Open Access Article
Open Access ArticleThe binding of key fishy off-flavor compounds to silver carp proteins: a thermodynamic analysis
Saiqi Gu ab,
Wangli Daia,
Yunqing Chongc,
Fei Lyua,
Xuxia Zhou*a and
Yuting Ding*a
ab,
Wangli Daia,
Yunqing Chongc,
Fei Lyua,
Xuxia Zhou*a and
Yuting Ding*a
aCollege of Food Science and Technology, Zhejiang University of Technology, Hangzhou 310014, China. E-mail: xzhou@zjut.edu.cn
bNinghai ZJUT Academy of Science & Technology, Ninghai, 315600, China
cCollege of Food Science and Biotechnology, Zhejiang Gongshang University, Hangzhou 310018, China
First published on 19th March 2020
Abstract
The binding of key fishy off-flavor compounds (KFOCs), heptanal, octanal, nonanal, and 1-octen-3-ol, to silver carp proteins (total myofibrillar protein, myosin, and actin) was studied. Myosin was the primary binding receptor for all four KFOCs, and it showed the strongest binding at neutral pH and at higher ionic strengths. Thermodynamic models were applied to evaluate the number of binding sites, the binding constant, and the Gibbs free energy for the binding of the KFOCs to myosin. Myosin had approximately 1.0 sites for binding with the three linear-chain aldehydes and about 1.6 sites for binding with 1-octen-3-ol. Moreover, myosin showed the highest affinity for 1-octen-3-ol, and both its binding constant and its number of binding sites with the three linear-chain aldehydes were negatively correlated with the chain length. For all four KFOCs, the trends of the Gibbs free energies were the opposite of those observed for the binding constant and the number of binding sites. These results may provide a theoretical basis for improving the deodorization efficiency of traditional surimi rinsing methods by adjusting the properties of the solutions used.
Introduction
Surimi is a tasteless and odorless fish-meat protein concentrate, which when further processed is an intermediate ingredient in surimi-based products such as kamaboko and crab-meat analogues. In general, the fish muscle is minced, washed to remove the fat and undesirable matters (such as blood, pigments and odorous substances), and mixed with cryoprotectants (such as sugar and/or sorbitol) to obtain surimi.1,2 In China, the traditional materials used for surimi production are various kinds of low-value freshwater fish, such as silver carp (Hypophthalmichthys molitrix). However, freshwater-fish surimi has a high concentration of fishy off-flavor compounds (FOCs) that are difficult to remove. The presence of these compounds is an important factor that determines the acceptability and consumption of surimi and has been considered one of the main constraints to the development of the surimi food industry.The traditional method of removing FOCs in surimi processing is rinsing.3 However, rinsing can only wash away some of the free FOCs. The main reason for this is the complexity of the FOCs and the binding of the FOCs to non-volatile food ingredients.4–6 Studies have shown that meat proteins can interact with volatile compounds, which would affect their release and the perception of quality in a significant manner.7 Recently, there has been increased interest in elucidating the interactions between flavor compounds and non-volatile food components, especially proteins.
These interactions have been studied for vegetable proteins (e.g. soy proteins)8 as well as skeletal peptides (e.g. carnosine and anserine) and sarcoplasmic proteins (e.g. myoglobin and β-lactoglobulin).4,9 There have also been several studies performed on the interactions between volatile compounds and myofibrillar proteins in duck and pork muscle.10,11 However, to the best of our knowledge, relatively few studies have been performed on the interactions between different types of natural myofibrillar proteins (e.g. myosin and actin) and FOCs which, thus far, have not been successfully removed during fish surimi processing.
The interactions between flavor compounds and proteins, or more specifically, the binding of flavor compounds to proteins are affected by both the physicochemical properties of the flavor compounds and the properties of the proteins (e.g. protein concentration, protein type, and conformation).12 The organic compounds responsible for the fishy off-flavor in freshwater fish products are widely reported to be geosmin,3,13 as well as volatile aldehydes and alcohols such as heptanal, octanal, nonanal, and 1-octen-3-ol.14,15 Our previous studies have shown that 13 volatile compounds with odor activity values greater than 1 (OAVs > 1) were the aroma-active compounds (AACs) that contribute primarily to the integral aroma profile of silver carp mince.3 In addition, we reported that the AACs—heptanal, octanal, nonanal, and 1-octen-3-ol—which displayed relatively high OAV values, make a major contribution to the overall fishy flavor of minced products of silver carp. Therefore, the above compounds were defined as the key fishy off-flavor compounds (KFOCs) of interest in the present study. The clarification of the interactions between fish meat proteins and these compounds is of particular importance for the removal of fishy off-flavor during fish surimi processing.
