 and 213 showed activities against Vibrio rotiferianus and shared the MIC value of 7.3 μg mL−1. Moreover, 213 and 214 exhibited equal anti-MRSA activities compared to positive control. Compounds 212 and 213 demonstrated significant cytotoxicity against the HepG.2 and HeLa cell lines, respectively.64
N-Glutarylchaetoviridins A–C (215–217), embedded with glutamine residues, were obtained from Chaetomium globosum HDN151398, a deep-sea sediment derived fungus collected in the South China Sea. Remarkably, 217 exhibited strong cytotoxicity against human cancer cell lines MGC-803 and HO8910 with the IC50 values of 6.6 and 9.7 μM, respectively.90 Sclerotiorin D and sclerketide C (222) sharing with the same structure and configuration, were simultaneously reported from the two strains of Penicillium sclerotiorum, CHNSCLM-0013 and OUCMDZ-3839, respectively.69,77 Compound 222 inhibited the NO production in the LPS-induced macrophage cell RAW 264.7 with the IC50 value of 2.7 μM and also suppressed the expression of iNOS and COX-2 in mRNA level.69,77 Bartalinia robillardoides LF550 metabolized a cytotoxic compound, isochromophilone X (223), which showed inhibitory activity against PDE4 (IC50 = 11.7 μM).78
In 2019, Tang's group described the isolation and characterization of nine new azaphilone alkaloids, penazaphilones A–I (224–232), which were isolated from the rice solid fermented culture of Penicillium sclerotiorum cib-411. Meanwhile, structures of 228 and 231 were confirmed by X-ray diffraction analysis. Cell viability assay indicated that 224, 228, 229, and 231 inhibited NO production with IC50 values ranging from 7.05 to 15.29 μM, without cytotoxicity towards RAW 264.7 cells (50.0 μM).91
Recently, a series of azaphilone pigments named atrorosins A, C–I, K–N, Q, R, T, V, W, and Y (235–252) have been isolated from the filamentous fungus Talaromyces atroroseus. Structually, these atrorosins shared a carboxylic acid group at C-3 and an amino acid tethered to the isochromene core.92
3. Chemical synthesis
Total synthesis could be the desirable alternatives to provide enough materials for further biological studies and determination of the absolute configurations. General synthetic protocol for azaphilones scaffolds had been concluded in 2013.3 Recently, several synthetic strategies of significance have been developed for these novel diversified azaphilones, which mainly focused on stereoselectivity,29 complementary biocatalysts,93 and the formation of angular lactone and chiral azaphilone dimers.94 In general, cazisochromene and dioxinone have been employed as important intermediates and precursors, whilst AzaH and AfoD have been applied in synthesis as common biocatalysts.93,95 The recently reported chemical synthetic study of azaphilones were summarized herein.3.1 The total synthesis of felinone A
Recently, Ito, et al. have reported the first total synthesis of felinone A (29a) and also revised the absolute configurations of natural felinone A (29).29 The synthesis of dihydropyrane ring was based on Shi asymmetric epoxidation of a bicyclic lactone, and intramolecular oxymercuration/demercuration.29The synthesis began with starting material propargyl alcohol (253), which underwent 2 steps to provide the precursor of enantioselective epoxidation, bicyclic lactone (254). Then 254 went through chemo- and enantio-selective epoxidation and gave the epoxide 255, and then gave 256 in four steps with high diastereoselectivity. Simultaneously, 256 went through reduction to afford 257, and then via Wittig reaction to afford alcohol 258 in high yield as an isomer. The cyclization of E-258 afford the hexahydroisochromene derivative (259). Selectively cleaved the acetonide group of 259 provided 260, which went through Ley oxidation to form 261. Treatment of the resulting ketone 261 with TBAF provided the target felinone A (29a) in middle yield (Scheme 1).
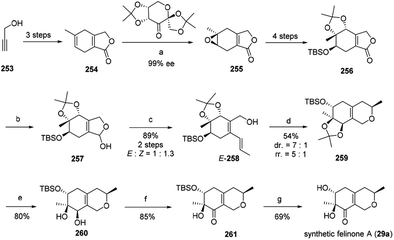 | ||
| Scheme 1 Synthesis of felinone A29 Reagents and conditions: (a) Oxone®, K2CO3, CH3CN-DMM-buffer, 98%, 87% ee, then recrystallization; (b) DIBALH, toluene; (c) Ph3PCH2CH3 Br, LHMDS THF; (d) Hg(OCOCF3)2, MeOH, then NaBH4; (e) TFA, CH2Cl2–H2O; (f) TPAP, NMO, CH2Cl2; (g) TBAF, THF. | ||
3.2 Total synthesis of trichoflectin, (S)-deflectin-1a, and lunatoic acid A
Total synthesis of trichoflectin (262), (S)-deflectin-1a (263), and lunatoic acid A (264) were accomplished using complementary biocatalysts.93The synthesis of trichoflectin (262) was initiated through a five-step route to enone 265. Dearomatization of 265 afforded azaphilone scaffold (R)-266 in high yield. Then, acylation of (R)-266 provided (S)-262. The enantiomeric tricycle was synthesized from the AfoD generated product (S)-266. Finally, it is proposed that the structure of the natural product should be revised to the R-configuration (Scheme 2A).
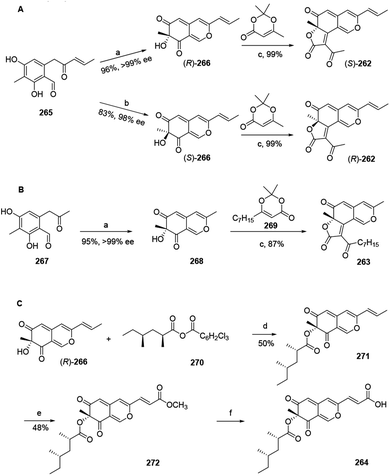 | ||
Scheme 2 Synthesis of trichoflectin, (S)-deflectin-1a, and lunatoic acid A by Pyser and his co-worker.93 Reagents and conditions: (a) AzaH (0.2 mol%), NADPH recycling system, KPi buffer, pH 8.0; (b) AfoD (0.8 mol%), NADPH recycling system, KPi buffer, pH 8.0; (c) Et3N, mol sieves, toluene, 110 °C; (d) DMAP, toluene, 110 °C; (e) Grubbs 2nd Gen, methyl acrylate, DCM, 45 °C; (f) LiOH, THF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, r.t. H2O, r.t. | ||
Methyl ketone 267 was dearomatized with AzaH to afford bicycle 268. Acylation and Knoevenagel condensation with the acylketene derived from precursor 269 furnished the desired butenolide to deliver (S)-deflectin-1a (263) in high yield (Scheme 2B).
Lunatoic acid A (264) was constructed from (R)-266, which went through Yamaguchi esterification with 270 to produce 271. Lunatoic acid A (264) was provided by saponification of methyl ester 272 using LiOH (Scheme 2C).
3.3 The first total synthesis of chaetoglobin A
The first total synthesis of chiral azaphilone dimer, chaetoglobin A (273), was finished in 12 steps by Kang and his co-workers in 2017.94 The vanadium-catalyzed atroposelective oxidative phenol coupling was a key step to form stereoaxis of chiral azaphilone dimer.94Optimization of Sonogashira coupling between iodide 274 and alkyne 275 afforded oxidative phenol coupling precursor 276 in 98% yield. Catalytic 276 with chiral vanadyl catalyst 277 led to the formation of dimer 278, which underwent the Vilsmeier–Haack formylation to provide 279 in 86% yield. Cycloisomerization of 279 afforded bicyclic dimer 280. And acetylation of 280 gave 281, which then afforded 282 after hydroxylation. Treatment of 282 with excess NH4OAc led to the nearly quantitative formation of chaetoglobin A (273) in 96% yield (Scheme 3).
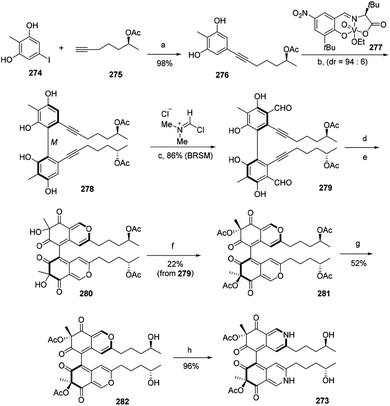 | ||
| Scheme 3 Synthesis of chaetoglobin A.94 Reagents and conditions: (a) PdCl2(PPh3)2, CuI, Et2NH, DMF, 65 °C, 18 h; (b) HOAc, O2, ClPh, 0 °C, 48 h, 67% (BRSM); (c) CH2Cl2, −35 °C, 12 h; (d) AgOTf, DCE/TFA, r.t., 1.5 h; (e) IBX, Bu4NI, r.t., 18 h; (f) Ac2O, DMAP, Et3N, CH2Cl2, −78 °C, 0.5 h; (g) Ti(Oi-Pr)4, THF, 50 °C, 24 h; (h) NH4OAc, CH2Cl2, r.t., 20 h. | ||
3.4 Total synthesis of chaetoviridins
The first synthesis of chaetoviridin A (283) had been achieved in 10 steps in 2017.95 Vanadium-catalyzed oxidative phenol coupling is a vital step in the formation of the axial chirality.95The synthesis of the key intermediate cazisochromene (290) started with chlorination of methyl atratate (284). Benzylic deprotonation of 285 yielded 286, which was activated as pentafluorophenol ester (287). Following Cossy's procedure of 287 formed the β-ketoester 288. Treating 288 via Horner–Wadsworth–Emmons reaction and lactonization led to the formation of 289. Lactone 289 was then reduced and oxidatively dearomatized to yield 290. Condensation of a chiral dioxin-4-one 291 to cazisochromene 290 formed angular lactone. (S,R)-dioxinone (291) and 290 were heated in toluene for 30 min before adding Et3N and then afforded 292, which was directly treated by HF/pyridine to give the (7S,4′S,5′R)-chaetoviridin A (283a) and (7R,4′S,5′R)-chaetoviridin A (283b) with 17% and 20% yield, respectively (Scheme 4).
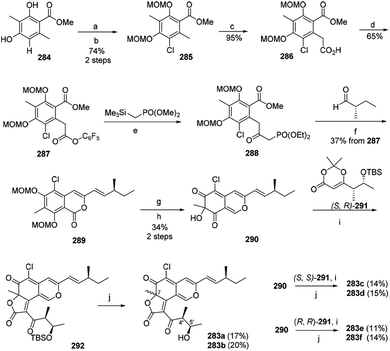 | ||
| Scheme 4 Synthesis of chaetoviridins.95 Reagents and conditions: (a) NCS; (b) NaH/MOMCl; (c) LDA/CO2; (d) C6F5OH, EDC/DMAP; (e) n-BuLi; (f) K2CO3, EtOH; (g) DIBAL-H; (h) IBX/TFA/H2O; (i) Et3N, toluene, reflux; (j) HF/pyridine. | ||
Starting from (S,S)-291, (7S,4′S,5′S)-chaetoviridin A (283c) and (7R,4′S,5′S)-chaetoviridin A (283d) were obtained with 14% and 15% yield, respectively. Whereas the other set of (7S,4′R,5′R)-chaetoviridin A (283e) and (7R,4′R,5′R)-chaetoviridin A (283f) were prepared starting from (R,R)-291 (Scheme 4).
3.5 Total synthesis of chlorofusin and its chromophore diastereomers
Yao and his group developed a newly stereo divergent total synthesis of chlorofusin (293a), along with seven chromophore diastereomers (293b–h) of 293a in enantiopure forms.96Synthesis of them started from a racemic azaphilone precursor 296, which was prepared from O-alkynylbenzaldehyde (294) via 3 steps. The site-selective chlorination from rac-296 to rac-297 was performed with SO2Cl2 in DCM. The reaction of rac-297 with (R)-(+)-4-methoxyl-α-methylbenzylamine led to the formation of two diastereomeric vinylogous γ-pyridones (4R)-298 and (4S)-298. Parallelly, removing the chiral auxiliary of (4R)-298 and (4S)-298 and then replacing them with allyl bromide led to the formation of (4R)-299 and (4S)-299 (Scheme 5).
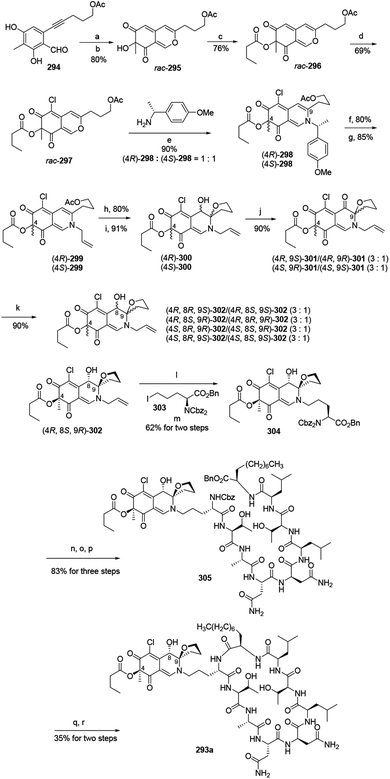 | ||
Scheme 5 Synthesis of chlorofusin and its chromophore diastereomers.96 Reagents and conditions: (a) AgNO3, TFA, ClCH2CH2Cl; (b) IBX, n-Bu4NI; (c) butytryl chloride pyridine, DMAP, DCM; (d) SO2Cl2, DCM; (e) aq. NaHCO3 (sat.), CH3CN, r.t.; (f) TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (v/v = 10 H2O (v/v = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), (i-Pr)3SiH; (g) allyl bromide Pd(PPh3)4, KF·Al2O3, DMF; (h) K2CO3, MeOH; (i) I2, NaHCO3, n-Bu4NI, CH3CN/H2O; (j) DMP, NaHCO3, DCM; (k) NaBH3CN, CeCl3·7H2O, MeOH, r.t.; (l) Pd(Ph3)4, NDMBA, DCM, MW; (m) KF·Al2O3, DMF; (n) Pd/C, H2, THF/DMF; (o) Cbz-OSu, NaHCO3, THF/H2O; (p) TFA·H2N-octapeptide-OBn 71, EDCl, HOBt, DlEA, DCM/DMF; (q) Pd/C, H2, THF/DMF; (r) EDCl, HOAt, NaHCO3, DMF. 1), (i-Pr)3SiH; (g) allyl bromide Pd(PPh3)4, KF·Al2O3, DMF; (h) K2CO3, MeOH; (i) I2, NaHCO3, n-Bu4NI, CH3CN/H2O; (j) DMP, NaHCO3, DCM; (k) NaBH3CN, CeCl3·7H2O, MeOH, r.t.; (l) Pd(Ph3)4, NDMBA, DCM, MW; (m) KF·Al2O3, DMF; (n) Pd/C, H2, THF/DMF; (o) Cbz-OSu, NaHCO3, THF/H2O; (p) TFA·H2N-octapeptide-OBn 71, EDCl, HOBt, DlEA, DCM/DMF; (q) Pd/C, H2, THF/DMF; (r) EDCl, HOAt, NaHCO3, DMF. | ||
In parallel, the reaction of acetate (4R)-299 and (4S)-299 with K2CO3 in MeOH at room temperature, followed by intramolecular iodoetherification and in situ hydrolysis carried out four inseparable diastereomers, (4R)-300 and (4S)-300, in high yields. Each mixture underwent Dess–Martin oxidation and yielded two separable single diastereomeric ketones (4R,9S)-301/(4R,9R)-301 and (4S,9R)-301/(4S,9S)-301, respectively. The reduction of ketones furnished eight fully functionalized chromophores, (4R,8R,9S)-302/(4R,8S,9S)-302, (4R,8S,9R)-302/(4R,8R,9R)-302, (4S,8R,9R)-302/(4S,8S,9R)-302, (4S,8R,9S)-302/(4S,8S,9S)-302 (Scheme 5).
(4R,8S,9R)-302 underwent removal of N-allylation and condensed with iodoornithine derivative 303 to generate the chromophore-ornithine derivative 304 in 62% yield by two steps. The precursor 305 was provided in 83% yield from 304 via three steps. Removal of both N-Cbz and benzyl ester groups of 305 afforded natural (4R,8S,9R)-chlorofusin (293a), with 35% yield in two steps (Scheme 5).
Starting from the corresponding chromophore diastereomers, (4R,8R,9S)-302, (4R,8S,9S)-302, (4R,8R,9R)-302, (4S,8R,9R)-302, (4S,8S,9R)-302, (4S,8R,9S)-302, (4S,8S,9S)-302, the parallel total synthesis of other seven chlorofusin chromophore diastereomers were formed through this reproducible route.96
4. Biosynthesis
In 2019, Chen and co-workers have presented an overview of a unified biosynthetic pathway with the diverse structures of the 111 Monascus azaphilones congeners.97 Generally, biosynthesis of azaphilones contain the polyketide pathway and the fatty acid synthesis pathway, while some may involve polyketide–amino acid mixed biosynthesis.4 The azaphilone polyketide is synthesized by an NR-fPKS with a reductive release domain, and the pyran ring cyclization is based on hydroxylation-mediated dearomatization of a benzaldehyde intermediate.48 The structural complexity of azapholones may result from interaction of biosynthesis with metabolic and chemical fluke. Herein, the recently proposed biosynthetic pathways of novel azaphilones were summarized.4.1 Biosynthesis of mycoleptone A
Andrioli proposed that mycoleptone A (44) was originated from the condensation of two austdiol units.15 It was assumed that two austdiol units underwent decarbonylation and reduction to afford two units, respectively, which then led to the formation of mycoleptone A by Friedel–Crafts alkylation (Scheme 6).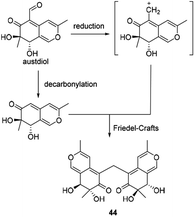 | ||
| Scheme 6 Biosynthesis of mycoleptone A.15 | ||
4.2 Biosynthesis of monasfluols A and B, MC-2 and MC-4
Balakrishnan and his co-workers thought that monasfluol A (306) and monasfluol B (84) were derived from the presumed intermediate 307 by nonenzymatic Knoevenagel condensation and reduction.46 In 2017, it was assumed that MC-2 (87) and MC-4 (88) might share similar biosynthetic pathway, both of which were catalyzed by Δmppc98 (Scheme 7).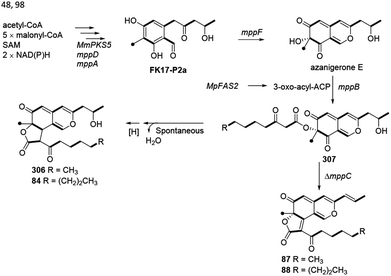 | ||
| Scheme 7 Biosynthesis of monasfluols A and B, MC-2 and MC-4.46,48,98 | ||
4.3 Proposed biosynthetic route to azaphilone 7
It was assumed that azaphilone 7 could be converted from the pentaketide, 2,4-dihydroxy-3-methyl-6-(2-oxopropyl)benzaldehyde, which started from acetate, 4 malonates, and methionine in the A. oryzae transformant harbouring pTA-tspks2 (ref. 18) (Scheme 8).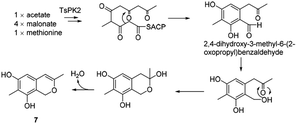 | ||
| Scheme 8 Proposed biosynthetic route to azaphilone 7.18 | ||
4.4 Postulated biogenetic pathway for penicilazaphilone E
Wang, et al. thought WB could be the precursor of penicilazaphilone E (173).76 Firstly, WB went through oxidization to provide intermediate 308. Then 308 underwent cyclization between 7′-methyl and 4′-OH to afford 173 in two different possible pathways (Scheme 9).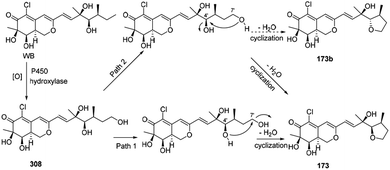 | ||
| Scheme 9 Postulated biogenetic pathway for penicilazaphilone E.76 | ||
4.5 Proposed biosynthesis of chaephilone B
The biogenetic pathway of chaephilone B (145) was suggested from the precursor chaetomugilin S, which was extensively investigated in C. globosum by C. Chen and co-workers.62 The pathway included two main steps of ring opening and oxidation (Scheme 10).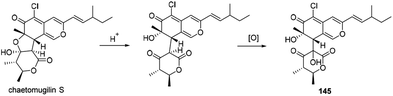 | ||
| Scheme 10 Proposed biosynthesis of chaephilone B.62 | ||
4.6 Biosynthesis of acetosellin
Acetosellin (309) was proposed from epicocconone (310) by the condensation of two polyketide-derived intermediates, 311 and 312. The reduction and subsequent cyclization of 310 formed the naphthopyran moiety of 309 (ref. 38) (Scheme 11).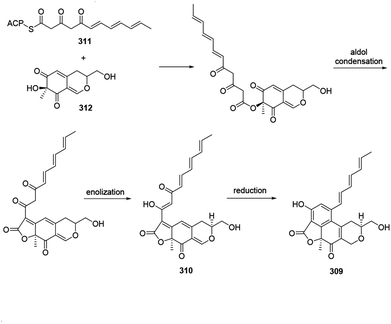 | ||
| Scheme 11 Biosynthesis of acetosellin.38 | ||
4.7 Biosynthesis of chaephilone C, chaetoviridides A and B
The biogenetic pathway of chaephilone C (146) was postulated to start from chaetoviridin A (313).64 Hydration of 313 formed intermediate 314, which then followed hydrolytic opening of the γ-lactone and post dehydration to obtain 146 (Scheme 12A). The biogenetic pathway of chaetoviridides A (212) and B (213) was assumed to include a Schiff base formation and dehydration reaction (Scheme 12B).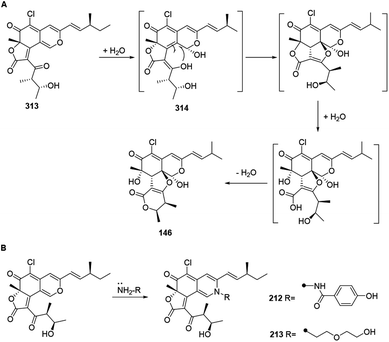 | ||
| Scheme 12 Biosynthesis of chaephilone C, chaetoviridides A and B.64 | ||
4.8 Proposed biosynthesis of colletotrichones A–C, and chermesinonen B
Colletotrichones A–C (54–56) and chermesinonen B (317) might proceed from a pentaketide 316 by several steps. 316 could be formed by the condensation of a 4-methyl-3-oxohexanoic acid and an isochromene analogue 31537 (Scheme 13).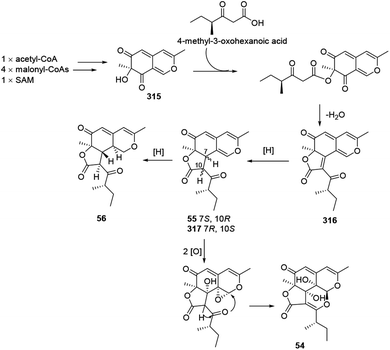 | ||
| Scheme 13 Proposed biosynthesis of colletotrichones A–C, and chermesinonen B.37 | ||
4.9 Postulated biosynthesis of coniellins A, H, and I
The azaphilone intermediate 318 was formed through esterification of a β-ketoacid to the polyketide chromophore and followed by a C-8/C-12 Knoevenagel cyclization and reduction. Elimination of intermediate 318 afforded coniellin A (57). Hydrolysis of intermediate 318 gave the product 319. Coniellin H (64) was derived from the intermediate 319 by elimination, [2 + 2] cycloaddition, and methylation. The pathway from 319 to coniellin I (65) involved esterification and the formation of hemiketal group35 (Scheme 14).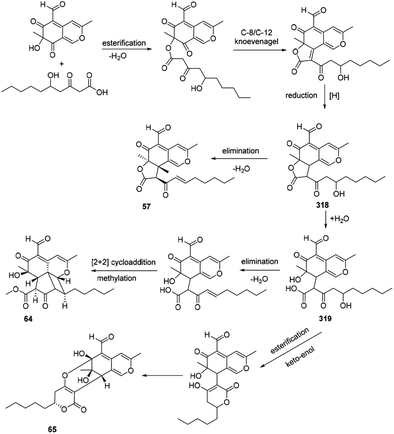 | ||
