Open Access Article
Open Access ArticleThree-dimensional cell printing of gingival fibroblast/acellular dermal matrix/gelatin–sodium alginate scaffolds and their biocompatibility evaluation in vitro
Peng Liu†
ac,
Qing Li†abc,
Qiaolin Yangd,
Shihan Zhanga,
Chunping Lina,
Guifeng Zhang*e and
Zhihui Tang *ac
*ac
aSecond Clinical Division, Peking University School and Hospital of Stomatology, Beijing, 100101, P. R. China
bCenter of Digital Dentistry, Peking University School and Hospital of Stomatology, Beijing, 100081, P. R. China
cNational Engineering Laboratory for Digital and Material Technology of Stomatology, Beijing, 100081, P. R. China. E-mail: 1210303145@bjmu.edu.cn
dDepartment of Orthodontics, Peking University School and Hospital of Stomatology, Beijing, 100081, P. R. China
eState Key Laboratories of Biochemical Engineering, Institute of Process Engineering, Beijing, 100190, P. R. China
First published on 21st April 2020
Abstract
Tissue engineering has emerged as a promising approach for soft tissue regeneration. Three-dimensional (3D) cell printing showed great potential for producing cell-encapsulated scaffolds to repair tissue defects. The advantage of 3D cell printing technology is precise cell loading in scaffolds to achieve tissue regeneration instead of only relying on the cells from surrounding tissue or blood. A new acellular dermal matrix/gelatin–sodium alginate (ADM/A/G) scaffold with living gingival fibroblasts was constructed by 3D cell printing technology for potential oral soft tissue regeneration in this study, and the biological characteristics of the 3D cell printing scaffolds were evaluated. The residue of nucleic acid and growth factors in ADM were detected. Three biomaterials were mixed at an appropriate radio with human gingival fibroblasts (hGFs) to prepare bioinks. Two kinds of layer scaffolds were fabricated by 3D cell printing technology. The mechanical strength and degradability of the scaffolds were determined by measuring their compressive modulus and mass loss. CCK-8 assay and calcein-AM/PI staining were conducted to detect the cell proliferation and viability in 3D cell printing scaffolds. The morphology of the hGFs in the scaffolds were observed using SEM and FITC-phalloidin staining. The expression of COL1A1, PECAM1, and VEGF-A of hGFs in the scaffolds were quantified by qRT-PCR. The gelatin–sodium alginate (A/G) scaffolds were used as control group in all experiments. Compared with the control group, 3D cell printing ADM/A/G scaffolds showed better mechanical strength and longer degradation time. The ADM/A/G scaffolds obviously had a better promotion effect on cell proliferation and viability. Most of the hGFs observed had a fully extended spindle morphology in the ADM/A/G scaffolds but oval morphology in the control group. The expression of COL1A1 was significantly higher than in the control group with time, and the expression of PECAM1 and VEGF-A was slightly higher in ADM/A/G scaffolds on day 14. 3D cell printing gingival fibroblast-ADM/A/G scaffolds showed excellent biological properties, which could be potentially useful in oral soft tissue regeneration.
1. Introduction
Oral soft tissue defects, due to aging, inflammation, tumor, trauma and other causes, have become a focus of problems that need to be solved in clinical medicine.1,2 Many surgical therapeutic approaches have been developed in an attempt to reconstruct oral soft tissue since the 1960s,3 for example, “denudation technique”, “periosteal retention” and “periosteal fenestration”, but these approaches have been abandoned because of pain, unpredictable results and severe post-operative complications.4,5 To date, the autogenous tissue graft procedure is considered as the gold standard in terms of predictable results and is the most satisfactory for soft tissue defects.6 However, limitations of these procedures have been exposed during long-term clinical application, such as insufficient tissue supply, pain and increased procedure time associated with a second surgical site, and possible unsatisfactory aesthetic outcome.7–10 For these reasons, clinicians have been interested in alternate materials and new technologies for soft tissue augmentation.Nowadays, a wide range of substitute materials such as collagen matrix, enamel matrix derivatives, fibroblast-derived dermal substitute, and platelet-rich fibrin have been investigated in clinic.11–14 However, these materials also present limitations in terms of controlling in vivo degradation and mechanical strength.15 Besides, they are mostly applied as membranes in form, showing a poor performance in soft tissue regeneration. Moreover, biomaterials in tissue engineering for soft tissue regeneration such as chitosan, hyaluronic acid, and silk matrix are also being widely explored in scientific researches.16–18 But these materials still have limited physically and mechanically characteristic for clinical application. In addition, the risk of inflammatory response and the immunogenicity caused by the source of the materials remain a major problem.19 Among alternate materials, acellular dermal matrix (ADM) represents one of the most widely used substitutes.20 ADM has been explored as alternative materials to achieve soft tissue regeneration because of its 3D porous structure, non-immunogenicity and good stability. ADM was originally applied in the repair of burned skin tissue.21 Since the 1990s, numerous studies have proved the effective role of ADM in soft tissue augmentation and relatively satisfied aesthetic coordination. However, compared with autologous tissue graft, problems such as technique sensitivity, unpredictable postoperative results and the expensive cost, are commonly encountered in clinical practice, limiting the scope of clinical application.22–26 The limitation of the application of ADM alone in clinic may due to that the alternative materials do not contain cells and cannot ensure the accurate position and distribution of cells from the surrounding tissue. Therefore, the research on clinical tissue increment of ADM needs more improvement.
In recent years, 3D cell printing technology has been attracting great interests in tissue engineering.27 Moreover, the cell printing technology could construct a three-dimensional scaffold suitable for tissue defect using bioinks containing composite hydrogels and living autologous cells.28 3D cell printing technology has achieved printing resolution in the micron-scale region and can precisely pattern the living cells and biomaterials at predefined positions. With the advantages of assembling multiple cell types along with different biomaterials in a layer-by-layer fashion and fabricating biomimetic microenvironment, it enjoys great potential in complicate tissue defect repairing.29,30 With the minimal invasive treatment development, surgery is increasingly demanding precision and personalization. The application of 3D cell printing scaffolds could reduce the surgical time, decrease the postoperative complications and promote the customized operation of periodontal surgery.
With the development of 3D cell printing technology, more and more biobased polymer materials have been utilized to improve the properties of scaffolds. Currently, sodium alginate (A) and gelatin (G) have been widely studied and applied in tissue engineering because of good biosafety and biocompatibility.31–33 Sodium alginate can provide three-dimensional growth space and meet the diversity requirement of morphology.34 The combination of sodium alginate and calcium ions can solidify the hydrogel to enhance the mechanical strength. Gelatin is a hydrolyzed product of collagen, which commonly serves as growth factor carrier and has good fluidity.35 Furthermore, gingival fibroblasts, derived from mesenchymal stem cells, is the main cell type forming gingival connective tissue and are closely related to the stability of gingival tissue and periodontal tissue regeneration. Gingival fibroblasts secrete extracellular matrix, such as collagen, hyaluronic acid, and elastic fiber, which contribute to periodontal tissue microenvironment stabilization and periodontal tissue repair.36 For the above reasons, gingival fibroblasts have been widely used in the researches on periodontal tissue engineering.37 Tissue engineering with seeding living gingival fibroblasts has already achieved initial success.13,38
In this study, a matrix material by mixing acellular dermal matrix, sodium alginate and gelatin were prepared to be the bioink. Human gingival fibroblasts (hGFs) were seeded in three-dimensional scaffolds with different layers using 3D cell printing technology. The mechanical and biological properties of the scaffolds were tested to clarify its biosafety and biocompatibility, which provide scientific basis for the potential application of oral soft tissue regeneration.
2. Experimental section
This study was approved by the Ethics Committee of Peking University School of Stomatology, Beijing, China (PKUSSIRB-201950166).2.1 Preparation of acellular dermal matrix
The epidermal and subcutaneous adipose tissue of porcine skin was cut out under sterile conditions. The rest dermal tissue was immersed in a decellularized mixture solution containing 2% NaOH and 2% Triton X-100 at a material-to-liquid radio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 (wt%). The mix was stirred at 4 °C for 12 hours, and the solution was changed every 2 hours. Then the solution was rinsed with PBS buffer until became neutral. After lyophilized, pulverized and filtered, the mix was stored at 4 °C for later use.
15 (wt%). The mix was stirred at 4 °C for 12 hours, and the solution was changed every 2 hours. Then the solution was rinsed with PBS buffer until became neutral. After lyophilized, pulverized and filtered, the mix was stored at 4 °C for later use.
2.2 Nucleic acid residue test of ADM
DNA was extracted using TIANamp genomic DNA kit (Tiangen, China) from 10 mg ADM under the same conditions (n = 5). DNA quantitation kit (Sigma-Aldrich, USA) was used to measure the absorbance values of standards and samples by fluorescence enzyme-labeling instrument, and the DNA content in the samples was calculated according to the linear regression curve equation of the standards.2.3 Enzyme-linked immunosorbent assay (Elisa)
The contents of fibroblast growth factor 2 (FGF-2), connective tissue growth factor (CTGF), vascular endothelial growth factor (VEGF) and transforming growth factor β1 (TGF-β1) in ADM were measured. Growth factor extraction: 1 mL buffer containing 2.5 mg mL−1 sodium heparin and 2 mol L−1 urea was added to 5 mg ADM (n = 5), then homogenized the mixture by shanking table at 4 °C for 24 h. The supernatant of mixture was collected by centrifuge (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 30 min).39 The OD values (450 nm) of samples and standards processed by ELISA kits (YMbio, China) were determined, and the concentrations of growth factors in the samples were calculated.
000 rpm, 30 min).39 The OD values (450 nm) of samples and standards processed by ELISA kits (YMbio, China) were determined, and the concentrations of growth factors in the samples were calculated.
2.4 Extraction and cultivation of hGFs
Healthy gingiva were collected from three patients (20–25 years of age, with informed consent) who was performed crown lengthening surgery in Department of Periodontology of Peking University School of Stomatology. The tissue block and enzymatic digestion methods were used for extracting gingival fibroblasts.40,41 The gingiva was gently shredded and digested in trypsin (Gibco, USA) for five minutes. The small pieces of tissue were then seeded onto a culture bottle and incubated in a growth medium (GM), which is composed of alpha minimum essential medium enriched with 10% fetal bovine serum (Gibco, USA) and 1% penicillin and streptomycin in the presence of 5% CO2 and a temperature of 37 °C. After every two days, the culture medium was changed. The passages 3–4 of hGFs were utilized for subsequent experiments.2.5 Preparation of the bioinks
The applicable concentration of hydrogel materials is typically 10% of the mass volume ratio.42 A 10% concentration ADM/A/G bioink was formulated in the study. 2% sodium alginate was selected for the balance of mechanical strength and cell viability.43 Based on the principle of using the largest proportion of ADM and obtaining suitable viscosity of the bioink, it was determined after repeated trials that the composition of bioink materials was ADM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) A
A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G = 6%
G = 6%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2%
2%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2% (wt%). The three materials were mixed with PBS buffer and placed in 37 °C for 4 hours to fully dissolve. The hydrogels containing only A
2% (wt%). The three materials were mixed with PBS buffer and placed in 37 °C for 4 hours to fully dissolve. The hydrogels containing only A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G = 2%
G = 2%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8% (wt%) was used as a control group.43 hGFs were gently mixed with the prepared two groups of hydrogels at the radio of 1 × 106 mL−1. Finally, the groups were divided into ADM-cell group, ADM-cell free group, AG-cell group and AG-cell free group (Table 1).
8% (wt%) was used as a control group.43 hGFs were gently mixed with the prepared two groups of hydrogels at the radio of 1 × 106 mL−1. Finally, the groups were divided into ADM-cell group, ADM-cell free group, AG-cell group and AG-cell free group (Table 1).
| Groups | Ratio of materials (wt%) | hGFs (cells per mL) |
|---|---|---|
| ADM-cell | ADM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) A A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G = 6% G = 6%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2% 2%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2% 2% |
1 × 106 mL−1 |
| AG-cell | A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G = 2% G = 2%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8% 8% |
1 × 106 mL−1 |
| ADM-cell free | ADM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) A A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G = 6% G = 6%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2% 2%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2% 2% |
None |
| AG-cell free | A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G = 2% G = 2%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8% 8% |
None |
2.6 3D cell printing
The 3D cells printer (Medprin, China) was used for building scaffolds. Two kinds of layers of 10 mm × 5 mm×2 mm cube model were designed by MP Bioprint 4.0 software according to the clinical requirement, the dense layers have small pores to ensure the strength of the scaffold, while the porous layers have lager pores, which is beneficial for culturing cells and blood vessels (Table 2). The model of the scaffold was shown in Fig. 1. The printing parameters of the 3D cells printer were determined after repeated printing trials, which ensured consistency and repeatability of 3D cell printing (Table 3). The printing scaffolds were cross-linked with 10% CaCl2 solution for 4–5 minutes immediately after printed.| Parameters | Length (mm) | Width (mm) | Height (mm) | Dense layers | Porous layers | ||||
|---|---|---|---|---|---|---|---|---|---|
| Fill density | Layer count | Grid distance | Fill density | Layer count | Grid distance | ||||
| Dimension | 10 | 5 | 2 | 50% | 4 | 290 μm | 30% | 4 | 410 μm |
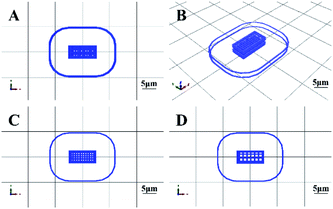 | ||
| Fig. 1 3D cell printing scaffold model designed by MP Bioprint 4.0 software. (A) The top view of the structure. (B) The side view of the structure. (C) The dense layer. (D) The porous layer. | ||
| Parameters | Temperature (°C) | Needle description (mm) | Layer height (mm) | Pressure (bar) | Speed (mm s−1) | Platform temperature (°C) |
|---|---|---|---|---|---|---|
| Reference value | 4–10 | 0.26 | 0.3 | 2.0–3.0 | 30 | 10 |
2.7 Mechanical strength of the 3D cell printing scaffolds
The compressive modulus of the scaffolds in ADM-cell free group and AG-cell free group (n = 5) were evaluated by dynamic thermomechanical analysis (TA Instruments, USA). The size of samples were measured using caliper. The scaffolds were then placed on the platform and loaded at a rate of 6.0 N min−1 until the scaffolds were crushed. The compressive modulus were recorded as the slope of the linear region of the stress–strain curves.2.8 Degradability in vitro of the 3D cell printing scaffolds
3D cell printing scaffolds in ADM-cell free group and AG-cell free group (n = 5) were first lyophilized, with the dry weight W0 recorded. Then the freeze-dried scaffolds were immersed in sterilized PBS buffer at 37 °C, the same method was used to record the dry weight (W1) of the scaffolds at each testing time point. The PBS buffer was changed every two days. The mass loss (ML) was calculated using the equation:
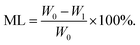 | (1) |
2.9 Scanning electron microscope (SEM)
At 7 days after printing, the scaffolds of ADM-cell group and AG-cell group were immersed in 4% paraformaldehyde for 15 minutes to fix the hGFs. Then the morphology of the freeze-dried scaffolds and hGFs were observed using SEM.2.10 hGFs proliferation and viability in the 3D cell printing scaffolds
The hGFs proliferation in the scaffolds of ADM-cell group and AG-cell group were detected using CCK-8 kit (Dojindo, Japan) at 1 day, 3 days, 5 days, 7 days and 9 days after printing. The hGFs viability and viable cell count in the scaffolds were examined in ADM-cell group and AG-cell group separately at 1 day and 7 days. The scaffolds were rinsed with PBS buffer 3 times and immersed in 2 mL solution containing 2 μmol L−1 calcein-AM (CAM) and 4.5 μmol L−1 propidium iodide (PI) at 37 °C for 45 minutes, then washed with PBS buffer again. The 2 mm thick scaffold was cut in the middle, and three different positions on the surface and in the middle were randomly selected for scanning. The hGFs in the scaffolds were observed at wavelengths of 490 nm (green, viable cells) and 545 nm (red, dead cells) with a confocal laser scanning microscope (CLSM, 10×). Different layers of the scaffolds were scanned and three-dimensionally images were reconstructed. Image pro plus 6.0 was used to count the number of viable and dead cells in both surface and middle of the scaffolds, and the hGFs viability within the 3D cell printing scaffolds in each group was calculated using the equation:
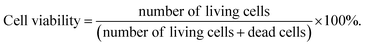 | (2) |
2.11 Analysis of hGFs morphology in the 3D cell printing scaffolds
The 3D cell printing scaffolds of ADM-cell group and AG-cell group cultured 7 days were immersed in 4% paraformaldehyde for 15 minutes to fix the hGFs and permeated with 0.1% Triton X-100 for 3–5 minutes. The hGFs in the scaffolds were stained by 50 μg mL−1 FITC-phalloidin staining solution (Sigma-Aldrich, USA) for 40 minutes at room temperature and DAPI solution (Solarbio, China) for 10 minutes. Then the cell morphology was observed under a confocal laser scanning microscope (75×).Cell morphology was evaluated using ImageJ software by two independent evaluators. The cell spreading area and perimeter were measured by the outline of the cell body and the lamellar pseudopod. Cell circularity was calculated from data based on the following equation:44
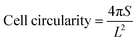 | (3) |
2.12 Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
The 3D cell printing scaffolds of ADM-cell group and AG-cell group cultured for 1 day, 7 days and 14 days were analyzed for gene expression (n = 3). Total RNA was extracted (Beyotime, China) and reverse transcribed (TaKaRa, Japan) according to the instructions of manufacturer. Real-time quantitative PCR were performed using SYBR Green Master Mix kit (Sigma-Aldrich, USA). The primers for COL1A1, PECAM1 and VEGF-A were synthesized by BGI, which were listed in Table 4. GAPDH was used as normalization.| Genes | Sequences |
|---|---|
| GAPDH | Forward: 5′-CGACAGCAGCCGCATCTT-3′ |
| Reverse: 5′-CCAATACGACCAAATCCGTTG-3′ | |
| COL1A1 | Forward: 5′-AGAGGAAGGAAAGCGAGGAG-3′ |
| Reverse: 5′-GGACCAGCAACACCATCTG-3′ | |
| PECAM1 | Forward: 5′-CCTCCAGCCCTAGAAGCCAATTA-3′ |
| Reverse: 5′-CTCAAAGACTGAGTCAGGCCAGTG-3′ | |
| VEGF-A | Forward: 5′-TCACAGGTACAGGGATGAGGACAC-3′ |
| Reverse: 5′-CAAAGCACAGCAATGTCCTGAAG-3′ |
2.13 Statistical analysis
IBM SPSS 20 software was used for statistical analysis. Data were expressed as mean ± standard deviation. Statistical methods included independent sample T test and one-way ANOVA. For all tests, statistical significance was accepted at p values lower than 0.05.3. Results
3.1 Characteristics of ADM
In order to determine whether ADM would cause immune rejection in the host, the amount of nucleic acid residue was detected. The linear regression curve equation of the standards was determined and the residue of nucleic acid in ADM was calculated according to the OD values of samples. The residue of nucleic acid in ADM was 26.53 ± 8.82 ng mg−1, which is lower than 50 ng mg−1, indicating that the host immune response upon implantation of the ADM can be avoided.45 The concentration of growth factors detected by Elisa were listed in Table 5. The growth factors of the ADM were obviously higher than previous reports, which could promote cells adhesion, proliferation, differentiation, extracellular matrix formation and vascular regeneration.39,46 The SEM results showed the structure of ADM was loose and irregular, the length of the material patch was 2.7–10.9 μm (Fig. 2).| Growth factors | Concentration (ng g−1) |
|---|---|
| FGF-2 | 35.75 ± 2.09 |
| CTGF | 604.35 ± 32.26 |
| VEGF | 39.25 ± 4.15 |
| TGF-β1 | 49.38 ± 6.06 |
 | ||
| Fig. 2 The morphology of ADM in SEM. The white lines marked the length of ADM patch in microstructure. | ||
3.2 The 3D cell printing scaffolds
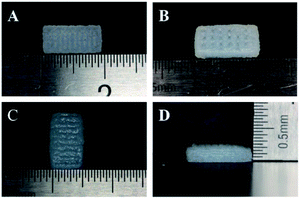
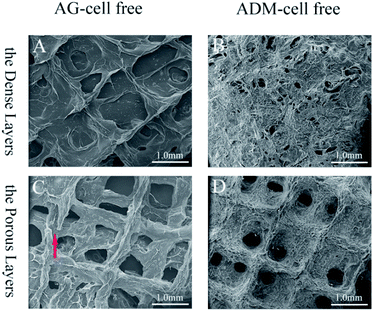
The tissue block and enzymatic digestion methods were used to isolate gingival fibroblasts and as reported in previous studies, most cells are spindle-shaped and some are polygonal in morphology (Fig. 3).40,41,47 Based on the methods and the typical spindle morphology of the cells, the isolated hGFs can be confirmed. The optimized formulation designed allowed plotting via the 3D cell printing technology with high shape fidelity. The 3D cell printing scaffolds displayed regular 10 mm × 5 mm × 2 mm cubic appearance with evenly distributed pores (Fig. 4). The scaffold was composed of two kinds of layers, the dense layers at the bottom (Fig. 4A) and the porous layers on the top (Fig. 4B).According to the data acquired by SEM, the pore diameter was 242.93 ± 90.31 μm in the dense layer and 372.99 ± 79.76 μm in the porous layer in ADM-cell free group, the pore diameter was 403.44 ± 48.09 μm and 543.77 ± 97.56 μm in the dense and porous layers respectively in AG-cell free group. The SEM results showed that the structure of the ADM-cell free group is more condense, the pore size was closer to the designed model. The fracture of strands could be found in the control group, which indicated that the structural stability is higher in ADM-cell free group (Fig. 5).
3.3 Mechanical strength of the 3D cell printing scaffolds
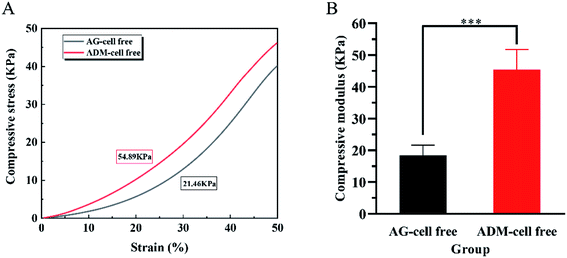
The compressive force was loaded on the scaffolds to obtain stress–strain curves in the elastic region (Fig. 6A). The compressive modulus of the scaffolds in two groups were calculated respectively. The compressive modulus of the ADM-cell free group was 45.38 ± 6.34 kPa, and the AG-cell free group was 18.48 ± 3.18 kPa. Two groups showed a statistically significant difference (p < 0.001) (Fig. 6B).3.4 Degradability of the 3D cell printing scaffolds in vitro
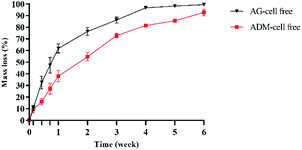
The degradation rates of the two groups of scaffolds in vitro were evaluated by weighing after freeze-dried (Fig. 7). The mass loss of AG-cell free group was higher than that of ADM-cell free group in all times. After 2 weeks of cell printing, the mass loss of ADM-cell free group was 54.65% ± 3.52%, which was less than 76.37% ± 3.33% of AG-cell free group (p < 0.001). After cell printing for 4 weeks, the AG-cell free group was almost completely degraded (96.72% ± 0.36%), while the scaffolds of ADM-cell free group existed for more than 6 weeks.3.5 hGFs proliferation and viability in the 3D cell printing scaffolds
The ability of hGFs proliferation in ADM-cell group and AG-cell group was evaluated by CCK-8 assay (Fig. 8). There was no significant difference in the quantity of cells between these two groups after 1 day of cell printing (p > 0.05). The OD values of the two groups increased by the cultural time. After 5 days of culturing, the quantity of cells in both groups increased significantly (p < 0.05), and difference between the two groups was statistically significant (p < 0.001).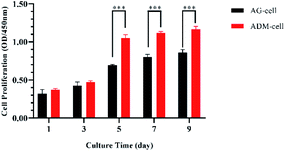 | ||
| Fig. 8 Cell proliferation (measured by CCK-8 assay) in AG-cell group and ADM-cell group cultured for day 1, 3, 5, 7 and 9. ***p < 0.001. | ||
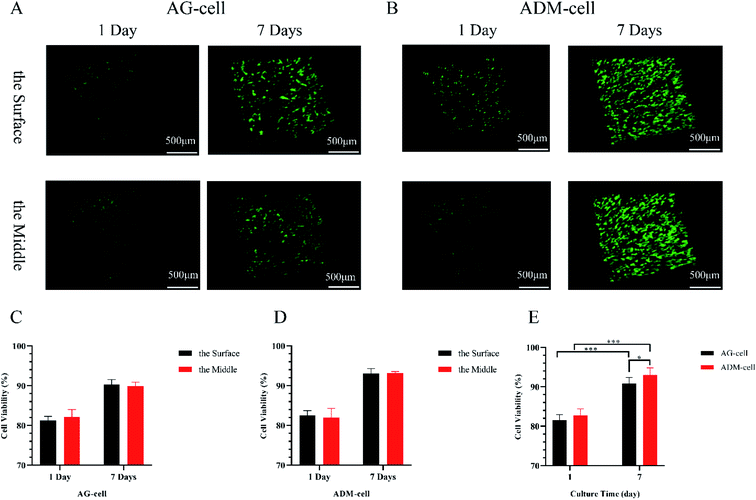
The pictures of CAM/PI staining at different levels of the scaffolds and time points were supplemented as follows and the difference in cell viability in different layers were checked (Fig. 9). The cells were evenly distributed in both groups. According to CAM/PI staining results, the amount of viable cells of both AG-cell group and ADM-cell group on the 1st day after cell printing were less than those on day 7, indicating once again that hGFs could proliferate in both scaffolds (Fig. 9A and B). In AG-cell group, the cell viability on day 1 was 81.25% ± 0.83% on the surface and 82.14% ± 1.50% on the middle layer. The cell viability on day 7 was 90.30% ± 1.02% on the surface and 89.90% ± 0.80% on the middle layer. In ADM-cell group, the cell viability on day 1 was 82.54% ± 0.97% on the surface and 81.94% ± 1.90% on the middle layer. The cell viability on day 7 was 93.09% ± 0.97% on the surface and 93.16% ± 0.32% on the middle layer. In both groups, no significant difference was found in the viability of cells on the surface and middle layers (p > 0.05) (Fig. 9C and D).
Comprehensive comparison of cell viability between AG-cell group and ADM-cell group was designed. Quantitative analysis showed that the cell viability in AG-cell group was 81.55% ± 1.35% on day 1 and 90.79% ± 1.60% on day 7. The cell viability in ADM-cell group was 82.71% ± 1.70% on day 1 and 93.01% ± 1.81% on day 7. The cell viability on day 7 was higher than that on day 1 in both groups (p < 0.001). Besides, the cell viability of ADM-cell group was higher than that in AG-cell group on day 7 (p < 0.05) (Fig. 9E).
3.6 The morphology of hGFs in the 3D cell printing scaffolds
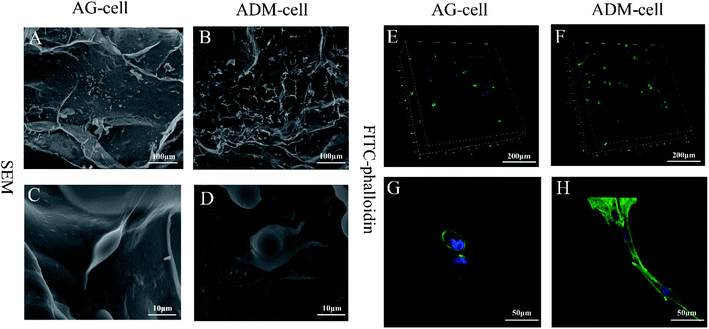
After 7 days of culturing, the images of SEM showed that the cells were successfully seeded on both groups of scaffolds and attached scaffolds well, while fully extended in ADM-cell group (Fig. 10A–D). The FITC-phalloidin staining showed the morphology of the adhered cells. The cells seeded on AG-cell group exhibited oval morphology, in contrast to the spindle morphology observed on ADM-cell group, which exhibited complete cell extension (Fig. 10E–H). hGFs in ADM-cell group had increased cell protrusions (Fig. 10D) and close intercellular contacts (Fig. 10H).In order to assess the spreading of cells in scaffold, the cell circularity was analyzed. The results showed that the cell circularity in the AG-cell group was 0.78 ± 0.18, and the cell circularity in the ADM-cell group was 0.55 ± 0.25 (Fig. 11). The cells in ADM-cell group had lower circularity versus AG-cell group (p < 0.01), confirming the better ability of ADM scaffold to support cell adhesion and spreading.
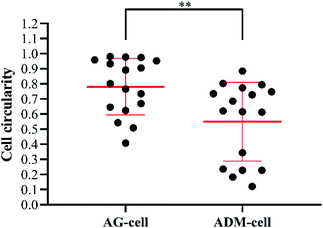 | ||
| Fig. 11 Cell circularity of hGFs in AG-cell group and ADM-cell group: closer to 0 more elongated; closer to 1 more rounded. Dots represent single cells from two independent experiments. **p < 0.01. | ||
3.7 3D cell printing scaffolds up-regulate the expression of COL1A1, PECAM1, VEGF-A in the hGFs
The expression of COL1A1, PECAM1, VEGF-A were detected after 1 day, 7 days and 14 days of culture. The relative COL1A1 expression in ADM-cell group was higher than that in AG-cell group after 7 days (p < 0.05), and it increased in ADM-cell group but decreased in AG-cell group on day 14 compared to that on day 7 (Fig. 12A). For the relative PECAM1 expression, there was a statistically significant difference for both groups between day 1 and day 14 (p < 0.01), and between ADM-cell group and AG-cell group on day 14 (p < 0.01) (Fig. 12B). Whereas statistically significant difference was showed in ADM-cell group only in the expression of VEGF-A between day 1 and day 14 (p < 0.05). And the expression of VEGF-A in ADM-cell group was higher than that in AG-cell after 14 days of culture (p < 0.05) (Fig. 12C).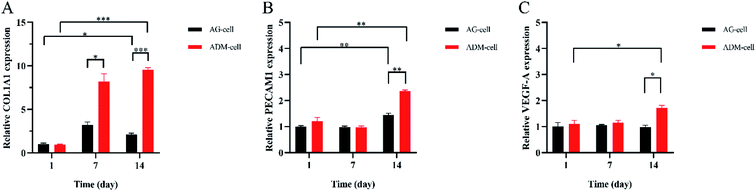 | ||
| Fig. 12 The relative expression of COL1A1 (A), PECAM1 (B) and VEGF-A (C) detected by qRT-PCR in different groups on day 1, 7 and 14. *p < 0.05. **p < 0.01. ***p < 0.001. | ||
4. Discussion
3D cell printing technology is emerging as a disruptive innovation for the tissue regeneration. Cells are the functional component of any tissue and/or organ. 3D cell printing provides a powerful tool to regulate the precise positioning of different cell types in very specific pattern, which is a great progress in tissue regeneration.48 In this study, the matrix were made of porcine acellular dermal matrix, sodium alginate and gelatin at an appropriate radio. The bioinks containing the matrix mentioned above and human gingival fibroblasts were prepared to construct a porous scaffold using 3D cell printing technology. Then the properties of this scaffold were tested for its clinical application. Gingival fibroblasts were chosen in this study because it plays an important role in synthesizing, renewing and constituting fibers of gingiva. Besides, gingival fibroblasts can promote gingiva repair by proliferating and secreting extracellular matrix including collagen, hyaluronic acid and fibers.36Biomaterials provide a fundamental interface to support cells and other living components, so the premise of constructing 3D cell printing scaffolds is selection and evaluation of biomaterials.49 The application of single hydrogel material usually have printing or biological problems.50,51 ADM retains the three dimensional structure of normal collagen, which can provide a stable scaffold for cell regeneration.52 The main components of ADM, including collagen, non-collagen glycoprotein and elastic fibers, are closely related to wounded tissue repair.53 Therefore, the combination of acellular dermal matrix, gelatin, and sodium alginate can integrate the advantages of each material, and make up for mechanical strength or biological defects.
The mechanical strength of the scaffolds is of great significance for cell survival and further clinical applications.54 Compared with AG-cell free group, ADM-cell free group had greatly enhanced mechanical strength due to its regular three-dimensional structure and fibrin-rich properties. Moreover, the ADM-cell free group had better stability than control group. The degradation time of ADM-cell free group was over 6 weeks, which is coordinate with periodontal soft tissue healing and regeneration. Scanning electron microscopy showed that ADM was loose and multilayered, and the porous structure of the 3D cell printing scaffolds provided sufficient space for cells adhesion, spreading, migration, proliferation and neovascularization. Cell proliferation and viability are the key indicators for evaluating the biocompatibility of the 3D cell printing scaffolds. CCK-8 assay and CAM/PI staining showed that gingival fibroblasts can well proliferate in both groups of scaffolds, but ADM-cell group obviously had better promotion effect on cell proliferation. CAM/PI staining results showed that the cell viability of AG-cell group and ADM-cell group on day 1 after cell printing were 81.55% ± 1.35% and 82.71% ± 1.70% respectively, and the viability of the two groups on day 7 were 90.79% ± 1.60% and 93.01% ± 1.81% respectively. The cell viability of the ADM-cell group was higher than that of AG-cell group, indicating that ADM have excellent effect on cell viability. Compared with other literature reported, the cell viability was higher.55,56 This high cell viability benefit from the utilization of gentle dispensing pressure and choice of the print needle aperture.
The images of SEM showed that most of the hGFs in ADM-cell group adhered to the fibrous pores of the scaffolds, which demonstrated that the ADM had sufficient cell-binding domains and excellent biocompatibility.56 Most of hGFs in ADM-cell group displayed spindle shape but oval morphology in AG-cell group using FITC-phalloidin staining, which further demonstrated that the porous structure in the ADM-cell group allowed the cells to fully extend. The differences in cell shape of hGFs between two groups indicated that the cells in ADM-cell group had better cell-substrate attachment, which were probably beneficial for intercellular communication.57
The qRT-PCR results showed that the expression of COL1A1 of the hGFs in the ADM-cell group was significantly increased compared with that of the control group. At the same time, the expression of cell adhesion (PECAM1) and angiogenesis-related genes (VEGF-A) also increased as the culture time increased, which was significantly higher than that of the control group. On one hand, it showed that hGFs had excellent adaptability in the scaffolds of ADM-cell group. On the other hand, the glycyl–histidine–lysine tripeptide released by the degradation of ADM can also promote the synthesis of collagen, mucopolysaccharide and proteoglycan, stimulate prolinease activity, and promote angiogenesis.58
In recent years, the study of periodontal soft tissue regeneration using gingival fibroblasts as seed cells combined with different biomaterials has achieved certain progress, such as chitosan, hyaluronic acid and so on.17,59 But compare to using 3D cell printing to precisely locate cells in tissue scaffolds, there are still some limitations to conquer in the properties of biomaterials and the structure of scaffolds.42,60 This study pioneered and investigated the performance and application potential of 3D cell printing gingival fibroblasts/acellular dermal matrix/gelatin–sodium alginate scaffolds. Overall, the 3D cell printing scaffolds owned excellent biocompatibility. However, the concrete effect of the scaffolds on oral soft tissue regeneration remains further in vivo research in the future.
5. Conclusion
3D cell printing gingival fibroblasts/acellular dermal matrix/gelatin–sodium alginate scaffolds showed satisfactory biological properties, which could be a potentially useful approach in oral soft tissue regeneration.Conflicts of interest
There are no conflicts of interest.Acknowledgements
This work was supported by the National Key Research and Development Program of China (2016YFC1102900, 2017YFA0701302).Notes and references
- A. F. Mavrogenis, V. G. Igoumenou, I. Ignatiadis, K. Mourouzis and S. G. Spyridonos, Injury, 2019, 50(suppl. 5), S117–S122 CrossRef PubMed.
- C. H. F. Hammerle and D. Tarnow, J. Clin. Periodontol., 2018, 45(suppl. 20), S267–S277 CrossRef PubMed.
- I. L. Franca-Grohmann, J. P. M. Sangiorgio, M. R. Bueno, M. Z. Casati and E. A. Sallum, J. Periodontol., 2019, 1–8 Search PubMed.
- E. R. Costich and S. P. Ramfjord, J. Periodontol., 1968, 39, 127–134 CrossRef CAS PubMed.
- S. F. Ramfjord and E. R. Costich, J. Periodontol., 1968, 39, 199–207 CrossRef CAS PubMed.
- L. Chambrone and D. N. Tatakis, J. Periodontol., 2015, 86, S8–S51 CrossRef PubMed.
- W. J. Brasher, T. D. Rees and W. A. Boyce, J. Periodontol., 1975, 46, 133–138 CrossRef CAS PubMed.
- A. R. Pack, W. M. Gaudie and A. M. Jennings, J. Periodontol., 1991, 62, 269–271 CrossRef CAS PubMed.
- T. J. Griffin, W. S. Cheung, A. I. Zavras and P. D. Damoulis, J. Periodontol., 2006, 77, 2070–2079 CrossRef PubMed.
- K. Bertl, M. Melchard, N. Pandis, M. Muller-Kern and A. Stavropoulos, Clin. Oral Invest., 2017, 21, 505–518 CrossRef PubMed.
- M. Sanz, R. Lorenzo, J. J. Aranda, C. Martin and M. Orsini, J. Clin. Periodontol., 2009, 36, 868–876 CrossRef PubMed.
- M. K. McGuire, E. T. Scheyer and M. Nunn, J. Periodontol., 2012, 83, 1353–1362 CrossRef PubMed.
- M. K. McGuire and M. E. Nunn, J. Periodontol., 2005, 76, 867–880 CrossRef PubMed.
- S. K. Agarwal, R. Jhingran, V. K. Bains, R. Srivastava, R. Madan and I. Rizvi, Eur. J. Dent., 2016, 10, 121–133 CrossRef PubMed.
- M. C. Bottino and V. Thomas, Front. Oral Biol., 2015, 17, 90–100 Search PubMed.
- K. Su and C. M. Wang, Biotechnol. Lett., 2015, 37, 2139–2145 CrossRef CAS PubMed.
- G. P. Prato, R. Rotundo, C. Magnani, C. Soranzo, L. Muzzi and F. Cairo, J. Periodontol., 2003, 74, 262–267 CrossRef PubMed.
- G. H. Altman, R. L. Horan, H. H. Lu, J. Moreau, I. Martin, J. C. Richmond and D. L. Kaplan, Biomaterials, 2002, 23, 4131–4141 CrossRef CAS PubMed.
- H. E. Jazayeri, M. Tahriri, M. Razavi, K. Khoshroo, F. Fahimipour, E. Dashtimoghadam, L. Almeida and L. Tayebi, Mater. Sci. Eng., C, 2017, 70, 913–929 CrossRef CAS PubMed.
- J. H. Fu, C. Y. Su and H. L. Wang, J. Evid. Base Dent. Pract., 2012, 12, 129–142 CrossRef.
- D. J. Wainwright and S. B. Bury, Aesthetic Surg. J., 2011, 31, 13S–23S CrossRef PubMed.
- P. C. Wei, L. Laurell, M. Geivelis, M. W. Lingen and D. Maddalozzo, J. Periodontol., 2000, 71, 1297–1305 CrossRef CAS PubMed.
- R. J. Harris, Int. J. Periodontics Restor. Dent., 2000, 20, 51–59 CAS.
- R. J. Harris, J. Periodontol., 2001, 72, 932–938 CrossRef CAS PubMed.
- M. Shanmugam, V. Sivakumar, V. Anitha and B. Sivakumar, J. Indian Soc. Periodontol., 2012, 16, 218–223 CrossRef CAS PubMed.
- S. I. Gallagher and D. C. Matthews, J. Indian Soc. Periodontol., 2017, 21, 439–448 Search PubMed.
- N. Mehrban, G. Z. Teoh and M. A. Birchall, Int. J. Bioprint., 2016, 2, 6–19 CAS.
- D. W. Hutmacher, M. Sittinger and M. V. Risbud, Trends Biotechnol., 2004, 22, 354–362 CrossRef CAS PubMed.
- N. E. Fedorovich, J. R. De Wijn, A. J. Verbout, J. Alblas and W. J. Dhert, Tissue Eng., Part A, 2008, 14, 127–133 CAS.
- M. K. Wlodarczyk-Biegun and A. Del Campo, Biomaterials, 2017, 134, 180–201 CrossRef CAS PubMed.
- J. Lewandowska-Lancucka, K. Mystek, A. Mignon, S. Van Vlierberghe, A. Latkiewicz and M. Nowakowska, Carbohydr. Polym., 2017, 157, 1714–1722 CrossRef CAS PubMed.
- S. Nemati, A. Rezabakhsh, A. B. Khoshfetrat, A. Nourazarian, C. Biray Avci, B. Goker Bagca, H. Alizadeh Sardroud, M. Khaksar, M. Ahmadi, A. Delkhosh, E. Sokullu and R. Rahbarghazi, Biotechnol. Bioeng., 2017, 114, 2920–2930 CrossRef CAS PubMed.
- M. Nabavinia, A. B. Khoshfetrat and H. Naderi-Meshkin, Mater. Sci. Eng., C, 2019, 97, 67–77 CrossRef CAS PubMed.
- B. Sibaja, E. Culbertson, P. Marshall, R. Boy, R. M. Broughton, A. A. Solano, M. Esquivel, J. Parker, L. De La Fuente and M. L. Auad, Carbohydr. Polym., 2015, 134, 598–608 CrossRef CAS PubMed.
- B. Huber, K. Borchers, G. E. Tovar and P. J. Kluger, J. Biomater. Appl., 2016, 30, 699–710 CrossRef CAS PubMed.
- A. Z. Rodrigues, P. T. Oliveira, A. B. Novaes Jr, L. P. Maia, S. L. Souza and D. B. Palioto, Braz. Dent. J., 2010, 21, 179–189 CrossRef PubMed.
- M. Mohammadi, R. Mofid and M. A. Shokrgozar, Acta Med. Iran., 2011, 49, 319–324 Search PubMed.
- T. G. Wilson Jr, M. K. McGuire and M. E. Nunn, J. Periodontol., 2005, 76, 881–889 CrossRef PubMed.
- Y. Zhang, L. Xia and J. Gao, Mater. Sci., 2018, 8, 595–602 Search PubMed.
- M. J. Somerman, S. Y. Archer, G. R. Imm and R. A. Foster, J. Dent. Res., 1988, 67, 66–70 CrossRef CAS PubMed.
- K. Tanaka, K. Iwasaki, K. E. Feghali, M. Komaki, I. Ishikawa and Y. Izumi, Arch. Oral Biol., 2011, 56, 380–388 CrossRef CAS PubMed.
- N. E. Fedorovich, W. Schuurman, H. M. Wijnberg, H. J. Prins, P. R. van Weeren, J. Malda, J. Alblas and W. J. Dhert, Tissue Eng., Part C, 2012, 18, 33–44 CrossRef CAS PubMed.
- X. F. Wang, P. J. Lu, Y. Song, Y. C. Sun, Y. G. Wang and Y. Wang, RSC Adv., 2016, 6, 6832–6842 RSC.
- M. Tozluoglu, A. L. Tournier, R. P. Jenkins, S. Hooper, P. A. Bates and E. Sahai, Nat. Cell Biol., 2013, 15, 751–762 CrossRef CAS PubMed.
- N. T. Remlinger, P. D. Wearden and T. W. Gilbert, J. Visualized Exp., 2012, e50059 Search PubMed.
- X. Lei, Y. Yang, G. Shan, Y. Pan and B. Cheng, Biomed. Mater., 2019, 14, 035004 CrossRef CAS PubMed.
- A. Mariotti and D. L. Cochran, J. Periodontol., 1990, 61, 103–111 CrossRef CAS PubMed.
- R. K. Birla and S. K. Williams, APL Bioeng., 2020, 4, 010903 CrossRef PubMed.
- L. Ning and X. Chen, Biotechnol. J., 2017, 12, 1–16 Search PubMed.
- S. V. Murphy and A. Atala, Nat. Biotechnol., 2014, 32, 773–785 CrossRef CAS PubMed.
- G. Gao, T. Yonezawa, K. Hubbell, G. Dai and X. Cui, Biotechnol. J., 2015, 10, 1568–1577 CrossRef CAS PubMed.
- D. J. Wainwright, Burns, 1995, 21, 243–248 CrossRef CAS PubMed.
- G. C. Gurtner, S. Werner, Y. Barrandon and M. T. Longaker, Nature, 2008, 453, 314–321 CrossRef CAS PubMed.
- G. Huang, L. Wang, S. Wang, Y. Han, J. Wu, Q. Zhang, F. Xu and T. J. Lu, Biofabrication, 2012, 4, 042001 CrossRef PubMed.
- M. Gruene, M. Pflaum, A. Deiwick, L. Koch, S. Schlie, C. Unger, M. Wilhelmi, A. Haverich and B. N. Chichkov, Biofabrication, 2011, 3, 015005 CrossRef CAS PubMed.
- L. Ning, H. Sun, T. Lelong, R. Guilloteau, N. Zhu, D. J. Schreyer and X. Chen, Biofabrication, 2018, 10, 035014 CrossRef PubMed.
- A. Sachar, T. A. Strom, S. San Miguel, M. J. Serrano, K. K. Svoboda and X. Liu, J. Tissue Eng. Regener. Med., 2014, 8, 862–873 CrossRef CAS PubMed.
- A. Simeon, Y. Wegrowski, Y. Bontemps and F. X. Maquart, J. Invest. Dermatol., 2000, 115, 962–968 CrossRef CAS PubMed.
- G. Lotfi, M. A. Shokrgozar, R. Mofid, F. M. Abbas, F. Ghanavati, A. A. Bagheban and R. P. Shariati, J. Periodontol., 2011, 82, 1367–1375 CrossRef CAS PubMed.
- F. Guillemot, A. Souquet, S. Catros, B. Guillotin, J. Lopez, M. Faucon, B. Pippenger, R. Bareille, M. Remy, S. Bellance, P. Chabassier, J. C. Fricain and J. Amedee, Acta Biomater., 2010, 6, 2494–2500 CrossRef CAS PubMed.
Footnote |
| † Peng Liu and Qing Li equally contributed to this article. |
| This journal is © The Royal Society of Chemistry 2020 |