A traditional method for evaluating the interactions between volatile compounds and proteins has been headspace GC-MS. More recently, solid-phase microextraction (SPME) has been widely used to trap free molecules and detect the concentration of the analytes.4 The headspace concentration of the analyte is related to its concentration in the fiber coating, which is based on the equilibrium between the polymeric coating, the headspace, and the matrix. The fiber/headspace partition coefficients can be determined from the forces that drive the analyte from the air to the fiber coating.
In the present study, we analyzed the effects of silver carp surimi proteins on the removal of four KFOCs (heptanal, octanal, nonanal, and 1-octen-3-ol), the effects of protein type, and the properties of the medium on the binding of KFOCs. For the first time, the binding behavior of these KFOCs to myosin was investigated via the analysis of thermodynamic models.
Materials and methods
Materials
Three batches of fresh silver carp (ten per batch) of commercial size (average weight approximately 1500 g) were obtained from the Zhaohui Agricultural Products Market in Xiacheng District, Hangzhou, Zhejiang province, China, in the middle of February 2019. The silver carp were transported alive to the laboratory in plastic bags containing local pond water, mixed together and labeled. All silver carp samples used in this study were killed by a percussive blow on the head with a 250 g hammer. Next, the foreign substances were washed away, the fish scales scraped off, and the head, net viscera and bones removed. The fish muscle was cut into 100 g portions, packaged in vacuum bags, and stored at −20 °C prior to further preparation of the fish proteins (total myofibrillar protein, myosin, and actin). The standard samples of the four KFOCs used in this study—heptanal, octanal, nonanal and 1-octen-3-ol (≥99.2%)—were obtained from Aladdin Industrial Corporation Shanghai, China.Methods of extraction and determination
Volatile-protein binding analysis
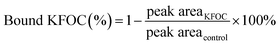 | (1) |
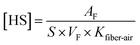 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 986, 38
986, 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 792, 41
792, 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 865 and 30
865 and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 560 for heptanal, octanal, nonanal and 1-octen-3-ol, respectively.
560 for heptanal, octanal, nonanal and 1-octen-3-ol, respectively.The free concentration (F) and the moles of each KFOC bound per mole of myosin (ν) in the aqueous phase were calculated using the equations reported by Pérez-Juan et al.:20
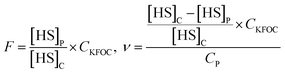 | (3) |
In above equations, [HS]C and [HS]P are the HS concentrations of each KFOC in the control and the protein (myosin) vials, respectively. CKFOC is the linear concentration of KFOC (0.1 mg L−1, 0.25 mg L−1, 0.5 mg L−1, 1.0 mg L−1, 2.5 mg L−1, and 5.0 mg L−1) added to the myosin solutions, and CP is the myosin concentration (5.0 g L−1). CKFOC and CP need to be converted into molar concentrations before calculation.
Non-covalent (reversible) binding of the KFOCs with proteins can be represented by the Scatchard equation,4 which can also be expressed in the form of a double-reciprocal equation (Klotz plot):21
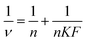 | (4) |
The binding parameters (n and K) are calculated from a plot of 1/ν versus 1/F, which should give a straight line with a slope of 1/nK and an intercept of 1/n. The equilibrium binding constant K, determined under experimental conditions as a function of temperature, facilitates the determination of the thermodynamic parameter the Gibb's free energy of binding (ΔG):
ΔG = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K K
| (5) |
Statistical analyses
The interactions between the proteins and the KFOCs were analyzed by an analysis of variance (ANOVA) using SPSS, version 22.0 (SPSS Inc., Chicago, IL, USA). The means were compared using Fisher's least significance difference (LSD) procedure (p < 0.05). All graphs were created using Origin 8.5 (Microcal Software, Inc., Northampton, MA, USA).Results and discussion
Standard curves for the KFOCs
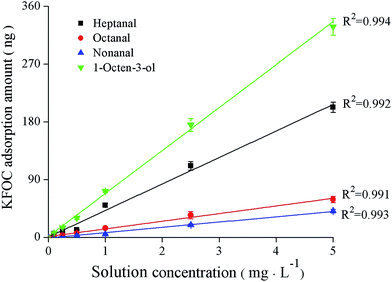
The standard curves showing the correlation between the GC responses (peak areas) and the quantities of the KFOCs (heptanal, octanal, nonanal, and 1-octen-3-ol) are shown in Fig. 1. A favorable linear relationship between the quantities of the KFOCs in the experimental ranges and the responses of the GC can be observed. This indicates that the GC-MS method can be successfully used for the subsequent determination of the linearity of the SPME extraction method.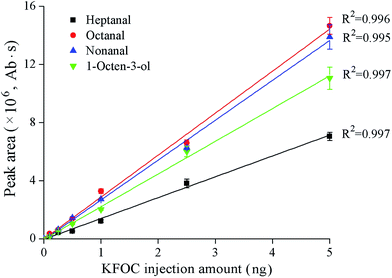 | ||
| Fig. 1 Linear correlations between the peak areas and the quantities of the four key fishy off-flavor compounds (KFOCs) obtained by liquid direct injection. | ||
Analysis of the linearity of the SPME extraction
The linearity of the KFOCs detected under equilibrium conditions can be observed in Fig. 2. After extraction equilibria were attained, the correlation coefficients between the SPME adsorption quantities of the KFOCs (obtained by Fig. 1) and their concentrations in solution were greater than 0.99. This indicates that the SPME technique can be used, under the conditions tested, to quantify changes in volatile concentrations, over the range of 0.1 mg L−1 to 5 mg L−1, of the four KFOCs.Binding of the KFOCs to the fish proteins
The percentages of the bound KFOCs in the headspaces of different binding solutions, each containing 5.0 g L−1 of fish protein, are shown in Table 1. As a control, the percentage of each bound KFOC in the absence of protein was set to 0%. The presence of total myofibrillar protein (TMP) and myosin produced a significant increase in the percentages of the bound KFOCs, reflecting that TMP and myosin were able to bind all the KFOCs tested. On the contrary, actin did not produce a remarkable increase in the percentages of bound KFOCs (p > 0.05). These differences in binding ability could be the result of differences in amino acid composition and protein structure. Gianelli et al.5 studied the binding of volatile compounds to peptides in skeletal muscle and concluded that the structures of the peptides had an important effect on their binding potential. Pérez-Juan et al.22 also studied the interactions of different types of proteins in post-rigor porcine muscle with volatile compounds and found that G-actin was unable to bind any of the assayed volatile compounds although the polymerized form (F-actin) bound higher quantities of the volatile compounds. It has been reported that changes in protein conformation affect the interactions between proteins and volatile compounds owing to modifications of the available protein binding sites.7| Protein | Percentage of bound KFOC/% | p-Value | |||
|---|---|---|---|---|---|
| Heptanal | Octanal | Nonanal | 1-Octen-3-ol | ||
| TMP | 48.744 ± 3.037Ab | 47.27 ± 2.914Ab | 47.015 ± 2.851Ab | 56.326 ± 3.301Bb | 0.017 |
| Myosin | 50.927 ± 3.265Ab | 50.402 ± 3.220Ab | 48.613 ± 3.007Ab | 63.704 ± 4.019Bc | 0.002 |
| Actin | 2.205 ± 0.417Aa | 2.644 ± 0.404Aa | 2.888 ± 0.394Aa | 3.207 ± 0.511Aa | 0.105 |
| p-Value | <0.001 | <0.001 | <0.001 | <0.001 | |
Although no significant differences were found between the binding properties of the three aldehydes studied (heptanal, octanal, and nonanal), total myofibrillar protein (TMP) and myosin bound more 1-octen-3-ol than the aldehydes under the same conditions. This might be the result of structural differences between the KFOCs. Wang et al.23 indicated that structural differences between the volatile aldehydes and ketones investigated in their study resulted in significant differences in protein binding. Zhou et al.24 pointed out that increases in the size of the alkyl groups strengthened hexanal binding with peptides. By comparison, myosin produced the highest percentages of bound KFOCs among all three fish proteins (Table 1). Therefore, myosin was used for further investigation of the effects of the characteristics of the reaction medium on binding.
Effects of the reaction medium on the binding of myosin to the KFOCs
The pH of the solution also affected the ability of myosin to bind the KFOCs (Fig. 3A). As the pH increased, binding was initially strengthened and then eventually weakened. The percentages of the bound KFOCs were basically highest at pH 7.0, indicating the strongest binding ability. This difference may be a result of the effect of pH on the solubility and conformation of the proteins.20 Xu et al.25 found that the solubility and turbidity of silver carp proteins declined with declining pH from 6.3 to 4.4 potentially making them less available to bind volatile organic compounds. Reiners et al.26 and Kühn et al.27 indicated that the adsorptive capacity of β-lactoglobulin to alcohols decreases with an increase of the pH value and that this might be attributed to the decrease of binding ability between hydrogen bonds and proteins under alkaline conditions.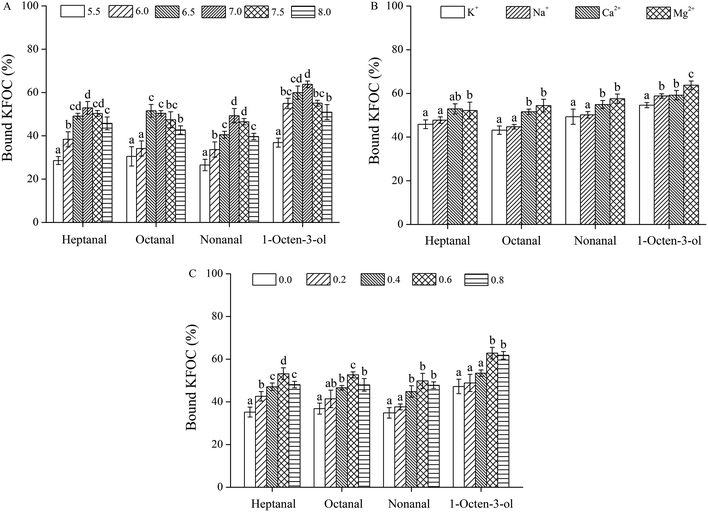 | ||
| Fig. 3 Effects of the characteristics of the medium on the binding of the four key fishy off-flavor compounds (KFOCs) to myosin: (A) pH; (B) salt type; (C) ionic strength. | ||
The ability of myosin to bind the KFOCs was influenced by different salt types as shown in Fig. 3B. Although K+ and Na+ were observed to have no significant effects on the binding of the three linear-chain aldehydes to myosin (p > 0.05), Na+ significantly promoted the adsorption of 1-octene-3-alcohol by myosin in comparison with K+ (p < 0.05). Compared to K+ and Na+, the presence of Mg2+ and Ca2+ both strengthened the binding of all four KFOCs to myosin. Moreover, the presence of Ca2+, when compared with Mg2+, strengthened the ability of myosin to bind heptanal but weakened the binding of the other three KFOCs. These results were basically consistent with a study by Pérez-Juan et al.,20 who reported that the presence of K+ and Na+ affected the adsorption of volatile compounds because of enhanced salting-out behavior, while Mg2+ and Ca2+ exhibited a weaker effect.
The ionic strength also affects the binding ability of myosin to the KFOCs (Fig. 3C). When the ionic strength was less than 0.6 M, the percentage of bound KFOCs increased with increasing ionic strength indicating that the capacity of myosin to bind KFOCs was strengthened as the ionic strength of the medium increased. However, when the ionic strength was greater than 0.6 M, the ability of myosin to bind heptanal and octanal was significantly decreased (p < 0.05), while the ability of myosin to bind nonanal and 1-octen-3-ol was not significantly affected (p > 0.05).
At lower ionic strengths, an increase in the salt concentration would likely promote protein solubility while at elevated ionic strengths the salts might compete with the abilities of the proteins to bind the volatile compounds, thus causing a decline in binding capacity.25 Pérez-Juan et al.20 reported that owing to salt solubilization, salt ions could significantly reduce the abilities of sarcoplasmic and myofibrillar proteins to bind branched aldehydes and other volatile compounds. Pighin et al.28 noted that the presence of salt ions affected the conformation of the proteins in their study. In addition, the structural characteristics of the volatile compounds could also affect the extent of binding with proteins.23 It has been reported that the existence and absence of some functional groups have been shown to significantly affect the extent of binding of proteins to volatile compounds.29
Characterization of the binding properties of myosin and the KFOCs
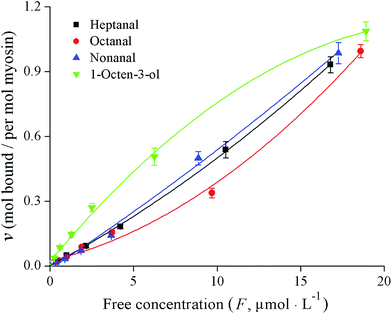
The binding properties of fish myosin with the four KFOCs were investigated. In previous studies, researchers found that binding curves are necessary to clarify the states of the binding sites.4 According to Klotz,21 a graph showing the number of moles bound plotted against the concentration of the free ligand on a logarithmic scale has three characteristics: an inflection point that appears at the half-maximum binding, an S-shaped curve that is symmetrical around the inflection point, and a plateau that is reached as the concentration of the free ligand approaches very large (infinite) values. In the present study, the binding curves of myosin and the four KFOCs (Fig. 4) were consistent with the behavior proposed by Klotz.21 The binding curve representing the data on a linear scale displayed a sigmoidal pattern, while that of the double-reciprocal plots were linear (Fig. 5), indicating the presence of non-covalent (reversible) binding sites.30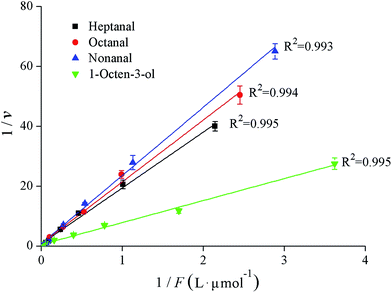 | ||
| Fig. 5 Klotz plots for the binding interactions between myosin and the four key fishy off-flavor compounds (KFOCs). | ||
The binding parameters for the reversible interactions between myosin and the KFOCs, including the number of binding sites (n) and the affinity constants (K), were obtained from the double-reciprocal plot (Fig. 5) and are shown in Table 2. The numbers of binding sites (n) for heptanal, octanal, and nonanal to myosin were all close to 1, indicating that myosin has only one binding site for the KFOCs. However, myosin had approximately 1.6 binding sites for 1-octen-3-ol. This difference was likely a result of the diversity in the structure of 1-octen-3-ol as compared with the other three KFOCs. In addition to a double bond, 1-octen-3-ol also possesses a hydroxyl group that is not present in the three straight-chain aldehydes. Chopin et al.1 also reported that the presence of a double bond in the aldehyde chain was necessary for binding to fish myosin. However, the influence of functional groups on the binding of volatile compounds with proteins requires further investigation.
| KFOC | pH | Binding parameters | ||
|---|---|---|---|---|
| n | K/(L mol−1) | ΔG/(kJ mol−1) | ||
| Heptanal | 7.0 | 1.063 ± 0.024a | 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 807 ± 1037c 807 ± 1037c |
−27.310 ± 0.046b |
| Octanal | 7.0 | 1.052 ± 0.022a | 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 261 ± 1116b 261 ± 1116b |
−27.074 ± 0.055c |
| Nonanal | 7.0 | 1.038 ± 0.025a | 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 473 ± 1085a 473 ± 1085a |
−26.859 ± 0.052d |
| 1-Octen-3-ol | 7.0 | 1.605 ± 0.032b | 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 487 ± 1551d 487 ± 1551d |
−28.622 ± 0.048a |
| p-Value | <0.001 | <0.001 | <0.001 | |
The analysis of the affinity constants (K) indicates that the affinity of myosin for 1-octen-3-ol was the highest among all four KFOCs (Table 2). Of the linear aldehydes, myosin exhibited a relatively high affinity constant for heptanal, presented a medium affinity constant for octanal, while it demonstrated a relatively low affinity constant for nonanal, indicating that the longer the chain length is, the lower the affinity. These results were in agreement with those of a study by Gianelli et al.,4 who indicated that the structure of the volatile compound could influence its binding affinity for peptides through steric effects. Chopin et al.1 not only reported that the presence of a double bond in the aldehyde chain is necessary for its binding to fish myosin, but also that binding occurs more quickly with long-chain aldehydes, presenting a double bond.
Thermodynamic data have been used to characterize the binding properties between proteins and ligands in several studies.31–33 Under the present experimental conditions, the values for the free energy of binding (ΔG) were similar for all of the KFOCs studied and were uniformly negative, indicating that the binding of all four KFOCs to myosin is spontaneous (Table 2).
The functional groups of the volatile compounds are important for protein binding. Damodaran et al.31 have used thermodynamic data to postulate the effects of the carbonyl bond type (2-heptanone, 2-octanone, 2-nonanone, 5-nonanone and nonanal) on their binding with soy protein. The authors have indicated that the increase in the free energy of binding is inversely proportional to its affinity. The same result was observed for the linear aldehydes (heptanal, octanal, and nonanal) in the present study. Damodaran et al.31 also indicated that the increase in free energy (ΔG) of binding with the chain length was consistent with the presence of hydrophobic interactions. However, this was not observed in the binding of the three linear aldehydes in the present study. Gianelli et al.4 also indicated that the variation in ΔG for the binding of hexanal and octanal to skeletal muscle peptides was inconsistent with their chain length. However, the interactions between volatile compounds and proteins or peptides, which involve reversible hydrogen bonding, hydrophobic bonding, ionic bonds, van der Waal's forces and irreversible covalent bonds, have not yet been clearly elucidated. More research is required to further clarify these mechanisms.
Conclusions
While actin had no significant effect on the four KFOCs studied (heptanal, octanal, nonanal, and 1-octen-3-ol), all four can be retained by fish total myofibrillar protein (TMP) and myosin solutions. Myosin was the primary receptor for the KFOCs. The binding capacities of the KFOCs to myosin were influenced by varying the pH, the presence of different ions, and the ionic strength to different degrees. At neutral pH and at high ionic strength, myosin showed stronger binding ability for the KFOCs. K+ and Na+ had a significant effect (p < 0.05) on the binding of 1-octene-3-ol to myosin but had no significant effect on the binding of heptanal, octanal, and nonanal. Compared to K+ and Na+, the presence of both Mg2+ and Ca2+ strengthened the binding of all four KFOCs to myosin. The binding parameters of myosin and the four KFOCs were determined using the Scatchard equation and a Klotz plot. The binding reactions between myosin and the four KFOCs were spontaneous (ΔG < 0). The number of binding sites for heptanal, octanal, and nonanal with myosin were all close to 1, while the number for 1-octen-3-ol with myosin was approximately 1.6. Moreover, myosin showed the highest affinity for 1-octen-3-ol, while the affinity of myosin for the three linear-chain aldehydes was negatively correlated with their chain lengths. These results indicate that differences in aroma retentions by silver carp muscle protein can result in different sensory perceptions of processed aquatic products, such as fish mince products. In addition, these results could provide a theoretical basis for improving the deodorization efficiency of traditional rinsing methods for surimi production by adjusting the characteristics of the media used.Compliance with ethical standards
This study was performed in strict accordance with the NIH guidelines for the care and use of laboratory animals (NIH Publication No. 85-23 Rev. 1985) and was approved by the Institutional Animal Care and Use Committee of the National Tissue Engineering Center (Shanghai, China).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R&D Program of China (2018YFD0901006), the National Science Foundation of China (31871869 and 31601500) and the Ningbo Natural Science Foundation (2017A610283).References
- C. Chopin, M. Kone and T. Serot, Food Chem., 2007, 105, 126–132 CrossRef CAS.
- Z. Wang, Y. Tan, N. Yang, Y. Jin, H. Sun and X. Xu, RSC Adv., 2019, 9, 33163–33169 RSC.
- X. Zhou, Y. Chong, Y. Ding, S. Gu and L. Liu, Food Chem., 2016, 207, 205–213 CrossRef CAS PubMed.
- M. P. Gianelli, M. Flores and F. Toldra, J. Agric. Food Chem., 2003, 51, 6828–6834 CrossRef CAS PubMed.
- M. P. Gianelli, M. Flores and F. Toldra, J. Agric. Food Chem., 2005, 53, 1670–1677 CrossRef CAS PubMed.
- N. Stapornkul, T. Prytkova and L. Were, Food Res. Int., 2016, 89, 1038–1045 CrossRef CAS.
- T. Lv, Y. Wang, D. D. Pan, J. X. Cao, X. Zhang, Y. Y. Sun, Y. J. Chen and Y. Liu, Food Chem., 2017, 214, 710–716 CrossRef CAS PubMed.
- A. Zhou, W. L. Boatright, L. A. Johnson and M. Reuber, J. Food Sci., 2002, 67, 142–145 CrossRef CAS.
- E. Guichard, Food Rev. Int., 2002, 18, 49–70 CrossRef CAS.
- Q. L. Yang, X. W. Lou, Y. Wang, D. D. Pan, Y. Y. Sun and J. X. Cao, Poult. Sci., 2017, 96, 1963–1969 CrossRef CAS PubMed.
- H. Shen, M. M. Zhao and W. Z. Sun, Food Chem., 2019, 287, 93–99 CrossRef CAS PubMed.
- M. Perez-Juan, M. Flores and F. Toldra, Food Chem., 2007, 105, 932–939 CrossRef CAS.
- X. J. Fu, Q. L. Lin, S. Y. Xu and Z. A. Wang, LWT--Food Sci. Technol., 2015, 61, 251–257 CrossRef CAS.
- Y. Q. Wang, M. Zhang, A. S. Mujumdar and K. J. Mothibe, Food Bioprocess Technol., 2013, 6, 1664–1680 CrossRef CAS.
- S. Yarnpakdee, S. Benjakul, S. Nalinanon and H. G. Kristinsson, Food Chem., 2012, 132, 1781–1788 CrossRef CAS.
- M. Nyaisaba, X. X. Liu, S. C. Zhu, X. J. Fan, L. L. Sun, S. Hatab, W. H. Miao, M. L. Chen and S. G. Deng, LWT--Food Sci. Technol., 2019, 106, 15–21 CrossRef.
- Y. Q. Ding, R. Liu, J. H. Rong and S. B. Xiong, Food Chem., 2014, 149, 237–243 CrossRef CAS PubMed.
- M. Perez-Juan, M. Flores and F. Toldra, Food Chem., 2007, 101, 1005–1011 CrossRef CAS.
- J. M. Walker, The protein protocols handbook, 2002, vol. 32, pp. 5–8 Search PubMed.
- M. Pérez-Juan, M. Flores and F. Toldra, Food Res. Int., 2007, 40, 687–693 CrossRef.
- I. M. Klotz, Science, 1982, 217, 1247–1249 CrossRef CAS PubMed.
- M. Pérez-Juan, M. Flores and F. Toldra, Food Chem., 2008, 108, 1226–1233 CrossRef.
- K. Wang and S. D. Arntfield, Food Chem., 2014, 157, 364–372 CrossRef PubMed.
- F. B. Zhou, M. M. Zhao, G. W. Su and W. Z. Sun, J. Agric. Food Chem., 2014, 62, 9544–9552 CrossRef CAS PubMed.
- Y. S. Xu, W. S. Xia, Q. X. Jiang and S. Q. Rao, Food Hydrocolloids, 2012, 27, 309–315 CrossRef CAS.
- J. Reiners, S. Nicklaus and E. Guichard, Lait, 2000, 80, 347–360 CrossRef CAS.
- J. Kühn, T. Considine and H. Singh, J. Food Sci., 2006, 71, R72–R82 CrossRef.
- D. G. Pighin, A. M. Sancho and C. B. Gonzalez, Meat Sci., 2008, 79, 549–556 CrossRef CAS PubMed.
- I. Martinez-Arellano, M. Flores and F. Toldra, Food Chem., 2016, 196, 9–16 CrossRef CAS PubMed.
- S. F. Okeefe, L. A. Wilson, A. P. Resurreccion and P. A. Murphy, J. Agric. Food Chem., 1991, 39, 1022–1028 CrossRef CAS.
- S. Damodaran and J. E. Kinsella, J. Agric. Food Chem., 1981, 29, 1253–1257 CrossRef CAS.
- X. R. Wang, X. Y. Xie, C. L. Ren, Y. Yang, X. M. Xu and X. G. Chen, Food Chem., 2011, 127, 705–710 CrossRef CAS PubMed.
- C. Chung, T. Rojanasasithara, W. Mutilangi and D. J. McClements, Food Chem., 2017, 218, 277–284 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |