Open Access Article
Open Access ArticleA rapid and reliable CE-LIF method for the quantitative analysis of miRNA-497 in plasma and organs and its application to a pharmacokinetic and biodistribution study†
Eunmi Ban,
Haejin Kwon and
Eun Joo Song *
*
College of Pharmacy and Graduate School of Pharmaceutical Sciences, Ewha Womans University, Seoul, 03760, Korea. E-mail: esong@ewha.ac.kr
First published on 18th May 2020
Abstract
MicroRNAs (miRNAs) are involved in the pathogenesis of many human diseases, and various miRNAs have been reported and developed as therapeutic candidates for treating various diseases. Various miRNA and carrier modification systems have been investigated for effective systemic miRNA delivery to cells, organs, and tissues of interest. Consequently, effective and reliable analytical methods of miRNAs are required for evaluating the pharmacokinetics and biodistribution of miRNAs as therapeutic candidates. The capillary electrophoresis with laser-induced fluorescence (CE-LIF) method has been recently reported as a promising and relatively rapidly developing tool with the potential to provide highly sensitive and specific analysis of biological molecules including miRNAs. Here, the CE-LIF method was used for application in the pharmacokinetic and distribution studies of miRNA-497 as a model miRNA for a lung target; miRNA-497 hybridized with 6-FAM-labeled DNA probes were separated using CE-LIF and detected within 6 min without any interference. This method showed a wide calibration range of 1.0–50 nM and 0.1–50 nM for plasma and the four organs, liver, spleen, lung, and kidney, respectively, with acceptable precision and accuracy. Using CE-LIF, the miRNA-497 level was evaluated in rat plasma and organs after intravenously administering 1 mg ml−1 of a miRNA-497 mimic. Hence, miRNA-497 displayed a relatively short half-life of 1.76 h and was delivered to the lungs but mainly accumulated in the liver and spleen. This study evaluated the pharmacokinetics and biodistribution of the miRNA-497 mimic using CE-LIF for the first time and suggested the need for further studies to extend the half-life and conduct lung-targeted delivery of miRNA-497.
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs (18–22 nucleotides long) that interact with their target mRNAs to regulate gene expression.1 Several miRNAs have been implicated in different physiological and pathological processes, including metabolism, proliferation, apoptosis, differentiation, development, viral infection, and cancer.2,3 Therefore, miRNAs have been widely investigated as therapeutic agents to treat various diseases.4 Therefore, to evaluate miRNAs for therapeutic purposes, the pharmacokinetics and distribution of miRNAs have been investigated after its administration.5,6 However, the delivery of miRNAs to target sites remains challenging because of poor cellular uptake and degradation by nucleases. In particular, in systemic administration, miRNAs broadly distribute into most organs or tissues via circulation, except in the central nervous system, and the amount of miRNA that accumulates differs greatly among different tissue types. Moreover, depending on the carrier type, the degree of miRNA accumulation in each organ or tissue varies. Consequently, various modification systems of miRNA and carriers have been investigated for effective systemic delivery of miRNAs to cells, organs, and tissues of interest. Therefore, pharmacokinetics and distribution evaluations have been conducted focusing on targeting and prolong circulation of miRNA. Mahato group showed prolonged the elimination half-life and enhanced delivery to pancreatic tumor of let-7b mimic using polymeric micelle through pharmacokinetics and distribution evaluation.7 And Yoshioka et al. designed heteroduplex oligonucleotide-anti-miR as a new technology comprising an anti-miR and its complementary RNA and demonstrated improvements of in vivo potency compared with the parent anti-miR through pharmacokinetics and distribution study.8MiRNA-497, a highly conserved miRNA located on chromosome 17p13.1, was recently found to play important inhibitory roles in malignancies by suppressing cancer cell proliferation and inducing apoptosis.9 Recent studies have shown that miRNA-497 is correlated with lung cancer.10,11 Besides, the studies of miRNA-497 as a therapeutic candidate have been conducted for treating lung cancer but little is known about its mechanism of action and the pharmacokinetics and biodistribution of circulating miRNA-497.
The analyses of circulating miRNA levels in plasma and organs have been mainly conducted using quantitative real-time PCR (qRT-PCR),12 enzyme-linked immunosorbent assay (ELISA),5 and liquid chromatography with tandem mass spectrometry (LC/MS/MS).6 More recently, capillary electrophoresis-laser induced fluorescence (CE-LIF) has been actively applied to analyze miRNA in biological samples.13–16 Krylov group showed accurate quantitative analysis of miRNAs from cells extracts using CE-LIF without interference by the sample matrix.13 Ban et al. have been successfully performed simultaneous detection of endogenous multiple miRNAs from cells using CE-LIF.15 Khan et al. showed that as low as 0.5 fM of miRNA in serum can be measured in a 20 min by CE-LIF without PCR amplification.16 Although qRT-PCR, ELISA, and LC/MS/MS have been previously used to evaluate pharmacokinetics and biodistribution, aspects such as extraction method, accuracy, and reproducibility remain a challenge in developing quantitative analysis methods for studying the pharmacokinetic and biodistribution of miRNAs. The qRT-PCR and ELISA system are very sensitive method for analyzing low levels of miRNAs in plasma but qRT-PCR analysis is interfered by the sample matrix or extraction reagents. The ELISA method has labor-and time intensive sample preparation process and relatively high variability. The LC/MS/MS method is a complicated sample extraction process and has the lowest sensitivity compared to other analytical methods. The CE-LIF method is a cost-effective and reliable tool with the potential to provide a highly sensitive, reproducible, and rapid analysis of miRNAs from biological samples without interference by the sample matrix.
Here, we first validated the CE-LIF method for its sensitivity, rapidity, and reliability in analyzing miRNA-497 levels in plasma and tissues. After intravenously administering miRNA-497 as a model therapeutic candidate to treat lung cancer, we also evaluated the pharmacokinetics and distribution of miRNA-497 by quantitatively analyzing its levels in plasma and organs, such as the liver, spleen, lung, and kidney, using the validated CE-LIF method.
Material and methods
Chemicals and materials
Fluorescence-labeled ssDNA oligonucleotide probe with 5′-carboxyfluorescein phosphoramidite (6-FAM) was purchased from Cosmogenetech (Seoul, Korea). Synthetic miRNA-497 (5′-CAGCAGCACACUGUGGUUUGUA-3′) and negative control mimic was purchased from Genolution (Seoul, Korea). TRIzol LS reagent and Cell/Tissue miRNA Purification Kit were obtained from Invitrogen (Carlsbad, CA, USA) and Genolution (Seoul, Korea), respectively. In vivo-jetPEI were purchased from Polyplus Transfection (Illkirch-Graffenstaden, France).Animal treatment, organ and blood collection
Sprague Dawley male rats (7 weeks old) were supplied by DooYeol Biotech (Seoul, Korea). All experimental protocols were approved by the Institutional Animal Care and Use Committee of Center at Woojung Bio (Suwon, Korea, Association for Assessment and Accreditation of Laboratory Animal Care-accredited facility) under approval number: WJIACUC 20181102. Rats were divided into two groups. The first group was treated with a single dose of 1 mg kg−1 miRNA-497 mimic loaded into in vivo-jetPEI while the second group was treated with 1 mg kg−1 negative control mimic loaded into in vivo-jetPEI. The solutions containing miRNA-497 or negative control mimic and in vivo jetPEI were mixed (N/P = 7) and incubated according to the manufacturer's protocol and intravenously injected via the tail vein at the dose of 1 mg kg−1 for each blood samples were collected from three rats per group per time point at 0, 1, 7.5, 15, 45, 90, 180, 360, 720, and 1440 min post-dose by retro-orbital puncture into EDTA tubes. Rats were also sacrificed on 0.25, 6, and 24 hours to collect the liver, spleen, lung, and kidney for analysis of miRNA-497. Thereafter, plasma was obtained from the blood sample by centrifugation at 3000 × g for 10 minutes at 4 °C. The resulting plasma was stored at −80 °C until ready to be used. Tissues were snap-frozen in the liquid nitrogen stored at −80 °C before analysis.RNA isolation
Total RNA from organs and plasma was isolated using Cell/Tissue miRNA Purification Kit (Genolution) and TRIzol LS Reagent (Thermo Fisher Scientific), respectively. Frozen organ samples were cut into small pieces in lysis buffer on ice to prevent thawing. RNA extractions from organs were performed according to the manufacturer's instructions. Extracted RNA was and stored at −80 °C. The total RNA concentration and quality were assessed using NanoDrop Lite Spectrophotometer 120 V (Thermo Fischer Scientific, MA, USA) at the absorbance of 260, and 280 nm.Plasma samples were mixed by TRIzol LS reagent (Invitrogen, Carlsbad, CA, USA), followed by chloroform. Each sample was vortexed for 30 seconds and incubated at RT for 5 minutes. Phase separation was performed by centrifuging the samples for 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 minutes at 4 °C. 400 μl of the aqueous phase was transferred to a new tube. 500 μl of isopropanol was added to the aqueous phase and added 5 mg ml−1 glycogen (Invitrogen, San Diego, CA, USA) and 10 μg ml−1 yeast tRNA (Sigma-Aldrich, St. Louis, MO, USA) for RNA precipitation. After precipitation of RNA incubate in −80 °C for 1 hour, centrifuge at 20
000 × g for 15 minutes at 4 °C. 400 μl of the aqueous phase was transferred to a new tube. 500 μl of isopropanol was added to the aqueous phase and added 5 mg ml−1 glycogen (Invitrogen, San Diego, CA, USA) and 10 μg ml−1 yeast tRNA (Sigma-Aldrich, St. Louis, MO, USA) for RNA precipitation. After precipitation of RNA incubate in −80 °C for 1 hour, centrifuge at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 minutes at 4 °C. The RNA pellet was dissolved with 30 μl of RNase-free water.
000 × g for 30 minutes at 4 °C. The RNA pellet was dissolved with 30 μl of RNase-free water.
Quantitative real-time PCR
Isolated RNA and miRNA were used for cDNA synthesis. The transcription was performed in a reaction mixture, containing 500 ng RNA samples using Mir-X™ miRNA first-strand synthesis and SYBR qRT-PCR kit from Clontech (Mountain View, CA, USA) according to the manufacturer's protocol. cDNA synthesis was conducted for 1 h at 37 °C and then terminated by heating at 85 °C for 5 min. cDNA was diluted 10 times and mixed with SYBR qPCR Mix containing 0.5 μl of 10 μM miRNA-497 specific primers and universal mRQ 3′ reverse primer from the Mir-X™ miRNA qRT-PCR SYBR kit. PCR was performed for 10 s at 95 °C; 5 s at 95 °C, 20 s at 60 °C for 40 cycles and finalized by a melting curve 5 s for each 0.5 °C. The CFX Connect™ real-time PCR detection system (Bio-Rad, Hercules, CA, USA) was performed for both cDNA and real-time PCR reactions. Finally, the samples were analyzed using the CFX Manager™ software (Bio-Rad).Capillary electrophoresis
The extracted miRNA was analyzed using the PA800 plus CE system (Beckman Coulter, Fullerton, CA, USA) with the LIF detector. Fluorescence was detected by excitation at 488 nm using a 3 mW argon-ion laser and emission through a 520 nm emission filter at a rate of 4 Hz. Separations were performed using an untreated capillary (Beckman Coulter), which had a 75 μm inner diameter and 50 cm length (40 cm effective length). Separations were performed using a running buffer of 100 mM Tris–borate (pH 10.0) containing 2.5 M urea at 35 °C by applying 20 kV while the sample compartment was maintained at 25 °C. Samples were introduced hydrodynamically at 0.5 psi for 9 s. Electropherograms were analyzed using 32 Karat software.Hybridization
For hybridization, 6-FAM-labeled DNA probes (1 nM) and synthetic miRNAs (1 nM) were mixed with the hybridization buffer (50 mM Tris–acetate, pH 8.0, containing 50 mM NaCl, 0.1 mM EDTA, and 1% Triton X-100). The labeled DNA probes were dissolved in elution buffer (10 mM Tris–Cl buffer, pH 8.5) from a QIAprep Spin Miniprep Kit (Qiagen, Crawley, UK). The samples were incubated in a thermal cycler (Eppendorf) for denaturation at 95 °C for 5 min, followed by a renaturation step at 40 °C for 15 min prior to introduction into the capillary.Stability test of miRNA by RNases in carrier
4 μg miRNA mimic was formulated with 0.64 μl in vivo-jet PEI carrier reagent to a 5% final glucose concentration,17 and then in incubated at 37 °C for 0.25, 1, 3, 6 and 24 h. The samples were analyzed by CE-LIF after extraction using TRIzol LS.Pharmacokinetic data analysis
Pharmacokinetic parameters were estimated using non-compartment analysis of the composite data with WinNonlin software version 6.0 (Pharsight, Mountain View, CA).Statistical analysis
Results are expressed as mean ± SD, unless otherwise indicated. Statistically significant difference between two groups was determined by two-tailed Student's t-test. A P value of 0.05 was taken as statistically significant.Results and discussion
Analysis of miRNA in plasma and organs using CE-LIF
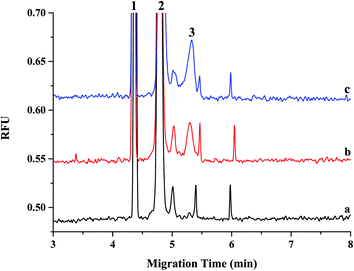
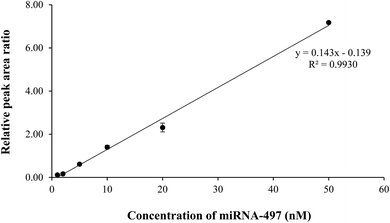
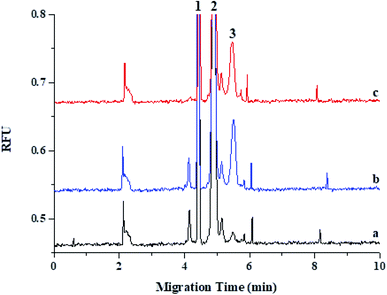
Currently, qRT-PCR is mainly used for analyzing the expression levels of miRNAs, although it has some limitations in quantifying miRNAs owing to its short length. Moreover, for conducting a reliable miRNA analysis, qRT-PCR demands high-purity RNA samples, but the purity of RNA extracted from matrices, such as plasma and tissue, depends on factors such as extraction method or sample type. Because of these limitations, many analytical systems such as LC/MS/MS and ELISA have been developed for detecting miRNA in biological fluids and tissues instead of qRT-PCR. However, these analytical systems have disadvantages such as an extensive extraction process or a low reproducibility. Recently, the CE-LIF system was used as an effective tool for analyzing miRNA, and many research groups, including ours, have been continuously developing this system for conducting miRNA analysis in biological fluids and cells. Here, we first applied CE-LIF system to quantitatively and reproducibly analyze miRNA levels in organs and plasma by evaluating the pharmacokinetics and biodistribution of miRNA-497 mimic as a therapeutic agent for treating lung cancer because its administration had shown a desirable therapeutic effect on a lung cancer model in our previous study.18 For validating CE-LIF-based miRNA analysis, spiked-in plasma with miRNA-497 mimic were extracted using Trizol LS reagent, and the extracted miRNA-497 was analyzed using CE-LIF. The migration time for miR-497 was estimated to be 5.4 min. Fig. 1 shows CE-LIF electropherograms representing the analysis of miRNA-497 levels in plasma. As shown in Fig. 1a and b, miRNA-497 showed good separation, and no interfering peaks from the endogenous plasma components, which were observed at the migration time of miRNA-497. Fig. 1c shows an electropherogram for a plasma sample obtained from a rat subject 15 min after administration of miRNA-497 mimic. The calibration curves for plasma miRNA-497 levels showed linearity in the range of 1–50 nM (R2 = 0.9930) and typical equations for the calibration curves were y = 0.1436x − 0.1391 (Fig. 2). The accuracy range and precision value at the three miRNA-497 concentrations was 81.0–113.4% and 2.57%, respectively. Likewise, homogenized mouse organ tissues, including those of the liver, spleen, lung, and kidney, were extracted using Tissue miRNA Purification Kit and analyzed using the CE-LIF system. Fig. 3 shows the typical CE-LIF electropherograms obtained for the lung samples. In Fig. 3, the peak miRNA-497 level in the lung sample collected at 15 min after intravenous (IV) administration of miRNA-497 mimic was 4.5-fold higher than that in the lung sample collected before administration of miRNA-497 mimic; however, the peak height in the former was similar to that in the lung sample spiked with 5 nM miRNA-497 mimic. ESI Fig. S1–S3† shows CE-LIF electropherograms representing miRNA-497 analysis in the liver, spleen, and lung. The migration time for miR-497 was estimated to range between 5.2 and 5.6 min, and on comparing its migration time according to tissue matrix, its migration time was found to be slower in tissues than in plasma. MiRNA-497 in four organs showed linear (R2 > 0.9990) concentration-dependent absorbance at concentration calibration lines (0.1–50 nM for the liver, 0.1–25 nM for the spleen, and 0.1–10 nM for the lung and kidney). Typical equations for the calibration curves for miRNA-497 in the liver, lung, kidney, and spleen were y = 0.1494x + 0.0250, y = 0.1564x + 0.1001, y = 0.1490x + 0.0476, and y = 0.1790x + 0.0730, respectively (ESI Fig. S4–S7†). The accuracy and precision range at the three miRNA-497 concentrations in the four organs was 83.5–108.9% and 0.81–4.48%, respectively.This validation procedure indicated that a fast and reliable analysis of miRNA-497 in plasma and organs can be conducted using the developed CE-LIF method. This method showed a 10-fold improvement in sensitivity over the LC/MS/MS method19 and was more precise than the ELISA method.20
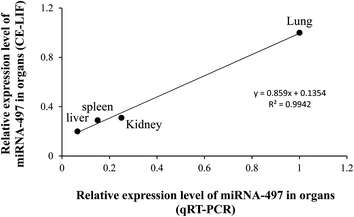
Additionally, to confirm that miRNA from biological samples, such as plasma and organs, could be accurately determined using the CE-LIF method, the intensity of endogenous miRNA-497 in the liver, spleen, lung, and kidney analyzed using CE-LIF was compared with that analyzed using qRT-PCR. The data obtained using CE-LIF were similar to those obtained using qRT-PCR, suggesting that the level of endogenous miRNA-497 was indeed the highest in the lung, as shown in Fig. 4. Thus, these two methods were well correlated (R2 = 0.9942); this also indicates that the CE-LIF system for evaluating miRNA-497 level is as efficient as the qRT-PCR method, which is currently considered as the most common and reliable method for evaluating miRNA expression level.
Pharmacokinetics and organ distribution of miRNA-497 after IV administration of miRNA mimic
The pharmacokinetics and biodistribution study of miRNA-497 was conducted after administering a single IV injection of 1 mg kg−1 of miRNA-497 mimic. For systemic in vivo delivery of the miRNA-497 mimic, a commercially available carrier, in vivo-jetPEI (Polyplus Transfection; Illkirch-Graffenstaden, France) based on cationic linear polyethyleneimine was used. The in vivo-jetPEI has been previously used for gene delivery in vivo studies and clinical trials, and many studies have demonstrated that this carrier can effectively deliver gene-based therapeutic agents with high stability to multiple organs without causing any toxicity.21 In the current study, before IV intravenously administration administering of miRNA-497 using in vivo jetPEI, the stability of miRNA-497 stability in plasma was tested. The result showed revealed that miRNA-497 mimic was protected from degradation by plasma nuclease for at least 6 h at 37 °C, from degradation by plasma nuclease as shown in Table 1.| % Remaining from initial concentration at a predetermined time (±SD) | ||||
|---|---|---|---|---|
| miRNA-497 | 0.25 h | 3 h | 6 h | 24 h |
| 98.4 ± 15.0 | 100.1 ± 4.8 | 85.5 ± 5.5 | 52.0 ± 2.8 | |
We next analyzed the pharmacokinetics following a single injection of 1 mg kg−1 of miRNA-497 mimic. The results indicated that the blood concentrations of miRNA-497 were ∼4105.6 nM at the earliest sampling time point (1 min) and then decreased to ∼28.4 nM within 15 min. Approximately 95.4% of the earliest measured concentration–time was eliminated from the blood during this time frame. Moreover, 3 h after IV injection, the plasma peak concentration of miRNA-497 was reduced to the quantification limit level and after 6–24 h, the miRNA was not detectable (Fig. 5). A non-compartmental fit of the blood concentration-versus-time curve produced an estimated half-life of miRNA-497 of 1.76 h and an AUC value of 9784.6 min nM (Table 2).
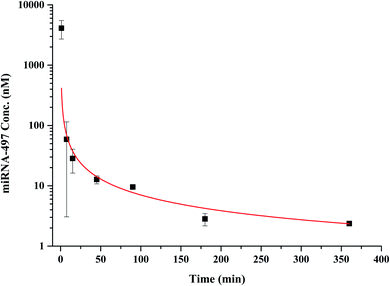 | ||
| Fig. 5 Plasma concentration–time profiles of miRNA-497 in rats following an iv injection (1 mg kg−1). Dots represent means of measured concentrations; bars represent SD (n = 3). | ||
| Parameters | miRNA-497 |
|---|---|
| a Data were fit to a non-compartmental PK model. Values represent means ± S.D generated from three rats. AUC(last), area under the plasma concentration–time curve from time zero to time of last measurable concentration; CL/F, apparent clearance; Cmax, peak plasma concentration; T1/2, plasma terminal half-life; Tmax, time to peak plasma concentration; Vz/F, apparent volume of distribution; Elimination K, elimination rate constant. | |
| AUC (last) (nM min) | 9784.56 ± 1561.66 |
| Cmax (nM) | 4105.57 ± 1135.50 |
| Tmax (min) | 1.0 |
| CL/F (L min−1 kg−1) | 0.01 ± 0.0016 |
| Vz/F (L kg−1) | 1.58 ± 0.59 |
| T1/2 (min) | 105.71 ± 23.57 |
| Elimination K (min−1) | 3.68 ± 2.40 |
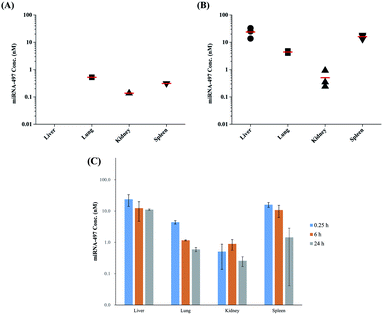
Preclinical development of synthetic miRNAs requires information on organ or tissue distribution, as with other small molecular drugs. MiRNAs as therapeutic candidates are currently delivered systemically using IV and subcutaneous injections, and these miRNAs distribute broadly distribute into most tissues, except the central nervous system. Among these such organs, the major sites of accumulation of miRNA accumulation s are the spleen and liver22 because, during blood circulation, a significant portion of drugs are taken up by the reticuloendothelial system (RES), such as including the spleen, liver, and lungs. However, the accumulation sites differ greatly according to miRNA types and injection site. Indeed, the organ peak concentration of miRNAs among in the treated organ miRNAs samples differed when for the three varieties of the injected synthetic miRNAs were injected.5 Moreover, some reports showed revealed that the accumulation organs in which accumulation of the tested miRNAs occurred were different by depending on injection sites when PEI was used as a carrier for gene delivery.23,24 The major sites of accumulation of Cel-miRNA-39 were the spleen and liver,25 while whereas DNA plasmids predominantly accumulated in the lungs26 when in vivo-jetPEI was used as a carrier for gene delivery and miRNA and DNA plasmids were administrated by IV injection intravenously. Thus, to develop miRNA-based therapeutics, it is important to check whether the administration of miRNA results in leads to its accumulation in the target organ. Therefore, in this study here, we analyzed the concentration of miRNA-497 concentration in the liver, spleen, kidney, and lungs as part of the typical RES, following after IV dose administering an IV dose of at 1 mg kg−1 whether miRNA-497 was accumulated in the lung. The analysis of the concentration of miRNA-497 concentration in tested organs was conducted using a validated CE-LIF approach. From these analyses, the an increase of in miRNA-497 peak was detected from the collected liver, spleen, and lung samples collected at 15 min after IV administration of miRNA-497 mimic compared to with the peak observed before its administration of miRNA-497 mimic. As shown in Fig. 6A and B, among the tested organs, the highest concentration of miRNA-497 concentration was the highest found in the liver followed by in the spleen and lung, and whereas it was the lowest level was observed in the kidney. In contrast, the highest endogenous level of miRNA-497 was found in the lung followed by in the spleen and kidney, and while the level of its level miRNA-497 was too low to be detected in the liver. Their results showed revealed that the administered injected miRNA-497 mainly accumulated in the liver, spleen, and lung. The level of miRNA-497 level in organs was also tested in a time-dependent manner, as shown in Fig. 6C. The concentration of miRNA-497 in the liver was the highest among the tested organs and was sustained maintained levels at that level for up to 24 h, while whereas the level of miRNA-497 concentration was not detected in plasma could not be detected at 24 h. In the lung, the miRNA-497 concentration peaked at 15 min, then after which it declined rapidly until 6 h post-injection and subsequently dropped slowly to the basal level thereafter for within 24 h. Based on this result, miRNA-497 could be delivered to the lung and used as a therapeutic agents for treating lung disease although the an appropriate formulation is needed for targeting the lungs. In the spleen, the miRNA-497 concentration peaked at 15 min, then after which it declined rapidly until within 24 h post-injection, but the concentration of miRNA-497 concentration at 24 h was higher than the basal level concentration; but however, a high inter-variation was observed among spleen samples was observed. Kidney had a distinctly different time course of miRNA-497 organ concentrations had a distinctly different time course in the kidney as opposed to the other organs: the concentrations of miRNA-497 peaked at 6 h, followed by a slow drop until for the next 24 h. However, the peak concentration was low, although detectable; and eventually, this result showed revealed that miRNA-497 is rarely delivered to the kidney. ESI Fig. S8–S11† showed the change in concentration of miRNA-497 concentration in the tested organs as a function of time. From these biodistribution studies, it was observed that the administered miRNA-497 was mainly delivered to the liver, lung, and spleen, and its level of miRNA-497 in the liver was maintained at for 6–24 h after the administration of miRNA-497 mimic.
Conclusion
In this study, as a candidate of a therapeutic agent for the treatment of lung cancer, the miRNA-497 level in plasma and organs was evaluated after intravenously administering 1 mg kg−1 of miRNA-497 mimic using in vivo-jetPEI as a carrier. Moreover, a fast and effective CE-LIF method was validated to analyze miRNA in plasma and organs. The result indicated that the half-life of miRNA-497 was 1.76 h and that miRNA-497 was delivered in the lung, liver, and spleen but was mainly accumulated in the liver and spleen.In conclusion, for the first time, the CE-LIF method was validated and applied to pharmacokinetics and biodistribution evaluation of miRNA. This CE-LIF method showed that a fast and reliable analysis of miRNA-497 in plasma and organs can be conducted. However, the addition of internal standard for reducing variation during extraction and analytical process is required for more precise and accurate quantitative analysis of miRNA. From the pharmacokinetics and biodistribution evaluation of miRNA-497, it was found that miRNA-497 was rapidly eliminated from the circulation and distributed in all the tested rat organs (liver, lung, kidney, and spleen); however, it was mainly accumulated in the liver and spleen after IV administration of miRNA-497 mimic. Therefore, further studies are needed to develop a formulation or chemical modification for the long biological half-life and the lung-targeted delivery of miRNA-497. Moreover, the pharmacodynamics and in vivo toxicity have to be determined as a therapeutic agent for lung cancer through further studies.
Ethics approval and consent
Institutional Animal Care and Use Committee of Center at Woojung Bio (Suwon, Korea, Association for Assessment and Accreditation of Laboratory Animal Care-accredited facility) under approval number: WJIACUC 20181102.Conflicts of interest
The authors declare that they have no conflicts of interest.Acknowledgements
This work was supported by the Ewha Womans University Research Grant of 2019. The Eunmi Ban was supported by RP-Grant 2019 of Ewha Womans University.References
- D. P. Bartel, Cell, 2004, 116, 281–297 CrossRef CAS PubMed.
- V. T. Nguyen, B. H. S. Le and Y. J. Seo, Analyst, 2019, 144, 3216–3220 RSC.
- X. Chen, D. Xie, Q. Zhao and Z. H. You, Briefings Bioinf., 2019, 20, 515–539 CrossRef CAS PubMed.
- X. Chen, N. N. Guan, Y. Z. Sun, J. Q. Li and J. X. Qu, Briefings Bioinf., 2020, 21, 47–61 Search PubMed.
- H. Wang, M. Chiu and Z. Xie, et al., Mol. Pharm., 2012, 9, 1638–1644 CrossRef CAS.
- M. E. G. Cantafio, B. S. Nielsen and C. Mignogna, et al., Mol. Ther.--Nucleic Acids, 2016, 5, e336 CrossRef.
- V. Kumar, V. Mundra, Y. Peng, Y. Wang, C. Tan and R. I. Mahato, Theranostics, 2018, 8, 4033–4049 CrossRef CAS PubMed.
- K. Yoshioka, T. Kunieda and T. Yokota, et al., Nucleic Acids Res., 2019, 47, 7321–7332 CrossRef CAS.
- L. Gu, H. Li and L. Chen, et al., Oncotarget, 2015, 6, 32545–32560 CrossRef.
- C. Huang, R. Ma, J. Yue, N. Li, Z. Li and D. Qi, Cell. Physiol. Biochem., 2015, 37, 342–352 CrossRef CAS.
- G. Nigita, R. Distefano and D. Veneziano, Sci. Rep., 2018, 8, 10222 CrossRef PubMed.
- K. Kelnar, H. J. Peltier, N. Leatherbury, J. Stoudemire and A. G. Bader, Anal. Chem., 2014, 86, 1534–1542 CrossRef CAS PubMed.
- L. Hu, A. S. Stasheuski and S. N. Krylov, et al., Anal. Chem., 2017, 89, 4743–4748 CrossRef CAS PubMed.
- E. Ban, D. K. Chae, Y. S. Yoo and E. J. Song, Anal. Bioanal. Chem., 2017, 409, 6397–6404 CrossRef CAS PubMed.
- E. Ban, D. K. Chae and E. J. Song, J. Chromatogr. A, 2013, 1315, 195–199 CrossRef CAS PubMed.
- N. Khan, J. Cheng, J. P. Pezacki and M. V. Berezovski, Anal. Chem., 2011, 83, 6196–6201 CrossRef CAS PubMed.
- J. You, Y. Li, N. Fang, B. Liu, L. Zu, R. Chang, X. Li and Q. Zhou, PLoS One, 2014, 9, e91827 CrossRef PubMed.
- D. K. Chae, J. Y. Park and E. J. Song, et al., Mol. Oncol., 2019, 13, 2663–2678 CrossRef CAS PubMed.
- M. Kullolli, E. Knouf and S. J. Pitteri, et al., J. Am. Soc. Mass Spectrom., 2014, 25, 80–87 CrossRef CAS PubMed.
- K. K. Chan, Z. Liu and G. Marcucci, et al., AAPS J., 2010, 12, 556–5568 CrossRef CAS PubMed.
- B. G. Nezami, S. M. Mwangi and J. E. Lee, et al., Gastroenterology, 2014, 146, 473–483 CrossRef CAS PubMed.
- D. B. Shushan, E. Markovsky, H. Gibori, G. Tiram, A. Scomparin and R. Satch-Fainaro, Drug Delivery Transl. Res., 2014, 4, 38–49 CrossRef PubMed.
- A. F. Ibrahim, U. Weirauch and M. Thomas, et al., Cancer Res., 2011, 71, 5214–5224 CrossRef CAS PubMed.
- K. Schlosser, M. Taha, J. Duncan and D. J. Stewart, Theranostics, 2018, 8, 1213–1226 CrossRef CAS PubMed.
- K. Schlosser, M. Taha, Y. Deng and D. J. Stewart, Pulm. Circ., 2017, 8, 1–4 Search PubMed.
- Y. K. Song, F. Liu F and D. Liu, Gene Ther., 1998, 5, 1531–1537 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra01213k |
| This journal is © The Royal Society of Chemistry 2020 |