| Scheme 14 Postulated biosynthesis of coniellins A, H, and I.35 | ||
4.10 Proposed biogenesis of thielavialides A–E
Biosynthetically, it was proposed that the biosynthetic pathway of thielavialides A–E (70–74) was started from pestafolide A (317).40 The probable biosynthetic pathway of thielavialides A–D (70–73) from 320 involved a favorskii-like rearrangement of its oxidation product, 5-dehydropestafolide A (321). Biosynthesis of thielavialide E (74) might involve a radical oxidative coupling of the intermediate 321 or its acetate (Scheme 15).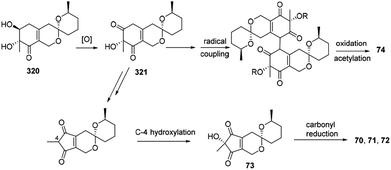 | ||
| Scheme 15 Proposed biogenesis of thielavialides A–E.40 | ||
4.11 Proposed biosynthetic relationships among dothideomynones D–F
Dothideomynone D (41) was assumed as a potential biosynthetic precursor of dothideomynones E and F (42–43).34 The condensation of acetyl-CoA and malonyl-CoA gave the linear intermediate, which followingly went through cyclization to form dothideomynone D (41). 41 underwent oxidation to afford the intermediate 322, which in turn employed malonyl-CoA as an extender to give 323. The reduction of the intermediate 323 could generate the intermediate 324. Cyclization of the intermediate 324 led to the formations of 42 and 43 (Scheme 16).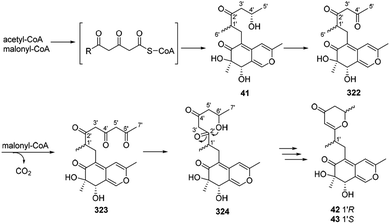 | ||
| Scheme 16 Proposed biosynthetic relationships among dothideomynones D–F.34 | ||
4.12 Proposed biosynthesis of cochliodone A and chaetoglobin A
Biosynthetic pathways of cochliodone A (325) and chaetoglobin A (326) were proposed in 2013.99 CHGG_10027 was confirmed its involvement in the biosynthesis of 325, which accepted acetyl-CoA as a starting unit and added four malonyl-CoA units to provide 6-methylorsellinic acid. Acetylation of the alcohol group on the ring system could be achieved by an O-acetyltransferase, and dimerization was likely performed by a fungal laccase-like CHGG_10025. A nonenzymatic reaction with ammonia of 325 could generate 326 (Scheme 17).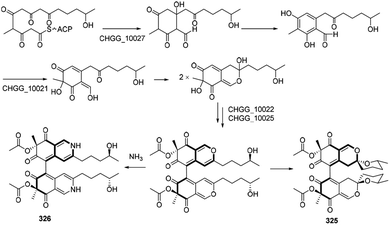 | ||
| Scheme 17 Proposed biosynthesis of cochliodone A and chaetoglobin A.99 | ||
4.13 Biosynthesis of 6′′-hydroxy-(R)-mitorubrinic acid, purpurquinone D (−)-mitorubrinic acid, (−)-mitorubrin, and purpurquinone A
Xiao, et al., hypothesized the biosynthesis of 6′′-hydroxy-(R)-mitorubrinic acid (109), purpurquinone D (110), (−)-mitorubrinic acid (327), (−)-mitorubrin (328), and purpurquinone A (329) were started from six acetate units through 2, 7-condensation.53 Besides, orselilinic acid was proposed to be involved in the pathway (Scheme 18).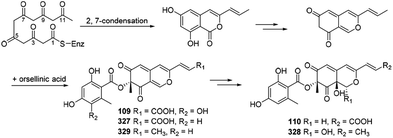 | ||
| Scheme 18 Biosynthesis of 6′′-hydroxy-(R)-mitorubrinic acid, purpurquinone D (−)-mitorubrinic acid, (−)-mitorubrin, and purpurquinone A.53 | ||
4.14 Possible biosynthesis of cazisochromene, chaetoviridin A, and chaetomugilin A
The biosynthesis of cazisochromene (290) was proposed to be derived from cazaldehyde A, which was the product of the interaction between HR-PKS and NR-PKS.100 290 was transferred to chaetoviridin A (330) by 8-CazF and chaetomugilin A (331) would be generated in turn100,101 (Scheme 19).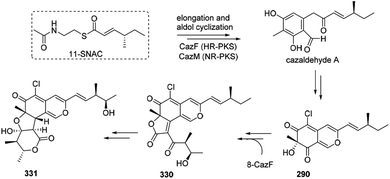 | ||
| Scheme 19 Possible biosynthesis of cazisochromene, chaetoviridin A, and chaetomugilin A.100 | ||
4.15 Plausible biosynthesis of citrifurans A–D
The plausible biosynthetic pathway for citrifurans A–D (24–27) was assumed by Yin, et al. in 2017.7 The presumed precursors 332 and 333 were originated from acetate and S-adenosyl methionine. Then decarbonylation of 332 and oxidation of 333 afforded intermediates 334 and 335, respectively. Afterward, deprotonation of 335 afforded two kinds of enolate anions, 336 and 337, which subsequently heterodimerized with 332 and 334 through a Michael addition reaction to form adducts 338 and 339, respectively. A subsequent intramolecular nucleophilic addition and reduction of 339 constructed a dihydropyran ring in citrifuran A (24), which further went via oxidization to form the epimers, citrifuran B (25) and citrifuran C (26). Similarly, the intramolecular dehydration of 339 could produce citrifuran D (27) (Scheme 20).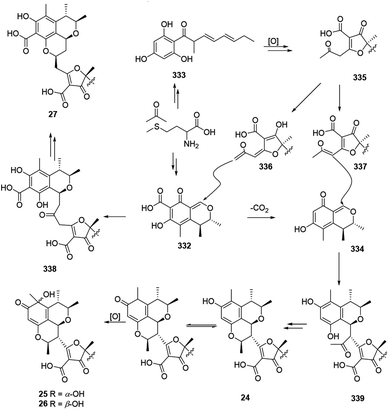 | ||
| Scheme 20 Plausible biosynthesis of citrifurans A–D.7 | ||
4.16 Postulated biosynthesis of sclerketides B–C
Plausible biosynthetic pathway of sclerketides B–C (174 and 222) was proposed in 2019.77 It was assumed that 340 could be generated by 4 mal-CoAs and SMA, following by the reduction, esterification, and condensation to yield 174 and 222 (Scheme 21).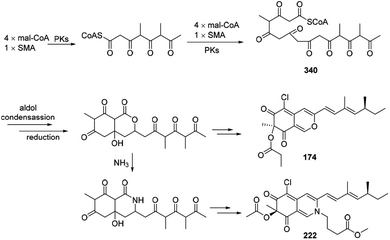 | ||
| Scheme 21 Postulated biosynthesis of sclerketides B and C.77 | ||
4.17 Plausible biosynthesis of bromophilones A–B
A plausible biosynthetic pathway for bromophilones A–B (177–178) was proposed by Frank, et al. in 2019.79The intermediate 341 went through reduction, enolization, and deprotonation, followed by isomerization to form the carbanion intermediate 342. Decarboxylation of vulvulic acid and followed by oxidation and isomerization resulted in formation of the para quinone methide intermediate 343. 177 and 178 were formed by a nucleophilic attack of the carbanion 342 to the carbocation 343 (Scheme 22).
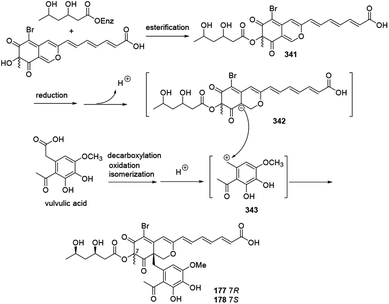 | ||
| Scheme 22 Plausible biosynthesis of bromophilones A and B.79 | ||
5. Conclusion
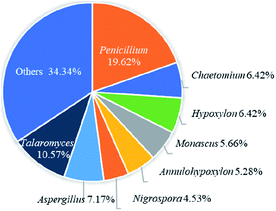
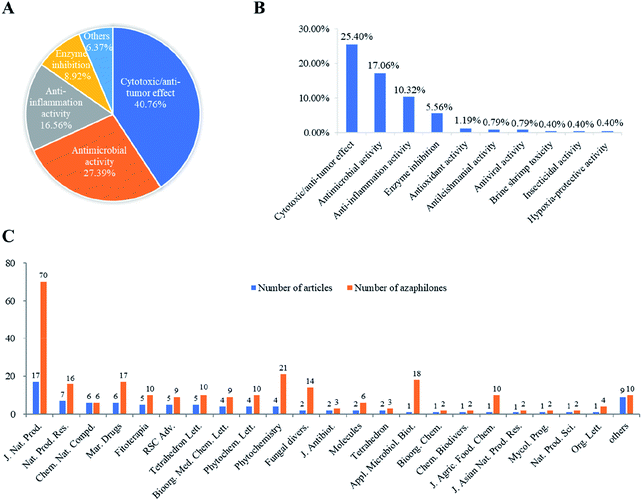
Recently, the fungal polyketides, azaphilones, have attracted continuous broad interests owing to their intriguing structural, biosynthetic, and pharmacological features. According to the literature survey from 2012 October to December of 2019, 252 newly reported azaphilones covered from 35 species, including 32 genera of fungi, 2 plants, and 1 bacterium, were summarized in this research. In terms of the taxonomy of the fungal sources, the genera of Penicillium (20%), Talaromyces (11%), and Aspergillus (7%) were the predominant producers of azaphilones (Fig. 14).Among these recently reported azaphilones, nearly 48% of them were discovered with a broad spectrum of bioactivities (122 of 252 compounds), including cytotoxic/anti-tumor effect (25%), antimicrobial activity (17%), anti-inflammatory activity (10%), enzyme inhibitions (5%), antioxidant activity (1%), antileishmanial activity (0.79%), antiviral activity (0.79%), brine shrimp toxicity (0.40%), insecticidal activity (0.40%), and hypoxia-protective activity (0.40%) (Fig. 15B). Moreover, cytotoxic/anti-tumor effect (40%), antimicrobial activity (27%), and anti-inflammatory activity (16%) were dominant in the above-mentioned bioactivities (Fig. 15A). In addition, a total of 88 articles involving 252 newly reported azaphilones have been published in 31 international journals during the past seven years. Amongst, 70 novel azaphilones (28%) in 17 articles (19%) were published in Journal of Natural Products, which was the most populous journal for recent reported azaphilones (Fig. 15C).
Based on the foregoing discussion, there are still some challenges in the chemical synthesis of azaphilones, such as stability of intermediates, stereoselectivity, reaction utilization, and overall yield. Thus, comprehensive and in-depth research would yet be needed in the synthesis and biosynthesis of these azaphilones with novel and complex structures. Notably, the semi-synthesis of naturally occurring azaphilones through metabolic engineering would accelerate the discovery of advanced intermediates and lead compounds.102 Considering the significant and broad biological activities of this class of metabolites, the azaphilone family would probably continue to draw attention in the chemical synthetic and biosynthetic processes. Collectively, this review would shed light on the further development in chemical and pharmacological investigations of azaphilones with clinically therapeutic applications.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was financially supported in part by the National Natural Science Foundation of China (No. 21772210, 81973235, 21977102), the Special Fund for Bagui Scholars of Guangxi (Yonghong Liu), the Scientific Research Foundation of Institute of Marine Drugs, Guangxi University of Chinese Medicine (2018ZD005-A01, 2018ZD005-A17), the Initial Scientific Research Foundation of Introduced Doctors in 2019 of Guangxi University of Chinese Medicine (2019BS021), and Guangdong Basic and Applied Basic Research Foundation (2019B151502042, 2018A0303130219).Notes and references
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2016, 79, 629–661 CrossRef CAS PubMed.
- H. Hussain, A. M. Al-Sadi, B. Schulz, M. Steinert, A. Khan, I. R. Green and I. Ahmed, Future Med. Chem., 2017, 9, 1631–1648 CrossRef CAS PubMed.
- J. M. Gao, S. X. Yang and J. C. Qin, Chem. Rev., 2013, 113, 4755–4811 CrossRef CAS PubMed.
- N. Osmanova, W. Schultze and N. Ayoub, Phytochem. Rev., 2010, 9, 315–342 CrossRef CAS.
- M. Chen, N. X. Shen, Z. Q. Chen, F. M. Zhang and Y. Chen, J. Nat. Prod., 2017, 80, 1081–1086 CrossRef CAS PubMed.
- T. X. Li, R. H. Liu, X. B. Wang, J. Luo, J. G. Luo, L. Y. Kong and M. H. Yang, J. Nat. Prod., 2018, 81, 1148–1153 CrossRef CAS PubMed.
- G. P. Yin, Y. R. Wu, M. H. Yang, T. X. Li, X. B. Wang, M. M. Zhou, J. L. Lei and L. Y. Kong, Org. Lett., 2017, 19, 4058–4061 CrossRef CAS PubMed.
- X. Luo, X. Lin, H. Tao, J. Wang, J. Li, B. Yang, X. Zhou and Y. Liu, J. Nat. Prod., 2018, 81, 934–941 CrossRef CAS PubMed.
- H. Yu, J. Sperlich, A. Mandi, T. Kurtan, H. Dai, N. Teusch, Z. Y. Guo, K. Zou, Z. Liu and P. Proksch, J. Nat. Prod., 2018, 81, 2493–2500 CrossRef CAS PubMed.
- D. Chen, S. Ma, L. He, P. Yuan, Z. She and Y. Lu, Tuberculosis, 2017, 103, 37–43 CrossRef CAS PubMed.
- W. Li, C. Lee, S. H. Bang, J. Y. Ma, S. Kim, Y. S. Koh and S. H. Shim, J. Nat. Prod., 2017, 80, 205–209 CrossRef CAS PubMed.
- Y. Liu, Q. Yang, G. Xia, H. Huang, H. Li, L. Ma, Y. Lu, L. He, X. Xia and Z. She, J. Nat. Prod., 2015, 78, 1816–1822 CrossRef CAS PubMed.
- D. Zhang, J. Zhao, X. Wang, L. Zhao, H. Liu, Y. Wei, X. You, S. Cen and L. Yu, J. Antibiot., 2018, 71, 905–907 CrossRef CAS PubMed.
- J. Y. Kim, E. E. Woo, L. S. Ha, D. W. Ki, I. K. Lee and B. S. Yun, Mycobiology, 2019, 47, 256–260 CrossRef PubMed.
- W. J. Andrioli, R. Conti, M. J. Araujo, R. Zanasi, B. C. Cavalcanti, V. Manfrim, J. S. Toledo, D. Tedesco, M. O. de Moraes, C. Pessoa, A. K. Cruz, C. Bertucci, J. Sabino, D. N. P. Nanayakkara, M. T. Pupo and J. K. Bastos, J. Nat. Prod., 2014, 77, 70–78 CrossRef CAS PubMed.
- F. Y. Du, X. M. Li, P. Zhang, C. S. Li and B. G. Wang, Mar. Drugs, 2014, 12, 2816–2826 CrossRef CAS PubMed.
- F. Lacatena, R. Marra, A. Piccolo, C. Digilio Maria, M. Lorito, F. Vinale, P. Mazzei, P. Mazzei, A. Piccolo, M. Giorgini, L. Woo Sheridan, M. Lorito, F. Vinale, L. Woo Sheridan, P. Cavallo and P. Cavallo, Molecules, 2019, 24, 750 CrossRef CAS PubMed.
- M. Hashimoto, D. Wakana, M. Ueda, D. Kobayashi, Y. Goda and I. Fujii, Bioorg. Med. Chem. Lett., 2015, 25, 1381–1384 CrossRef CAS PubMed.
- R. S. Orfali, A. H. Aly, W. Ebrahim, Rudiyansyah and P. Proksch, Phytochem. Lett., 2015, 13, 234–238 CrossRef CAS.
- X. H. Li, X. H. Han, L. L. Qin, J. L. He, Z. X. Cao, D. L. Guo, Y. Deng and Y. C. Gu, Nat. Prod. Res., 2019, 33, 1870–1875 CrossRef CAS PubMed.
- Y. Hsiao, M. J. Cheng, H. S. Chang, M. D. Wu, S. Y. Hsieh, T. W. Liu, C. H. Lin, G. F. Yuan and I. S. Chen, Nat. Prod. Res., 2016, 30, 251–258 CrossRef CAS PubMed.
- M. J. Cheng, M. D. Wu, H. Y. Chan, Y. C. Cheng, J. J. Chen, I. S. Chen, Y. S. Su and G. F. Yuan, Chem. Nat. Compd., 2017, 53, 44–47 CrossRef CAS.
- M. J. Cheng, M. D. Wu, Y. C. Cheng, J. J. Chen, H. Y. Chan, Y. L. Chen, I. S. Chen, P. H. Wu and G. F. Yuan, Chem. Nat. Compd., 2016, 52, 634–636 CrossRef CAS.
- W. Guo, D. Li, J. Peng, T. Zhu, Q. Gu and D. Li, J. Nat. Prod., 2015, 78, 306–310 CrossRef CAS PubMed.
- S. Boonyaketgoson, K. Trisuwan, B. Bussaban, V. Rukachaisirikul and S. Phongpaichit, Tetrahedron Lett., 2015, 56, 5076–5078 CrossRef CAS.
- S. P. Zhang, R. Huang, F. F. Li, H. X. Wei, X. W. Fang, X. S. Xie, D. G. Lin, S. H. Wu and J. He, Fitoterapia, 2016, 112, 85–89 CrossRef CAS PubMed.
- J. Kornsakulkarn, S. Saepua, R. Suvannakad, S. Supothina, N. Boonyuen, M. Isaka, S. Prabpai, P. Kongsaeree and C. Thongpanchang, Tetrahedron, 2017, 73, 3505–3512 CrossRef CAS.
- Z. Fan, Z. H. Sun, H. X. Liu, Y. C. Chen, H. H. Li and W. M. Zhang, J. Asian Nat. Prod. Res., 2016, 18, 1024–1029 CrossRef CAS PubMed.
- H. Abe, H. Tango, T. Kobayashi and H. Ito, Tetrahedron Lett., 2017, 58, 4296–4298 CrossRef CAS.
- J. Arunpanichlert, V. Rukachaisirikul, S. Phongpaichit, O. Supaphon and J. Sakayaroj, Nat. Prod. Res., 2016, 30, 46–51 CrossRef CAS PubMed.
- Y. S. Chen, M. J. Cheng, Y. Hsiao, H. Y. Chan, S. Y. Hsieh, C. W. Chang, T. W. Liu, H. S. Chang and I.-S. Chen, Helv. Chim. Acta, 2015, 98, 1167–1176 CrossRef CAS.
- J. Wang, G. Bai, Y. Liu, H. Wang, Y. Li, W. Yin, Y. Wang and F. Lu, Chem. Lett., 2015, 44, 1148–1149 CrossRef CAS.
- R. T. Hewage, T. Aree, C. Mahidol, S. Ruchirawat and P. Kittakoop, Phytochemistry, 2014, 108, 87–94 CrossRef CAS PubMed.
- K. Wijesekera, C. Mahidol, S. Ruchirawat and P. Kittakoop, Bioorg. Med. Chem., 2017, 25, 2868–2877 CrossRef CAS PubMed.
- H. Yu, J. Sperlich, S. P. Hofert, C. Janiak, N. Teusch, F. Stuhldreier, S. Wesselborg, C. Wang, M. U. Kassack, H. Dai, Z. Liu and P. Proksch, Fitoterapia, 2019, 137, 104249 CrossRef CAS PubMed.
- C. Huo, X. Lu, Z. Zheng, Y. Li, Y. Xu, H. Zheng and Y. Niu, Phytochemistry, 2019, 170, 112224 CrossRef PubMed.
- W. X. Wang, S. Kusari, H. Laatsch, C. Golz, P. Kusari, C. Strohmann, O. Kayser and M. Spiteller, J. Nat. Prod., 2016, 79, 704–710 CrossRef CAS PubMed.
- P. Hufendiek, S. S. M. Stoelben, S. Kehraus, N. Merten, H. Harms, M. Cruesemann, I. Arslan, M. Guetschow, T. Schneider and G. M. Koenig, Planta Med., 2017, 83, 1044–1052 CrossRef CAS PubMed.
- M. J. Cheng, M. D. Wu, H. Y. Chan, H. S. Chang, H. C. Wu, J. J. Chen, G. F. Yuan, J. R. Weng, C. T. Chang and H. C. Lin, Chem. Nat. Compd., 2019, 55, 79–81 CrossRef CAS.
- E. M. K. Wijeratne, P. Espinosa-Artiles, R. Gruener and A. A. L. Gunatilaka, J. Nat. Prod., 2014, 77, 1467–1472 CrossRef CAS PubMed.
- N. M. Tran-Cong, A. Mandi, T. Kurtan, W. E. G. Muller, R. Kalscheuer, W. Lin, Z. Liu and P. Proksch, RSC Adv., 2019, 9, 27279–27288 RSC.
- H. C. Wu, M. J. Cheng, M. D. Wu, J. J. Chen, Y. L. Chen and H. S. Chang, Phytochem. Lett., 2019, 31, 242–248 CrossRef CAS.
- M. D. Wu, M. J. Cheng, Y. J. Yech, Y. L. Chen, K. P. Chen, P. H. Yang, I. S. Chen and G. F. Yuan, Nat. Prod. Res., 2013, 27, 1145–1152 CrossRef CAS PubMed.
- M. J. Cheng, M. D. Wu, H. Y. Chan, Y. C. Cheng, J. J. Chen, Y. L. Chen, I. S. Chen and G. F. Yuan, Chem. Nat. Compd., 2015, 51, 1091–1093 CrossRef CAS.
- M. J. Cheng, M. D. Wu, H. Y. Chan, J. J. Chen, Y. C. Cheng, Y. L. Chen, I. S. Chen and G. F. Yuan, Chem. Nat. Compd., 2016, 52, 231–233 CrossRef CAS.
- B. Balakrishnan, C. C. Chen, T. M. Pan and H. J. Kwon, Tetrahedron Lett., 2014, 55, 1640–1643 CrossRef CAS.
- Y. Hu, Y. Zhou, Z. Mao, H. Li, F. Chen and Y. Shao, AMB Express, 2017, 7, 166 CrossRef PubMed.
- B. Bijinu, J. W. Suh, S. H. Park and H. J. Kwon, RSC Adv., 2014, 4, 59405–59408 RSC.
- A. A. Stierle, D. B. Stierle, T. Girtsman, T. C. Mou, C. Antczak and H. Djaballah, J. Nat. Prod., 2015, 78, 2917–2923 CrossRef CAS PubMed.
- E. Kuhnert, F. Surup, E. B. Sir, C. Lambert, K. D. Hyde, A. I. Hladki, A. I. Romero and M. Stadler, Fungal Diversity, 2015, 71, 165–184 CrossRef.
- F. Surup, A. Narmani, L. Wendt, S. Pfuetze, R. Kretz, K. Becker, C. Menbrives, A. Giosa, M. Elliott, C. Petit, M. Rohde and M. Stadler, Fungal Diversity, 2018, 92, 345–356 CrossRef.
- Y. Nalli, D. N. Mirza, Z. A. Wani, B. Wadhwa, F. A. Mallik, C. Raina, A. Chaubey, S. Riyaz-Ul-Hassan and A. Ali, RSC Adv., 2015, 5, 95307–95312 RSC.
- Z. E. Xiao, S. E. Lin, C. Tan, X. Huang, Z. She, Y. Lu and L. He, Mar. Drugs, 2015, 13, 366–378 CrossRef PubMed.
- P. Kalansuriya, Z. G. Khalil, A. A. Salim and R. J. Capon, Tetrahedron Lett., 2019, 60, 151157 CrossRef.
- D. L. Zhao, C. L. Shao, Q. Zhang, K. L. Wang, F. F. Guan, T. Shi and C. Y. Wang, J. Nat. Prod., 2015, 78, 2310–2314 CrossRef CAS PubMed.
- E. B. Sir, E. Kuhnert, F. Surup, K. D. Hyde and M. Stadler, Mycol. Prog., 2015, 14, 28 CrossRef.
- J. G. Luo, Y. M. Xu, D. C. Sandberg, A. E. Arnold and A. A. L. Gunatilaka, J. Nat. Prod., 2017, 80, 76–81 CrossRef CAS PubMed.
- Y. Shao, H. Yan, T. Yin, Z. Sun, H. Xie, L. Song, K. Sun and W. Li, J. Antibiot., 2020, 73, 77–81 CrossRef CAS PubMed.
- Y. Ma, H. Cai, M. Du, F. Cao and H. Zhu, Gaodeng Xuexiao Huaxue Xuebao, 2017, 38, 1963–1967 CAS.
- J. Ren, S. S. Ding, A. Zhu, F. Cao and H. J. Zhu, Chem. J. Chin. Univ., 2017, 80, 2199–2203 CAS.
- F. Cao, Z. H. Meng, X. Mu, Y. F. Yue and H. J. Zhu, J. Nat. Prod., 2019, 82, 386–392 CrossRef CAS.
- C. Chen, J. Wang, H. Zhu, J. Wang, Y. Xue, G. Wei, Y. Guo, D. Tan, J. Zhang, C. Yin and Y. Zhang, Chem. Biodiversity, 2016, 13, 422–426 CrossRef CAS PubMed.
- U. J. Youn, T. Sripisut, E. J. Park, T. P. Kondratyuk, N. Fatima, C. J. Simmons, M. M. Wall, D. Sun, J. M. Pezzuto and L. C. Chang, Bioorg. Med. Chem. Lett., 2015, 25, 4719–4723 CrossRef CAS PubMed.
- W. Wang, Y. Liao, R. Chen, Y. Hou, W. Ke, B. Zhang, M. Gao, Z. Shao, J. Chen and F. Li, Mar. Drugs, 2018, 16, 61 CrossRef PubMed.
- Z. X. Zhang, X. Q. Yang, Q. Y. Zhou, B. Y. Wang, M. Hu, Y. B. Yang, H. Zhou and Z. T. Ding, Molecules, 2018, 23, 1816 CrossRef PubMed.
- Q. Y. Zhou, X. Q. Yang, Z. X. Zhang, B. Y. Wang, M. Hu, Y. B. Yang, H. Zhou and Z. T. Ding, Fitoterapia, 2018, 130, 26–30 CrossRef CAS PubMed.
- Y. M. Wu, Q. Y. Zhou, X. Q. Yang, Y. J. Luo, J. J. Ojan, S. X. Liu, Y. B. Yang and Z. T. Ding, J. Agric. Food Chem., 2019, 67, 11877–11882 CrossRef CAS.
- S. M. Kim, S. Son, J. W. Kim, E. S. Jeon, S. K. Ko, I. J. Ryoo, K. S. Shin, H. Hirota, S. Takahashi, H. Osada, J. H. Jang and J. S. Ahn, Nat. Prod. Sci., 2015, 21, 231–236 CrossRef CAS.
- Q. Jia, Y. Du, C. Wang, Y. Wang, T. Zhu and W. Zhu, Mar. Drugs, 2019, 17, 260 CrossRef CAS PubMed.
- C. Hemtasin, S. Kanokmedhakul, P. Moosophon, K. Soytong and K. Kanokmedhakul, Phytochem. Lett., 2016, 16, 56–60 CrossRef CAS.
- J. Li, X. Yang, Y. Lin, J. Yuan, Y. Lu, X. Zhu, J. Li, M. Li, Y. Lin, J. He and L. Liu, Fitoterapia, 2014, 97, 241–246 CrossRef CAS PubMed.
- Q. Guo, L. Dong, X. Zang, Z. Gu, X. He, L. Yao, L. Cao, J. Qiu and X. Guan, Nat. Prod. Res., 2015, 29, 2000–2006 CrossRef CAS PubMed.
- B. B. Gu, Y. Wu, J. Tang, W. H. Jiao, L. Li, F. Sun, S. P. Wang, F. Yang and H. W. Lin, Tetrahedron Lett., 2018, 59, 3345–3348 CrossRef CAS.
- S. Son, S. K. Ko, J. W. Kim, J. K. Lee, M. Jang, I. J. Ryoo, G. J. Hwang, M. C. Kwon, K. S. Shin, Y. Futamura, Y. S. Hong, H. Oh, B. Y. Kim, M. Ueki, S. Takahashi, H. Osada, J. H. Jang and J. S. Ahn, Phytochemistry, 2016, 122, 154–164 CrossRef CAS PubMed.
- S. L. Zhou, M. Wang, H. G. Zhao, Y. H. Huang, Y. Y. Lin, G. H. Tan and S. L. Chen, Arch. Pharmacal Res., 2016, 39, 1621–1627 CrossRef CAS PubMed.
- C. Y. Wang, J. D. Hao, X. Y. Ning, J. S. Wu, D. L. Zhao, C. J. Kong, C. L. Shao and C. Y. Wang, RSC Adv., 2018, 8, 4348–4353 RSC.
- Z. Liu, P. Qiu, H. Liu, J. Li, C. Shao, T. Yan, W. Cao and Z. She, Bioorg. Chem., 2019, 88, 102973 CrossRef CAS PubMed.
- N. Jansen, B. Ohlendorf, A. Erhard, T. Bruhn, G. Bringmann and J. F. Imhoff, Mar. Drugs, 2013, 11, 800–816 CrossRef CAS PubMed.
- M. Frank, R. Hartmann, M. Plenker, A. Mandi, T. Kurtan, F. C. Ozkaya, W. E. G. Mueller, M. U. Kassack, A. Hamacher, W. Lin, Z. Liu and P. Proksch, J. Nat. Prod., 2019, 82, 2159–2166 CrossRef CAS PubMed.
- N. Panthama, S. Kanokmedhakul, K. Kanokmedhakul and K. Soytong, Arch. Pharmacal Res., 2015, 38, 585–590 CrossRef CAS PubMed.
- F. Surup, K. I. Mohr, R. Jansen and M. Stadler, Phytochemistry, 2013, 95, 252–258 CrossRef CAS PubMed.
- E. Kuhnert, F. Surup, S. Halecker and M. Stadler, Phytochemistry, 2017, 137, 66–71 CrossRef CAS PubMed.
- M. Chen, Y. Y. Zheng, Z. Q. Chen, N. X. Shen, L. Shen, F. M. Zhang, X. J. Zhou and C. Y. Wang, J. Nat. Prod., 2019, 82, 368–374 CrossRef CAS PubMed.
- L. Wang, F. Li, C. Y. Yang, A. A. Khan, X. Liu and M. K. Wang, Fitoterapia, 2014, 99, 92–98 CrossRef CAS PubMed.
- F. Cao, J. K. Yang, Y. F. Liu, H. J. Zhu and C. Y. Wang, Nat. Prod. Res., 2016, 30, 2448–2452 CrossRef CAS.
- L. H. Zhang, Y. Long, X. L. Lei, J. Y. Xu, Z. J. Huang, Z. G. She, Y. C. Lin, J. Li and L. Liu, Phytochem. Lett., 2016, 18, 180–186 CrossRef CAS.
- M. Venkatachalam, M. Zelena, F. Cacciola, L. Ceslova, E. Girard-Valenciennes, P. Clerc, P. Dugo, L. Mondello, M. Fouillaud, A. Rotondo, D. Giuffrida and L. Dufosse, J. Food Compos. Anal., 2018, 67, 38–45 CrossRef CAS.
- X. Li, Y. Tian, S. X. Yang, Y. M. Zhang and J. C. Qin, Bioorg. Med. Chem. Lett., 2013, 23, 2945–2947 CrossRef CAS.
- D. R. McMullin, M. W. Sumarah, B. A. Blackwell and J. D. Miller, Tetrahedron Lett., 2013, 54, 568–572 CrossRef CAS.
- C. Sun, X. Ge, S. Mudassir, L. Zhou, G. Yu, Q. Che, G. Zhang, J. Peng, Q. Gu, T. Zhu and D. Li, Mar. Drugs, 2019, 17, 253 CrossRef CAS PubMed.
- J. L. Tang, Z. Y. Zhou, T. Yang, C. Yao, L. W. Wu and G. Y. Li, J. Agric. Food Chem., 2019, 67, 2175–2182 CrossRef CAS PubMed.
- T. Isbrandt, G. Tolborg, A. Odum, M. Workman and T. O. Larsen, Appl. Microbiol. Biotechnol., 2020, 104, 615–622 CrossRef CAS PubMed.
- J. B. Pyser, S. A. Baker Dockrey, A. R. Benitez, L. A. Joyce, R. A. Wiscons, J. L. Smith and A. R. H. Narayan, J. Am. Chem. Soc., 2019, 141, 18551–18559 CrossRef CAS PubMed.
- H. Kang, C. Torruellas, Y. E. Lee and M. Kozlowski, Org. Lett., 2018, 20, 5554–5558 CrossRef CAS PubMed.
- M. Makrerougras, R. Coffinier, S. Oger, A. Chevalier, C. Sabot and X. Franck, Org. Lett., 2017, 19, 4146–4149 CrossRef CAS PubMed.
- H. B. Qiu, W. J. Qian, S. M. Yu and Z. J. Yao, Tetrahedron, 2015, 71, 370–380 CrossRef CAS.
- W. Chen, F. Chen, Y. Feng and I. Molnar, Nat. Prod. Rep., 2019, 36, 561–572 RSC.
- B. Balakrishnan, S. H. Park and H. J. Kwon, Appl. Biol. Chem., 2017, 60, 437–446 CrossRef CAS.
- T. Nakazawa, K. I. Ishiuchi, M. Sato, Y. Tsunematsu, S. Sugimoto, Y. Gotanda, H. Noguchi, K. Hotta and K. Watanabe, J. Am. Chem. Soc., 2013, 135, 13446–13455 CrossRef CAS PubMed.
- J. M. Winter, D. Cascio, D. Dietrich, M. Sato, K. Watanabe, M. R. Sawaya, J. C. Vederas and Y. Tang, J. Am. Chem. Soc., 2015, 137, 9885–9893 CrossRef CAS PubMed.
- J. M. Winter, M. Sato, S. Sugimoto, G. Chiou, N. K. Garg, Y. Tang and K. Watanabe, J. Am. Chem. Soc., 2012, 134, 17900–17903 CrossRef CAS PubMed.
- T. Asai, K. Tsukada, S. Ise, N. Shirata, M. Hashimoto, I. Fujii, K. Gomi, K. Nakagawara, E. N. Kodama and Y. Oshima, Nat. Chem., 2015, 7, 737–743 CrossRef CAS PubMed.
- M. J. Cheng, M. D. Wu, Y. L. Chen, I. S. Chen, Y. S. Su and G. F. Yuan, Chem. Nat. Compd., 2013, 49, 249–252 CrossRef CAS.
- Y. H. Zhang, X. Y. Peng, L. X. Feng, H. J. Zhu, F. Cao and C. Y. Wang, Nat. Prod. Res., 2019 DOI:10.1080/14786419.2019.1669028.
- R. Paranjape Smita, P. Riley Andrew, D. Somoza Amber, C. E. Oakley, C. C. Wang Clay, E. Prisinzano Thomas, R. Oakley Berl, T. C. Gamblin, R. Paranjape Smita, P. Riley Andrew, D. Somoza Amber, C. E. Oakley, C. C. Wang Clay, E. Prisinzano Thomas, R. Oakley Berl and T. C. Gamblin, ACS Chem. Neurosci., 2015, 6, 751–760 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |