Open Access Article
Open Access ArticleResidue-based propensity of aggregation in the Tau amyloidogenic hexapeptides AcPHF6* and AcPHF6†
Abha Dangi‡
abc,
Abhishek Ankur Balmik‡cd,
Archana Kisan Ghorpadeab,
Nalini Vijay Gorantlacd,
Shweta Kishor Sonawanecd,
Subashchandrabose Chinnathambi *cd and
Udaya Kiran Marelli
*cd and
Udaya Kiran Marelli *abc
*abc
aCentral NMR Facility, CSIR-National Chemical Laboratory, Dr HomiBhabha Road, 411008 Pune, India
bDivision of Organic Chemistry, CSIR-National Chemical Laboratory, Dr HomiBhabha Road, 411008 Pune, India
cAcademy of Scientific and Innovative Research (AcSIR), 110025 New Delhi, India
dNeurobiology Group, Division of Biochemical Sciences, CSIR-National Chemical Laboratory, Dr HomiBhabha Road, 411008 Pune, India
First published on 21st July 2020
Abstract
In Alzheimer's disease and related tauopathies, the aggregation of microtubule-associated protein, Tau, into fibrils occurs via the interaction of two hexapeptide motifs PHF* 275VQIINK280 and PHF 306VQIVYK311 as β-sheets. To understand the role of the constituent amino acids of PHF and PHF* in the aggregation, a set of 12 alanine mutant peptides was synthesized by replacing each amino acid in PHF and PHF* with alanine and they were characterized by nuclear magnetic resonance (NMR) spectroscopy, circular dichroism (CD), transmission electron microscopy (TEM) and ThS/ANS fluorescence assay. Our studies show that while the aggregation was suppressed in most of the alanine mutant peptides, replacement of glutamine by alanine in both PHF and PHF* enhanced the fibrillization.
Conformational misfolding of proteins leading to protein aggregates, also known as amyloids, is a prime trait of neurodegenerative diseases such as Alzheimer's (AD) and Parkinson's disease (PD).1,2 Amyloid protein aggregates are also observed in other disease conditions such as diabetes and arthritis.1,3–7 Deposition of amyloids in AD leads to irreversible cell death and chronic dementia, resulting in the loss of vital bodily functions, severe behavioural disorders and ultimately death. The search for an effective cure for AD is in persistent progress and interfering with the protein deposition mechanisms in neuronal cells and tissues is a rational target8–13 for the majority of studies focused on the drug discovery research for AD.
Two proteins, amyloid beta (Aβ) and Tau, have been identified to form protein deposits linked with AD. Aβ are various small length protein fragments derived from amyloid precursor protein (APP), among which Aβ-42 has been proven to be toxic and form amyloid plaques.8,14 However, in recent times, increasing evidence for the pathological role of Tau protein in AD was found.15–19 Tau, a microtubule-binding protein, forms neurofibrillary tangles (NFTs) in AD via soluble oligomers, which are stabilized by beta-sheet interactions. Tau is 441 amino acids long with an N-terminus region, a proline rich region, a key microtubule-binding domain consisting of 3 or 4 repeat regions depending on the Tau isoform, and a C-terminus domain.20 A high resolution cryo-EM structure of Tau filaments from the brain of an AD patient was recently established and highlighted the presence of beta sheet structures stabilized by hydrogen bonds and hydrophobic interactions.21 Interestingly, the cryo-EM structure of Tau filaments obtained from the brain of a patient with Pick's disease revealed a different fold for Tau, suggesting disease specific conformational misfolding of Tau.22
Two hexapeptide regions, 275VQIINK280 and 306VQIVYK311, residing in the second and third repeat units of Tau, respectively, form the core beta sheet structure in Tau, a structural feature responsible for Tau oligomerization and the formation of NFTs.16,23–26 Synthetic analogues of both these hexapeptides, Ac-VQIINK-NH2 (AcPHF6*, 1; PHF stands for paired helical filaments) and Ac-VQIVYK-NH2 (AcPHF6, 1′), which has acetyl protection on the N-terminus and amidation on the C-terminus, are known to form structurally similar fibrils akin to those of Tau. Due to this structural mimicry, AcPHF6 and AcPHF6* stand out as model peptides to study the promotion or inhibitory effect of various ligands on Tau aggregation27–29 and for the development of any peptide based Tau aggregation inhibitors.30–39 Previous studies have shown that even shorter fragments from AcPHF6 are also capable of fibril formation but with a lower rate of polymerization and only upon nucleation with known fibrils.40 The aggregation properties in lysine mutants of AcPHF6 were explored by the Goux research group and the results showed variable fibril formation tendency for the mutants subject to buffer and salt conditions.40,41 Recently, Chemerovski-Glikman et al. demonstrated that proline mutants of AcPHF6 inhibited the aggregation of AcPHF6.42
In general, AcPHF6* remains less explored compared to AcPHF6, although the recent studies attribute a higher role to 275VQIINK280 in Tau aggregation.27 Despite several studies on these hexapeptides, to our knowledge, the contribution of individual amino acids in AcPHF6 and AcPHF6* towards Tau aggregation has not been examined hitherto, which information is fundamentally important for the design of Tau aggregation inhibitors. Hence, in the present study, the residue specific propensity of amino acids constituting AcPHF6* and AcPHF6 in their aggregation is investigated. For our study, alanine mutation peptides of AcPHF6* and AcPHF6 were synthesized, and their conformational and aggregation properties were characterized using various biophysical and biochemical techniques.
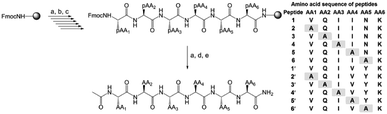
A set of twelve peptides consisting of parent peptides AcPHF6* (1) and AcPHF6 (1′) and their sequential single site alanine mutant peptides (2–6 and 2′–6′) were synthesized by employing Fmoc-based solid phase peptide synthesis using rink amide MBHA resin (Fig. 1). All of the peptides were subsequently purified by reverse phase semi preparative HPLC for further studies. Although mutant peptides of 1 and 1′ in which lysine is substituted by alanine were synthesized, they could not be purified because of partial solubility in an acetonitrile–water combination and other buffers.
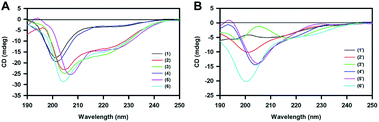
Under normal physiological conditions, Tau protein exists in a random coil conformation or a natively disordered state.43,44 CD spectroscopic study of all of the twelve peptides was carried out at 1 mg mL−1 concentration in plain 5 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (Fig. 2) under non-aggregating conditions. All of the peptides exhibited a similar pattern of CD absorption with only negative mode absorption. Peptides 1 and 4 had similar CD spectra with negative absorption at ∼203 nm and a very weak shoulder at around 225 nm, whereas 2, 3, 5 and 6 exhibited slightly intense CD spectra compared to 1 and 4, with negative absorption at 205 nm and a weak negative shoulder at around 225 nm. This shift for 1 and 4 could be due to the presence of mixed secondary structures. Overall, the described CD analysis is mostly indicative of a random coil structure.40,41 Hence, a possible random coil population may be inferred for all of the peptides 1–6. A similar trend was observed for the conformation of the peptides in the 1′–6′ series, wherein the 2′, 4′, 5′ and 6′ peptides exhibited negative absorptions at around ∼203–205 and ∼225 nm, with the latter being weak in intensity. A slight rightward shift for 4′ and 5′ compared to 2′ and 6′ could indicate a possible mixed population of random coils and beta sheets. Peptides 1′ and 3′ did not have a characteristic CD pattern corresponding to any of the standard peptide secondary structural scaffolds. The signatures obtained for peptides 1′ and 3′ indicate a structural orientation towards β-sheets.
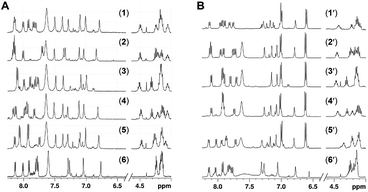
1H NMR spectra for the compounds 1–6 and 1′–6′ were recorded in DMSO-d6 (Fig. 3) and phosphate buffer (see the ESI†) at 298 K at 5 mM concentration. The chemical shift dispersion of the amide and alpha proton regions in 1H NMR reflect the folded secondary structural nature of the linear peptides. It is evident from the overlay of these regions for peptides 1–6 and 1′–6′ (Fig. 3, ESI Fig. 1–24†) that none of the peptides have significant chemical shift dispersion, either for amide or alpha protons. For all of the peptides, the amide and alpha proton chemical shifts were found to be distributed within a range of 0.2–0.5 ppm and have severe overlaps, supporting the absence of a well folded conformation among these peptides. This is in concurrence with the CD spectroscopic studies, which indicated a random coil structure for 1–6 and 1′–6′ in monomeric form. 1H NMR based hydrogen bonding coefficients (calculated based on the change in the chemical shift over a temperature range; see the ESI†) for the amide NHs conveys information about the involvement of NHs in hydrogen bonding or solvent shielding. Constituent backbone amide NH resonances from all of the peptides have shown high temperature coefficients (in the range of −3.4 to −7.6 ppb K−1; see the ESI†), implying their non-involvement in strong hydrogen bonding. This complements the lack of chemical dispersion for amide and alpha protons, further supporting the absence of a robust conformation for any of the twelve peptides, 1–6 and 1′–6′. Therefore, based on the above results it can be stated that mutation with alanine neither promoted a robust helical or beta sheet conformtion nor brought any significant changes in the original conformtion of peptides 1–6 and 1′–6′.
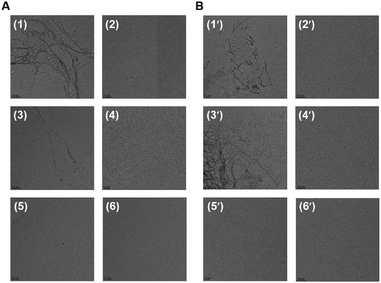
The self-aggregation properties of peptides 1–6 and 1′–6′ were characterized by TEM analysis (Fig. 4). The aggregation for all of the peptides was set up at 100 μM concentrations in 20 mM BES buffer at pH 7.4 with 25 μM heparin as an inducer. After incubation for 168 h at 37 °C, TEM images were recorded for 2 μM peptides on carbon coated copper grids. The parent peptides, AcPHF6* (1) and AcPHF6 (1′) formed aggregates as expected. Out of ten mutant peptides, peptides 3 and 3′ both with glutamine substituted by alanine showed filamentous aggregates similar to 1 and 1′, while no aggregates were observed for the rest of the peptides.
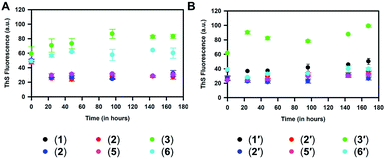
Aggregation of the 1–6 peptides was also monitored by ThS and ANS fluorescence. It was observed that the substitution of valine or isoleucine with alanine in 1 diminished the aggregation propensity of hexapeptides as compared to the native peptide 1. However, substituting glutamine with alanine in peptide 3 increased the aggregation propensity (Fig. 5A), while a marginally higher aggregation propensity was also observed for 6, in which alanine is substituted for asparagine. On comparing them at 168 h, 3 and 6 exhibited 83 and 60 au fluorescence, respectively, compared to 30 au for 1 (ESI Fig. 25C†). Similar results were observed for the aggregation assay carried out for the 1′–6′ peptides and their mono-substituted variants (Fig. 5B). The aggregation propensity was found to be suppressed for 2′, 4′, 5′ and 6′ when compared to 1′ and 3′. The aggregation propensity of 3′ was found to be greatly increased compared to all hexapeptides. At 168 h, 3′ exhibited an absolute fluorescence of 100 au, compared to 1′ where a fluorescence value of 48 au was obtained (ESI Fig. 25D†). In agreement with this, peptides 1′ and 3′, which showed high aggregation propensity in the ThS fluorescence assay, did not present any prominent signature for random coiling but showed orientation towards β-sheet structure in their CD spectra (Fig. 2). However, the ANS fluorescence assay for monitoring hydrophobicity changes over time did not show any significant difference between 1 and its variants (ESI Fig. 25E†). 1′ and its variants in the ANS fluorescence assay followed the pattern of the corresponding ThS assay, suggesting that glutamine to alanine substitution was associated with increased hydrophobicity (ESI Fig. 25F†). Overall, the current aggregation assay suggests that substitution of glutamine to alanine in 1 and 1′ elevates the aggregation propensity, while alanine replacements for the majority of all the other residues suppress the aggregation propensity.
Thus, it is interesting to note that out of the first five residues in 1 and 1′ (namely VQIIN in 1 and VQIVY in 1′) substitution of any amino acid except for glutamine with alanine nullified their aggregation properties. Conversely, it may be stated that except for glutamine (and asparagine as per the fluorescence assay results), the rest of the residues (V and I in 1, and V, I and Y in 1′) have a higher propensity to aggregate than alanine when present in 1 and 1′. Interestingly, in one of the recent studies where proline, a beta sheet breaker, is substituted for each amino acid in 1′, it was reported that the peptide with glutamine to proline substitution inhibited the aggregation properties of 1′ while the mutant peptide itself was non-aggregating in nature.42 We speculate that the glutamine in the PHF peptides might be in a conformationally favorable position for beta sheet formation and aggregation, which was disturbed by the cyclically constrained amino acid proline but not by an alanine in our case.
Furthermore, the aggregated 1–6 and 1′–6′ peptides after 168 h were studied in terms of their toxicity in neuro2a cells (see the ESI, Fig. 26†). The aggregated 1–6 peptides did not show toxicity to neuro2a cells up to 5 μM concentration.
In conclusion, Alzheimer's disease is characterized by the deposition of intracellular Tau aggregates and extracellular amyloid-beta plaques. Various treatment strategies have been designed to inhibit or revert this protein aggregation. In particular, PHF6* and PHF6 peptide based inhibition therapies have gained focus due to their structural mimicry to Tau aggregation. To understand the aggregation nature of these two peptides at an individual residue level, here we have synthesized alanine mutant libraries of AcPHF6* and AcPHF6, and characterized them using various biochemical and biophysical methods. The results showed that all of the mutant peptides, except for those with glutamine to alanine substitution (3 and 3′), did not aggregate as per the general conditions used for the parent peptides 1 and 1′. While asparagine to alanine mutation (6) showed aggregation propensity in fluorescence assays, no aggregates were observed in the TEM images. Therefore, it was inferred that except for glutamine, the residues V and I in AcPHF6* 1, and V, I and Y in AcPHF6 1′ impart a higher aggregation propensity than alanine in the parent peptides. These results are also in alignment with the higher beta sheet propensity of alanine compared to Q but lower than that of V, I and Y.45,46 Mutational studies using other amino acids that have higher beta sheet/alpha-helical propensities may provide more details on the conformational and aggregation properties of PHF peptides. Overall, alanine substitution in PHF peptides reduces their aggregation propensity without any major change in their conformation. Studying the aggregation properties of these alanine based PHF peptide derivatives provides important and site specific insight into the aggregation behavior of PHFs, i.e. PHF6 and PHF6*. Hence, this strategy can be potentially employed to selectively control the aggregation in PHFs via the rational choice of alanine mutations. While further extended studies are in progress in our research group, we hope that the current results can form the basis for designing PHF based Tau aggregation inhibitors using suitable mutations.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
UKM thanks CSIR-NCL for an internal grant (CSIR-NCL MLP031026). SC thanks CSIR-NCL for an internal grant (CSIR-NCL MLP029526). AD and NVG thank the CSIR, AB thanks CSIR-SPMF and SKS thanks DBT for research fellowships.Notes and references
- C. Soto and S. Pritzkow, Nat. Neurosci., 2018, 21, 1332–1340 CrossRef CAS PubMed.
- C. A. Ross and M. A. Poirier, Nat. Med., 2000, 10, S10 CrossRef PubMed.
- D. Eisenberg and M. Jucker, Cell, 2012, 148, 1188 CrossRef CAS PubMed.
- Y. S. Eisele, C. Monteiro, C. Fearns, S. E. Encalada, R. L. Wiseman, E. T. Powers and J. W. Kelly, Nat. Rev. Drug Discovery, 2015, 14, 759 CrossRef CAS PubMed.
- G. G. Glenner and C. W. Wong, Biochem. Biophys. Res. Commun., 1984, 122, 1131 CrossRef CAS PubMed.
- C. L. Masters, G. Simms, N. A. Weinman, G. Multhaup, B. L. McDonald and K. Beyreuther, Proc. Natl. Acad. Sci. U.S.A., 1985, 82, 4245 CrossRef CAS PubMed.
- M. Goedert, Science, 2015, 349, 1255555 CrossRef PubMed.
- J. Hardy and D. J. Selkoe, Science, 2002, 297, 353–356 CrossRef CAS PubMed.
- D. M. Holtzman, J. C. Morris and A. M. Goate, Sci. Transl. Med., 2011, 3(77), 77sr1 Search PubMed.
- C. Li and J. Götz, Nat. Rev. Drug Discovery, 2017, 16, 863 CrossRef CAS PubMed.
- M. Necula, R. Kayed, S. Milton and C. G. Glabe, J. Biol. Chem., 2007, 282, 10311 CrossRef CAS PubMed.
- M. Pickhardt, M. von Bergen, Z. Gazova, A. Hascher, J. Biernat, E.-M. Mandelkow and E. Mandelkow, Curr. Alzheimer Res., 2005, 2, 219 CrossRef CAS PubMed.
- K. L. Sciarretta, D. J. Gordon and S. C. Meredith, Methods Enzymol., 2006, 413, 273 CAS.
- M. P. Murphy and H. LeVine III, J. Alzheim. Dis., 2010, 19, 311 Search PubMed.
- K. S. Kosik, C. L. Joachim and D. J. Selkoe, Proc. Natl. Acad. Sci. U.S.A., 1986, 83, 4044 CrossRef CAS PubMed.
- M. Von Bergen, P. Friedhoff, J. Biernat, J. Heberle, E.-M. Mandelkow and E. Mandelkow, Proc. Natl. Acad. Sci. U.S.A., 2000, 97, 5129 CrossRef CAS PubMed.
- M. Goedert, F. Clavaguera and M. Tolnay, Trends Neurosci., 2010, 33, 317 CrossRef CAS PubMed.
- S. J. Jackson, C. Kerridge, J. Cooper, A. Cavallini, B. Falcon, C. V. Cella, A. Landi, P. G. Szekeres, T. K. Murray and Z. Ahmed, J. Neurosci., 2016, 36, 762 CrossRef CAS PubMed.
- P. Friedhoff, M. von Bergen, E.-M. Mandelkow and E. Mandelkow, Biochim. Biophys. Acta, Mol. Basis Dis., 2000, 1502, 122 CrossRef CAS.
- E.-M. Mandelkow and E. Mandelkow, Cold Spring Harbor Perspect. Med., 2012, 2, a006247 Search PubMed.
- A. W. Fitzpatrick, B. Falcon, S. He, A. G. Murzin, G. Murshudov, H. J. Garringer, R. A. Crowther, B. Ghetti, M. Goedert and S. H. Scheres, Nature, 2017, 547, 185 CrossRef CAS PubMed.
- B. Falcon, W. Zhang, A. G. Murzin, G. Murshudov, H. J. Garringer, R. Vidal, R. A. Crowther, B. Ghetti, S. H. Scheres and M. Goedert, Nature, 2018, 561, 137 CrossRef CAS PubMed.
- M. von Bergen, S. Barghorn, L. Li, A. Marx, J. Biernat, E.-M. Mandelkow and E. Mandelkow, J. Biol. Chem., 2001, 276, 48165 CrossRef CAS PubMed.
- P. Friedhoff, J. Biernat, J. Heberle, E.-M. Mandelkow and E. Mandelkow, Proc. Natl. Acad. Sci. U.S.A., 2000, 97, 5129 CrossRef PubMed.
- H. Inouye, D. Sharma, W. J. Goux and D. A. Kirschner, Biophys. J., 2006, 90, 1774 CrossRef CAS PubMed.
- M. von Bergen, S. Barghorn, J. Biernat, E.-M. Mandelkow and E. Mandelkow, Biochim. Biophys. Acta, Mol. Basis Dis., 2005, 1739, 158 CrossRef CAS PubMed.
- P. Seidler, D. Boyer, J. Rodriguez, M. Sawaya, D. Cascio, K. Murray, T. Gonen and D. Eisenberg, Nat. Chem., 2018, 10, 170 CrossRef CAS PubMed.
- M. Perez, I. Santa-María, E. Tortosa, R. Cuadros, M. Del Valle, F. Hernández, F. J. Moreno and J. Avila, J. Neurochem., 2007, 103, 1447 CrossRef CAS PubMed.
- T. Mohamed, T. Hoang, M. Jelokhani-Niaraki and P. P. Rao, ACS Chem. Neurosci., 2013, 4, 1559 CrossRef CAS PubMed.
- J. Zheng, A. M. Baghkhanian and J. S. Nowick, J. Am. Chem. Soc., 2013, 135, 6846 CrossRef CAS PubMed.
- A. Belostozky, M. Richman, E. Lisniansky, A. Tovchygrechko, J. H. Chill and S. Rahimipour, Chem. Commun., 2018, 54, 5980 RSC.
- M. Frenkel-Pinter, M. Richman, A. Belostozky, A. Abu-Mokh, E. Gazit, S. Rahimipour and D. Segal, Chem.–Eur. J., 2016, 22, 5945 CrossRef CAS PubMed.
- C. K. Wang, S. E. Northfield, Y. Huang, M. C. Ramos and D. J. Craik, Eur. J. Med. Chem., 2016, 109, 342 CrossRef CAS PubMed.
- C. Dammers, D. Yolcu, L. Kukuk, D. Willbold, M. Pickhardt, E. Mandelkow, A. H. Horn, H. Sticht, M. N. Malhis and N. Will, PLoS One, 2016, 11, e0167432 CrossRef PubMed.
- S. A. Sievers, J. Karanicolas, H. W. Chang, A. Zhao, L. Jiang, O. Zirafi, J. T. Stevens, J. Münch, D. Baker and D. Eisenberg, Nature, 2011, 475, 96 CrossRef CAS PubMed.
- J. Zheng, C. Liu, M. R. Sawaya, B. Vadla, S. Khan, R. J. Woods, D. Eisenberg, W. J. Goux and J. S. Nowick, J. Am. Chem. Soc., 2011, 133, 3144 CAS.
- V. G. KrishnaKumar, A. Paul, E. Gazit and D. Segal, Sci. Rep., 2018, 8, 71 CrossRef PubMed.
- A. Paul, S. Kalita, S. Kalita, P. Sukumar and B. Mandal, Sci. Rep., 2017, 7, 40095 CrossRef CAS PubMed.
- T. D. Vaden, S. A. Gowers, T. S. De Boer, J. D. Steill, J. Oomens and L. C. Snoek, J. Am. Chem. Soc., 2008, 130, 14640 CrossRef CAS PubMed.
- W. J. Goux, L. Kopplin, A. D. Nguyen, K. Leak, M. Rutkofsky, V. D. Shanmuganandam, D. Sharma, H. Inouye and D. A. Kirschner, J. Biol. Chem., 2004, 279, 26868 CrossRef CAS PubMed.
- F. A. Rojas Quijano, D. Morrow, B. M. Wise, F. L. Brancia and W. J. Goux, Biochemistry, 2006, 45, 4638 CrossRef CAS PubMed.
- M. Chemerovski-Glikman, M. Frenkel-Pinter, R. Mdah, A. Abu-Mokh, E. Gazit and D. Segal, Chem.–Eur. J., 2017, 23, 9618 CrossRef CAS PubMed.
- A. L. Fink, Curr. Opin. Struct. Biol., 2005, 15, 35 CrossRef CAS PubMed.
- S. Jeganathan, M. von Bergen, E.-M. Mandelkow and E. Mandelkow, Biochemistry, 2008, 47, 10526 CrossRef CAS PubMed.
- D. L. Minor Jr and P. S. Kim, Nature, 1994, 367, 660 CrossRef CAS PubMed.
- K. Fujiwara, H. Toda and M. Ikeguchi, BMC Struct. Biol., 2012, 12, 18 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details, HRMS, HPLC, cell morphology and cell viability data. See DOI: 10.1039/d0ra03809a |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2020 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: