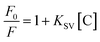Open Access Article
Open Access ArticleAn optical sensor for selective detection of phenol via double cross-linker precipitation polymerization
Xiaodong Lv *a and
Peng Gaob
*a and
Peng Gaob
aSchool of Electrical Engineering and Control Science, Nanjing Tech University, Nanjing 211899, China. E-mail: lvlvxiaodong@126.com
bSchool of Electrical Engineering, Tongling University, Tongling 244000, China
First published on 3rd July 2020
Abstract
Based on the electron-transfer mechanism between the template and quantum dots (QDs), an optical sensor was structured. It is a phenol sensor, which has the room-temperature phosphorescence (RTP) property of Mn-doped ZnS QDs and high selectivity of molecular imprinted polymers (MIPs). On the surface of the silane modified Mn-doped ZnS QDs, the phenol sensor was prepared by double cross-linker precipitation polymerization in the absence of any stabilizer and additive. Double cross-linkers, divinylbenzene (DVB) and ethyleneglycol dimethacrylate (EGDMA), make a great contribution to the imprinted polymerization with hydrogen-bonding interaction. Then, as a result of the functional monomer, methacrylic acid (MAA), a carboxylic acid was grafted onto the surface of ZnS QDs:Mn@MIPs. Under optimal conditions, the phenol determination experiment had a linear range of 5.0 to 55 μmol L−1 with a correlation coefficient of 0.9984, and a high imprinting factor (IF) of 3.43. In addition, the prepared ZnS QDs:Mn@MIPs were successfully used to detect phenol in real water samples. Therefore, this work provided a highly selective and sensitive RTP probe for phenol determination.
Introduction
A sensor1–3 is a new generation of the intelligent information exchange hub, which enables the mutual conversion of digital and analog signals, due to its miniaturization and integration. Its aim is to parameterize a model by collecting, processing and using data. Quantum dots4–6 (QDs) are a kind of photoluminescence nanomaterial, also called semiconductor nanocrystals, fluorescence QDs and phosphorescence QDs, for example. Based on the optical quenching phenomenon of electron-transfer induction between a template and QDs, QDs, the nucleus sensing matrix, were used as an optical sensor for template determination,7–9 including ions, small molecules and biological macromolecules.Compared with traditional fluorescence QDs,10,11 such as CdSe QDs, CdS QDs and so on, phosphorescence QDs12 have attracted more attention in recent years. As was known to all, ZnS QDs13–15 are one of phosphorescence QDs and have the room-temperature phosphorescence (RTP) property. Because of the case for spin-forbidden transition and intersystem crossing from excited triplet state to ground state, ZnS QDs have longer lifetime than other fluorescence QDs and are not vulnerable to scattered light and short-lived autofluorescence. Due to these extraordinary characters above, ZnS QDs are worth giving a higher priority. R. N. Bhargava16 proposed that prepared Mn-doped ZnS QDs, with sizes varying from 3.5 to 7.5 nm, had the ability to yield a photoluminescent quantum efficiency 18%. So, it is essential to develop a mathematical model based on RTP quenching of Mn-doped ZnS QDs for template target quantitative estimation. Then it is a possibility of taking steps to bridge a direct linear relationship between template target and Mn-doped ZnS QDs, so as to estimate unknown concentration of template target in real samples. However, when unknown samples were with the attendance of several structurally similar to the template target, the selectivity of a human-made optical sensor of Mn-doped ZnS QDs was weakened. Hence, its shortcoming still needs to be avoided. As a result, the tight coupling with molecular imprinted technology17–19 (MIT) is a consequent trend and it is an expecting action.
During these years, MIT, as a valid path, has been played a key role in guidance of synthesis and applications of molecular imprinted polymers (MIPs). MIPs go through an experience of pre-polymerization and copolymerization among three unites: template target, cross-linkers and functional monomers. MIPs with tailored recognition sites, stem from the removal of template target. There are so many recognition sites of MIPs, which are multiple complementary to template target in space and chemical groups, in many circumstances. Then, MIPs have a predetermined selectivity, serving different template targets for various purposes. As a consequence, MIPs have been widely applied in many fields,20–24 including chemical sensor, cell labelling, target analysis and so on. But the recognition dynamic is lamed by the reasons of three-dimensional highly cross-linked polymeric structure and residual impurities. To overcome these disadvantages above, double cross-linkers precipitation polymerization25–28 has been taken into consideration and it is beneficial to the improvement of the cleanliness of MIPs. In addition, double cross-linkers precipitation polymerization contributes to simplified aftertreatment, due to auto-precipitation of synthetic polymers. Therefore, it is necessary to make the most of double cross-linkers precipitation polymerization, which can build a template target sensor.
The traditional industries, such as metallurgical coal and coking, have brought us poisonous wastewater pollutants, in which phenolic wastewater29 is one of them. In particular, on the account of phenol residues in lakes, soils and so on, it has a serious impact on the human health. So, how to effectively measure phenol residues is a valuable application of engineering and its aim is to reach a balance between economic development and natural environment. To date, many approaches30,31 have been applied to detect phenol, for instance, electrochemistry and chromatography. By contrast, an optical sensor, based on QDs@MIPs, is more worth studying, because of low cost, fast response and optical stability. It is important to develop a phenol sensor.
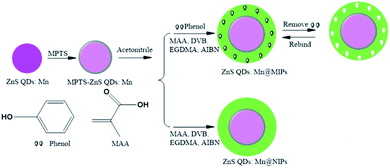
In this work, a phenol sensor, ZnS QDs:Mn@MIPs, was synthesized by double cross-linkers precipitation polymerization in the absence of any stabilizer and additive. On the surface of MPTS-ZnS QDs:Mn, a phenol sensor was obtained, which uses divinylbenzene (DVB) and ethyleneglycol dimethacrylate (EGDMA) as double cross-linkers, methacrylic acid (MAA) as functional monomer and 2,2′-azobisisobutyronitrile (AIBN) as initiator, respectively. In particular, double cross-linkers make great contributions to imprinted polymerization in hydrogen-bonding interaction. Then, as a result of the functional monomer (MAA), carboxylic acid was grafted onto the surface of ZnS QDs:Mn@MIPs, which contribute to improve binding kinetics and capacity between recognition sites and template target phenol. Moreover, the morphology and RTP of a phenol sensor were investigated and successfully used for phenol determination in real water samples.
Experimental
Reagents and chemicals
All reagents used in this work were analytical grade purity. ZnSO4·7H2O, MnCl2·4H2O, Na2S·9H2O, ethanol, N2, 3-mercaptopropyltriethoxysilane (MPTS), acetonitrile, divinylbenzene (DVB), ethyleneglycol dimethacrylate (EGDMA), methacrylic acid (MAA), 2,2′-azobisisobutyronitrile (AIBN), phenol, catechol, resorcinol and 2,6-dichlorophenol were all purchased from Aladdin Reagent Co., Ltd. (Shanghai, China). Double distilled water (DDW) was used throughout.Instrument
The morphology was obtained by transmission electron microscope (TEM, JEOL, JEM-2100). The phosphorescence measures were carried out by a Cary Eclipse spectrofluorometer (USA), equipped with a plotter unit and a quartz cell. Infrared spectra (4000 to 400 cm−1) were gained by using KBr disks with a Nicolet NEXUS-470 FTIR apparatus (USA).Synthesis of MPTS-ZnS QDs:Mn
MPTS-ZnS QDs:Mn was synthesized according to the previously published method.32,33 Under the protection of N2, ZnSO4·7H2O (2.013 g), MnCl2·4H2O (0.126 g) and DDW (40 mL) were mixed in a three-necked flask, with ultrasonic dispersion. At the room temperature, 97.6 g L−1 Na2S·9H2O of 10 mL DDW was dissolved step-by-step and stirring for 30 min. Next, 11.7 mL L−1 MPTS of 10 mL ethanol was added and stirring for 20 h again. MPTS-ZnS QDs:Mn, pale pink powder, was synthesized, through simplified aftertreatment (centrifugal separation, washing and drying).Synthesis of ZnS QDs:Mn@MIPs
To a single-necked flask, MPTS-ZnS QDs:Mn (30 mg) and acetonitrile (15 mL) were dissolved, with ultrasonic dispersion. MAA (60 μL), EGDMA (200 μL), DVB (100 μL), AIBN (10 mg) and phenol (5 mg) were mixed step-by-step and stirring for 29 h. The mixtures were at 50 °C for 5 h and at 60 °C for 24 h again. ZnS QDs:Mn@MIPs was synthesized, through simplified aftertreatment (centrifugal separation, washing and drying). As a comparison without adding template target phenol, ZnS QDs:Mn@NIPs was synthesized at the same condition.Measurement procedure
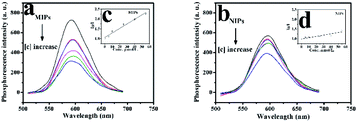
Firstly, ZnS QDs:Mn@MIPs and ZnS QDs:Mn@NIPs were dispersed in DDW to gain the stock solution (100 mg L−1). Phenol, catechol, resorcinol and 2,6-dichlorophenol were dissolved in DDW to get the target solution (10 mol L−1), separately. In the experiments, all the phosphorescence measures were performed under the same conditions: the slit widths of the excitation and emission were both 10 nm, the photomultiplier tube voltage was set at 720 V, and the excitation wavelength was set at 330 nm (curve 2) with the recording phosphorescence spectra range of 500 to 700 nm (curve 3), in Fig. 1. Based on previous work,34 there are two UV absorption characteristic peaks (204 nm and 270 nm) of phenol with different absorbance values (4.0 and 2.0), respectively, which do not overlap with emission spectra range of MPTS-ZnS QDs:Mn. Thus, it indicated that RTP quenching was non-radiative energy transfer.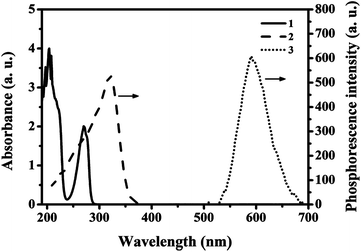 | ||
| Fig. 1 UV absorption spectra of the phenol (curve 1), RTP excitation (curve 2) and emission (curve 3) spectra of the MPTS-ZnS QD:Mn. | ||
Results and discussion
Preparation and characterization of ZnS QDs:Mn@MIPs
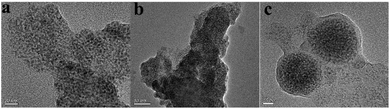
As was shown in Fig. 2, ZnS QDs:Mn@MIPs were synthesized via double cross-linkers precipitation polymerization. First of all, Mn-doped ZnS QDs were modified by MPTS. By harnessing the Si–O chain in MPTS, MPTS-ZnS QDs:Mn have better aqueous RTP stability. Then, pooling double cross-linkers (DVB and EGDMA) provides hydrogen-bonding interaction for imprinted polymerization. In the presence of double cross-linkers (DVB and EGDMA), functional monomer (MAA) and initiator (AIBN), carboxylic acid was grafted onto the surface of ZnS QDs:Mn@MIPs, which boosts binding kinetics and capacity between recognition sites and template target phenol. So, after the removal of template target phenol, ZnS QDs:Mn@MIPs with a lot of recognition sites, would show great selectivity and sensitivity. This phenomenon could be explained in carboxylic acid and hydroxyl interaction.As was shown in Fig. 3, the TEM images of (a) ZnS QDs:Mn, (b and c) ZnS QDs:Mn@MIPs were recorded. ZnS QDs:Mn had an even distribution in size, about 2.5 nm. By contrast with Fig. 2a, there were a large number of polymers, which indicated the existence of MIPs layer, in Fig. 3b. The diameter of ZnS QDs:Mn@MIPs was approximately 25 nm, in Fig. 3c. These phenomena were the same as earlier prediction and the experiment was feasible.
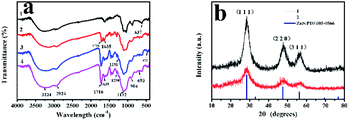
As was shown in Fig. 4a, FT-IR spectra of ZnS QDs:Mn (curve 1), MPTS-ZnS QDs:Mn (curve 2), ZnS QDs:Mn@MIPs (curve 3) and ZnS QDs:Mn@NIPs (curve 4) were recorded. The characteristic peak of 637 cm−1 was in line with sulfuret of ZnS QDs. The characteristic peak of 1112 cm−1 was Si–O–Si stretching vibration and other characteristic peaks of 693 cm−1 and 472 cm−1 were Si–O vibration. Some characteristic peaks of 1298 cm−1 and 1191 cm−1 were C–O–C stretching vibration, owing to the existence of double cross-linkers (DVB and EGDMA). Moreover, these peaks of 3224 cm−1, 1718 cm−1 and 1635 cm−1 were O–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–N stretching vibration. Because of the comprehensive effect of double cross-linkers and functional monomer (MAA), carboxylic acid was grafted onto the surface of ZnS QDs:Mn@MIPs. All the results suggest that ZnS QDs:Mn@MIPs have been successfully prepared.
O and C–N stretching vibration. Because of the comprehensive effect of double cross-linkers and functional monomer (MAA), carboxylic acid was grafted onto the surface of ZnS QDs:Mn@MIPs. All the results suggest that ZnS QDs:Mn@MIPs have been successfully prepared.
As was shown in Fig. 4b, XRD patterns of MPTS-ZnS QDs:Mn (curve 1) and ZnS QDs:Mn@MIPs (curve 2) were recorded. There were three obvious peaks (1,1,1), (2,2,0) and (3,1,1), which were in line with ZnS standard PDF card (PDF#05-0566). These results revealed that cubic crystal structures of ZnS were found in ZnS QDs:Mn@MIPs and double cross-linkers precipitation polymerization did not change original crystalline structure. By contrast with the curve 1, the characteristic diffraction peaks of the curve 2 weakened in general, certifying MIPs layer.
Effect of time, solution concentration and pH
As was shown in Fig. 5a, stability of RTP intensities of MPTS-ZnS QDs:Mn (curve 1), ZnS QDs:Mn@MIPs (curve 2) were measured. There were 12 repeated RTP intensities tests at different intervals during 48 h. The results were that ZnS QDs:Mn@MIPs had no difference in original stability RTP intensities of MPTS-ZnS QDs:Mn.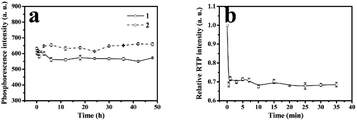 | ||
| Fig. 5 (a) Stability of RTP intensities of MPTS-ZnS QDs:Mn (curve 1), ZnS QDs:Mn@MIPs (curve 2); (b) effect of different interval detection time on the relative RTP intensity of ZnS QDs:Mn@MIPs. | ||
As usual, phosphorescent quenching would spend some time to reach a relatively stable state. As was shown in Fig. 5b, relative RTP intensities of ZnS QDs:Mn@MIPs at different interval detection time were recorded. There were 12 repeated RTP intensities tests at different intervals, with adding a certain concentration of phenol. When the detection time was from 0 to 5.0 min, the response level of ZnS QDs:Mn@MIPs was very high on account of double cross-linkers precipitation polymerization. Without any stabilizer and additive, this polymerization process avoided some problems of residual impurities and product extraction. Finally, the 20 min was considered as detection time of phenol sensor.
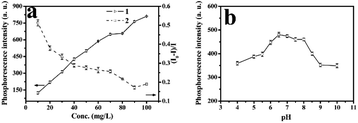
As was showed in Fig. 6a, various RTP intensities of ZnS QDs:Mn@MIPs were recorded, with adding a certain amount of phenol. Higher or lower concentrations of ZnS QDs:Mn@MIPs would lead to incomplete or excessive RTP quenching. So, a balance between linear range and sensitivity of the phenol sensor would be reached. When the concentration of ZnS QDs:Mn@MIPs was from 10 to 20 mg L−1, lower RTP intensity (from 123.46 a.u. to 219.89 a.u.) of ZnS QDs:Mn@MIPs obtained better sensitivity (from 0.375 to 0.513) and vice versa. Therefore, 38 mg L−1 was chosen as relative the optimal concentration value of ZnS QDs:Mn@MIPs.
As was shown in Fig. 6b, RTP intensities of ZnS QDs:Mn@MIPs at different pH values were recorded. When pH was from 6.0 to 8.0, the RTP intensity of ZnS QDs:Mn@MIPs reached a relative stable state. Then, pH 6.5 was used as an optimal experiment pH environment.
ZnS QDs:Mn@MIPs with the template target of different concentrations
As was shown in Fig. 7, the phenol sensor, based on RTP intensity of ZnS QDs:Mn@MIPs, was further studied. The mixtures of phenol and ZnS QDs:Mn@MIPs were kept for 20 min at room temperature. ZnS QDs:Mn@MIPs with the same operation, were a comparison. The RTP quenching relationship of the phenol sensor could be followed by the Stern–Volmer equation:35,36F and F0 were RTP intensity of ZnS QDs:Mn@MIPs or @NIPs in the case of presence or absence of template target, respectively. KSV is the Stern–Volmer quenching constant and [C] is the concentration of template target. With linearly increasing concentration of template target phenol, RTP intensities of ZnS QDs:Mn@MIPs decreased correspondingly. By calculation, KSV,MIPs of ZnS QDs:Mn@MIPs was 2.305 104 M−1 and linear range of standardization curve was 5.0 to 55 μmol L−1 with a correlation coefficient of 0.9984; by contrast, KSV,NIPs was 0.672 104 M−1 with a correlation coefficient of 0.9932. Imprinting factor (KSV,MIPs/KSV,NIPs IF) was 3.43 and the limit of detection (LOD, 3σ/k) of ZnS QDs:Mn@MIPs was 2.13 μmol L−1, in which σ was the standard deviation of six times tests and k was the slope of the calibration line. It would be found that double cross-linkers precipitation polymerization has a direct impact on linear quenching of the phenol sensor.
Selectivity of ZnS QDs:Mn@MIPs
Several phenol analogues, structurally similar to template target phenol, were used as obstacles to assess the selectivity of ZnS QDs:Mn@MIPs. As was shown in Fig. 7, the line of quench efficiency for ZnS QDs:Mn@MIPs was: phenol (0.560) > resorcinol (0.331) > catechol (0.324) > 2,6-dichlorophenol (0.301). The quench efficiency differences for four phenols between ZnS QDs:Mn@MIPs and @NIPs were: 0.251, 0.027, 0.030 and 0.017, respectively, because of the attendance of tailored recognition sites. Hence, the prepared phenol sensor had a better selectivity (Fig. 8).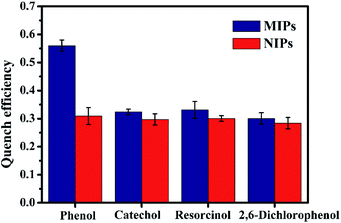 | ||
| Fig. 8 The selectively of ZnS QDs:Mn@MIPs and ZnS QDs:Mn@NIPs with addition of several phenol analogues (5 mg L−1). | ||
Application in real samples
In real water samples, ZnS QDs:Mn@MIPs were selected so as to evaluate the feasibility of phenol sensor, as was shown in Table 1. These samples were spiked with standard phenol (from 5.0 to 30 μmol L−1) without no other phenol analogues The results were found that the RSD was lower than 3.7% and the recovery was from 101.4% to 107.2%, indicating that the ZnS QDs:Mn@MIPs were great stability, sensitivity and selectivity probes for determination phenol in real samples.| Sample | Concentration Taken (μmol L−1) | Found (μmol L−1) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| a Average of three measurements. | ||||
| 1 | 5 | 5.41 | 107.2 | 3.7 |
| 2 | 10 | 10.23 | 104.6 | 2.9 |
| 3 | 15 | 15.19 | 102.3 | 1.6 |
| 4 | 20 | 20.35 | 103.1 | 2.3 |
| 5 | 30 | 30.26 | 101.4 | 1.4 |
Conclusions
In summary, via double cross-linkers precipitation polymerization, a phenol sensor was built without any stabilizer and additive. It was beneficial to the improvement of polymers cleanliness and avoided residual impurities which would have negative impacts on response level of the optical sensor. In the presence of double cross-linkers (DVB and EGDMA), functional monomer (MAA) and initiator (AIBN), ZnS QDs:Mn@MIPs had excellent sensitivity and selectivity for the template target phenol. In particular, double cross-linkers were responsible for imprinted polymerization in hydrogen-bonding interaction and functional monomer provided a channel to promote carboxylic acid to graft onto the surface of ZnS QDs:Mn@MIPs, in order to improve binding kinetics and capacity between recognition sites and template target phenol. Finally, this work gave a novel path of synthesis of MIPs for template target phenol.Conflicts of interest
There are no conflicts to declare and there are no potential interests conflicts.Acknowledgements
This work was financially supported by the University Natural Science Foundation of Anhui Province (No. KJ2018A0476).References
- Y. S. Suh, Sensors, 2019, 19(8), 1924 CrossRef PubMed.
- B. D. Majumder, J. K. Roy and S. Padhee, IEEE Sensor. J., 2019, 19, 1204–1214 Search PubMed.
- L. Sanford and A. Palmer, Enzymes as Sensors, 2017, 589, 1–49 CrossRef CAS PubMed.
- Y. Chung, J. Choi and H. S. Sim, J. Korean Phys. Soc., 2018, 72, 1454–1466 CrossRef.
- C. J. van Diepen, P. T. Eendebak, B. T. Buijtendorp, U. Mukhopadhyay, T. Fujita, C. Reichl, W. Wegscheider and L. M. K. Vandersypen, Appl. Phys. Lett., 2018, 113(3), 033101 CrossRef.
- D. Kang, M. B. Kumar, C. Son, H. Park and J. Park, Appl. Sci., 2019, 9(21), 4661 CrossRef CAS.
- S. S. Nath, D. Chakdar, G. Gope and D. K. Avasthi, J. Nanoelectron. Optoelectron., 2008, 3, 180–183 CrossRef.
- J. Hu, Y. Huang, Q. Zhu, Y. Lin, K. Xu, F. Ding, C. Zhang and P. Luo, J. Nanoelectron. Optoelectron., 2019, 14, 972–977 CrossRef.
- H. Pandey, P. Khare, S. Singh and S. P. Singh, Mater. Chem. Phys., 2020, 239 Search PubMed.
- J. Yang, Z. Zhang and G. Yan, Sensor. Actuator. B Chem., 2018, 255, 2339–2346 CrossRef CAS.
- L. J. Tian, N. Q. Zhou, X. W. Liu, X. Zhang, T. T. Zhu, L. L. Li, W. W. Li and H. Q. Yu, Sci. Rep., 2017, 7, 2048 CrossRef PubMed.
- W. Liu, H. Li, Y. Wei and C. Dong, RSC Adv., 2017, 7, 26930–26934 RSC.
- T. Chen, L. Li, G. Xu, X. Wang, J. Wang, Y. Chen, W. Jiang, Z. Yang and G. Lin, Front. Pharmacol., 2018, 9, 763 CrossRef PubMed.
- R. Li, H. Sun, S. Wang, Y. Wang and K. Yu, J. Agric. Food Chem., 2018, 66, 814–821 CrossRef CAS PubMed.
- M. Lim, W. Lee, G. Bang, W. J. Lee, Y. Park, Y. Kwon, Y. Jung, S. Kim and J. Bang, Nanoscale, 2019, 11, 10463–10471 RSC.
- R. N. Bhargava, D. Gallagher, X. Hong and A. Nurmikko, Phys. Rev. Lett., 1994, 72, 416–419 CrossRef CAS.
- O. S. Ahmad, T. S. Bedwell, C. Esen, A. Garcia-Cruz and S. A. Piletsky, Trends Biotechnol., 2019, 37, 294–309 CrossRef CAS PubMed.
- M. Komiyama, T. Mori and K. Ariga, Bull. Chem. Soc. Jpn., 2018, 91, 1075–1111 CrossRef CAS.
- M. Fang, L. Zhou, H. Zhang, L. Liu and Z. Y. Gong, Food Chem., 2019, 274, 156–161 CrossRef CAS PubMed.
- Y. Zhang, J. Zhang and Q. Liu, Sensors, 2017, 17(7), 1567 CrossRef PubMed.
- Y. Saylan, F. Yilmaz, E. Ozgur, A. Derazshamshir, H. Yavuz and A. Denizli, Sensors, 2017, 17(4), 898 CrossRef PubMed.
- N. Idil, M. Hedstrom, A. Denizli and B. Mattiasson, Biosens. Bioelectron., 2017, 87, 807–815 CrossRef CAS.
- R. Xing, Y. Wen, H. He, Z. Guo and Z. Liu, Trac. Trends Anal. Chem., 2019, 110, 417–428 CrossRef CAS.
- E. Yilmaz, B. Garipcan, H. K. Patra and L. Uzun, Sensors, 2017, 17(4), 691 CrossRef PubMed.
- M. Qiao, X. Lei, Y. Ma, L. Tian, X. He, K. Su and Q. Zhang, Nano Res., 2018, 11, 1500–1519 CrossRef CAS.
- B. Huang, F. Bai, X. L. Yang and W. Q. Huang, Chin. J. Polym. Sci., 2010, 28(2), 277–285 CrossRef CAS.
- X. H. Jiang, Z. M. Liu and W. P. Tu, Acta Polym. Sin., 2012, 6, 633–639 CrossRef.
- O. L. J. Virtanen, M. Kather, J. Meyer-Kirschner, A. Melle, A. Radulescu, J. Viell, A. Mitsos, A. Pich and W. Richtering, ACS Omega, 2019, 4, 3690–3699 CrossRef CAS PubMed.
- M. Y. Kilic, W. H. Abdelraheem, X. He, K. Kestioglu and D. D. Dionysiou, J. Hazard. Mater., 2019, 367, 734–742 CrossRef CAS PubMed.
- W. Ma, Y. Han, C. Xu, H. Han, W. Ma, H. Zhu, K. Li and D. Wang, Bioresour. Technol., 2018, 251, 303–310 CrossRef CAS.
- N. Zhan, F. Guo, Q. Tian, Z. P. Yang and Z. Rao, Anal. Lett., 2018, 51, 955–970 CrossRef CAS.
- X. Wu, X. Lv, J. Wang, L. Sun and Y. Yan, Anal. Methods, 2017, 9, 4609–4615 RSC.
- X. Wei, Z. Zhou, T. Hao, H. Li and Y. Yan, RSC Adv., 2015, 5, 19799–19806 RSC.
- X. Lv and P. Gao, RSC Adv., 2020, 10, 17906–17913 RSC.
- N. Azum, M. A. Rub and A. M. Asiri, J. Chem. Thermodyn., 2019, 128, 406–414 CrossRef CAS.
- T. N. J. I. Edison, R. Atchudan, J. J. Shim, S. Kalimuthu, B. C. Ahn and Y. R. Lee, J. Photochem. Photobiol. B Biol., 2016, 158, 235–242 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |